Abstract
Zika virus (ZIKV) is involved in the etiology of serious nervous system pathologies. Currently, there are no specific and effective vaccines or antiviral drugs to prevent the diseases caused by ZIKV. This study aimed to assess the activity of flavonoids present in crude hydroethanolic extract (CHE) and fractions obtained from B. holophylla leaves against ZIKV. O-glycosylated flavonoids were characterized by high-performance liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS/MS). The cytotoxic concentration and the effective concentration for 50% of the cells (CC50 and EC50, respectively) were determined, and the selectivity index (SI) was calculated. Molecular networks were constructed based on the chemical composition of the samples and global antiviral activity data using the Global Natural Products Social Molecular Networking (GNPS) platform. Protein–ligand docking was performed in the NS2B-NS3 protease, NS3 helicase, and NS5 methyltransferase of the ZIKV. CHE showed greater antiviral activity at a multiplicity of infection (MOI) of 1.0, with an EC50 of 11.93 µg/mL, SI = 13.38, and reduced cytopathic effects. Molecular networks indicated that O-glycosylated flavonoids are responsible for the activity against ZIKV, being quercetin-O-deoxyhexoside more selective and effective. Molecular docking confirmed the inhibitory activity of quercetin-O-deoxyhexoside, which showed an affinity for the tested targets, especially for NS2B-NS3 protease. The results showed that B. holophylla has flavonoids with potential for future therapeutic applications against ZIKV.
1. Introduction
Zika virus (ZIKV) is an arbovirus of the Flavivirus genus (Flaviviridae), first isolated in 1947, in rhesus monkeys residing in Uganda’s Zika Forest [1,2]. Its transmission is mainly vectorial and occurs through the bite of Aedes spp. mosquitoes, but it can also be transmitted to humans by non-vector mechanisms, such as blood transfusions and sexual contact, being the only arbovirus with this capacity [3,4]. Epidemics caused by ZIKV emerged with a pattern familiar to other epidemics and then spread rapidly, with important consequences for public health. ZIKV infections caused a worldwide panic in May 2015, especially in South American countries such as Brazil, which experienced a wide ZIKV outbreak, with approximately 440,000 to 1.3 million cases mainly in northeastern Brazil [5]. Generally, ZIKV infections are asymptomatic; however when symptomatic, the clinical symptoms are usually mild, with low-grade fever (<37.9 °C), maculopapular rashes, arthralgia, and non-purulent conjunctivitis [6,7]. ZIKV is involved in the etiology of serious nervous system pathologies, such as the occurrence of microcephaly in neonates, Guillain-Barré syndrome in adults, and macular atrophy [8,9]. However, there are still no specific, safe, and effective drugs or vaccines to protect against ZIKV infection. There are only palliative treatments and symptom management [4,10,11]. NS2B-NS3 protease is essential for ZIKV replication, designating it as a potential target for treatment [10].
Bauhinia holophylla is a native species from the Brazilian Cerrado used in folk medicine to treat infections, pain, and diabetes [12,13]. Flavonoids from B. holophylla showed a significant antiviral effect against the Dengue virus, which also belongs to the Flaviviridae family [14]. Ribeiro et al. [15] and Marena et al. [16] reported the absence of cytotoxic and mutagenic effects, as well as antimicrobial effects of hydroalcoholic leaf extract, suggesting that B. holophylla has potential as an herbal medicine.
Flavonoids correspond to an important group of plant-derived secondary metabolites found abundantly in the Bauhinia spp. They have well-reported antiviral effects [11,14,17,18,19]. Studies have demonstrated the antiviral potential of flavonoids in the early stages of infection and viral replication [14]. They have shown to inhibit gene expression and translation, thereby preventing the production of viral proteins [20]. In addition, flavonoids exhibit an immunomodulatory action, increasing the host’s immune response against viral infections [21,22]. These actions effectively hinder viral replication and reduce viral load within the host [18,23]. Flavonoids have a relatively low toxicity profile compared to conventional antiviral drugs, making these compounds promising candidates for treating viral infections [18,24]. Several studies report the effect of different flavonoids against ZIKV, such as naringenin [11,25], baicalein and baicalin [7], hesperitin and hesperedin [26], isoquercitrin [27], among others.
New drugs based on natural bioactive metabolites can be discovered by in silico analyses. Molecular networks correspond to a computational tool that correlates molecules based on their fragmentation patterns, connecting their mass spectra [28]. This strategy has been used in several areas, such as drug research, metabolomics, and forensic toxicology [29,30]. Global Natural Products Social Molecular Networking (GNPS) [31] is a platform used to analyze large data sets obtained by mass spectrometry, from fragmented ions, through grouping and annotating ion species within LC-MS data [30]. GNPS encompasses two main workflows for generating molecular networks: Classical Molecular Networking (CLMN) and Feature-Based Molecular Networking (FBMN) [32]. CLMN was initially introduced as a rapid and efficient method to visualize large data sets, utilizing only MS2 spectra. FBMN, building upon CLMN, incorporates both MS1 (such as isotopic pattern and retention time) and MS2 data for enhanced reproducibility and accuracy in molecular network creation. FBMN can be process LC-MS2 data using tools such as MZmine2, enabling the integration of chemical and biological data through information contained in the metadata [33].
Possible intermolecular interactions between bioactive metabolites and specific ZIKV proteins can also be identified through molecular docking simulations [7]. Docking assays search for new compounds against relevant therapeutic targets with known three-dimensional structures, making it possible to estimate the ability of compounds to bind to the target, determine in which region of the target the interactions occur, and identify the types of interactions formed [34,35]. Molecular docking can help estimate a compound’s ability to bind to a target, perform structure-based virtual screening of compounds, and identify a series of targets for which the ligands show good complementarity (target fishing) [36,37].
Recently, our research team demonstrated that flavonoids from B. holophylla showed a significant antiviral effect against serotype 2 of Dengue virus [14]. Consequently, this study aimed to assess the antiviral activity of flavonoids from B. holophylla leaves against ZIKV. Molecular networks were constructed using the GNPS platform, and protein–ligand docking simulations were performed to correlate possible molecular interactions between the O-glycosylated flavonoids and their antiviral activity.
2. Materials and Methods
2.1. Chemicals
The chemicals were purchased from different manufacturers: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Ludwig, Porto Alegre, Brazil), fetal bovine serum (Gibco®, São Paulo, Brazil), penicillin, and streptomycin (Cultilab, Campinas, Brazil). Dimethylsulphoxide, Dulbecco’s Modified Eagle Medium, Leibovitz-L-15 medium, and Amphotericin B were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
2.2. Plant Material, Extraction of Crude Extracts and Fractions, and Chemical Characterization
The crude hydroethanolic extract (CHE) and ethyl acetate (EtOAc), dichloromethane (DCM), and hydroethanolic (HE) fractions were prepared from leaves of Bauhinia holophylla (Bong.) Steud. (Fabaceae: Cercidoideae) collected in the Brazilian Cerrado in Ijaci, Southern Minas Gerais State, Brazil (21°09′97″ S and 44°55′65″ W GRW, at 835 m altitude) (SISBIO No. 24542-3, IBAMA Registration: 5042260). Fertile samples were collected, and the vouchers were identified by Andreia Fonseca Silva at the PAMG Herbarium (PAMG 57021), at the Agricultural Research Company of Minas Gerais (EPAMIG). This work has access permission for plant genetic heritage components (No. 010500/2014-6/CNPq/CGEN/MMA) and is registered on the SisGen Platform (Registration number A12A940), according to the Brazilian Biodiversity Law (13.123/2015). The plant material was dried in a ventilated oven (TE-394/500L, Tecnal; Piracicaba, Brazil) at 40 °C for 24 h, and pulverized in a knife mill (SL-31, Solab; Piracicaba, Brazil). The obtaining of the crude extract and fractions, and the chemical characterization by high-performance liquid chromatography, coupled with high-resolution mass spectrometry (LC-HRMS/MS), were performed and described by da Fonseca et al. [38]. CHE and fractions were dissolved in a 10% dimethylsulfoxide (DMSO) aqueous solution (v/v) to obtain the stock solution (10 mg/mL). A concentration of 0.5% DMSO was used as the vehicle or negative control since concentrations lower than 2% do not cause cytotoxic effects in the experiments [39,40].
2.3. Cell Line and Virus
African green monkey kidney epithelial cells (Vero, ATCC CCL-81) were used for cytotoxicity and antiviral assays. Cells were cultured at 37 °C in a 5% CO2 environment (Forma series II 3110 water jacket CO2 incubator, Thermo Fisher ScientificTM, Marietta, GA, USA) on Dulbecco’s Modified Eagle’s Medium (DMEM), adding 5% of heat-inactivated fetal bovine serum and supplemented with 100 μg/mL penicillin, 100 U/mL streptomycin, and 2.5 μg/mL amphotericin B. ZIKV (PE 243/2015 strain) (GenBank accession number KX197192.1) was propagated in Aedes albopictus clone C6/36 cells and titrated using the Dulbecco’s plaque formation test [41]. The supernatant from the infected cells was isolated and stored at −80 °C.
2.4. Determination of Cytotoxicity by the MTT Assay
The cytotoxicity of the CHE and EtOAc, DCM, and HE fractions was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric method [42]. Vero cells in 96-well plates were treated with serial dilutions of samples (3.12–200 μg/mL) [14] in a total of 100 μL of growth medium for 48 h at 37 °C with 5% CO2. Each well received 25 μL of freshly prepared 2 mg/mL MTT solution and was incubated at 37 °C for 3 h. Then, 100 μL of 0.5% DMSO was added to each well to dissolve the purple formazan product, and the plates were incubated at 37 °C for 5 min to remove any air bubbles. MTT signals were measured at 492 nm (PowerWave HT Microplate Spectrophotometer, BioTelTM, Winooski, VT, USA). Each concentration was tested in triplicate and in three independent experiments. Cell proliferation and viability were compared to untreated cells (negative control), and the cytotoxic concentration for 50% of cells (CC50) was determined [43] using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA).
2.5. Global Antiviral Assay
Based on the cytotoxicity results, concentrations close to CC50, or non-cytotoxic, were selected for global antiviral assays. Thus, the reduction in cell viability in this experiment must be associated with the effect of the virus on the cell. To achieve this, 90% confluent Vero cells in 96-well plates (1 × 104 cells/well) were treated with different concentrations of CHE and EtOAc, DCM, and HE fractions (3.12–100 μg/mL) and infected with ZIKV at MOI 1.0 and 5.0 [44]. Cells and virus were separately pretreated with the samples for 30 min at 37 °C and then transferred to the cell plate, and incubated at 37 °C, 5% CO2, for 48 h. After 48 h, the supernatants were collected, and images were recorded using an EVOSmicroscope (EVOS® FL Auto Imaging System, Thermo Fisher ScientificTM, Carlsbad, CA, USA). Cell viability was determined using the MTT assay at 492 nm and the percentage of viable cells was determined as described in Equation (1). Subsequently, these results were used to determine the 50% effective concentration (EC50). That is, the concentration of the extract that resulted in viral infection protection of 50% of the cells was determined by non-linear regression analysis using GraphPad Prism 7.0 software. The selectivity index (SI) was calculated as a selection criterion and was expressed by the CC50/EC50 ratio [45].
where A, B, and C represent the absorbances at 490 nm of the wells, in which treated and infected (A), untreated and infected (B), and untreated and uninfected (C) cells are present, respectively.
2.6. Global Natural Products Social Molecular Networking (GNPS)
LC-HRMS/MS data obtained from B. holophylla leaves [38] in positive and negative ionization modes and antiviral activity data corresponding to EC50 of the samples in MOI 1.0 and 5.0 were used for the construction of molecular networks. Product ion spectra arising from the LC-HRMS/MS analysis of the CHE and fractions from B. holophylla were processed using MZmine 2.53 features, including mass detection, chromatogram builder, chromatogram deconvolution, isotopic peaks grouper, and alignment. The processed data were converted to the mzML format with MSConvertGUI (64-bit) V.3 software (ProteoWizard; https://proteowizard.sourceforge.io/, accessed on 18 April 2024) [46] and uploaded on the GNPS web platform “https://gnps.ucsd.edu (accessed on 11 January 2024)” [47] using the FBMN workflow, with parameters set for high-resolution data. Data were visualized by Cytoscape 3.7.0 software. The annotation of chemical compounds was performed by manual interpretation of MS/MS spectra, compared to the databases inside GNPS.
2.7. Molecular Docking
ZIKV protein crystal structures were obtained from the Protein Data Bank (PDB) as follows: NS2B-NS3 protease (ZIKVPro, PDB: 5H4I), NS3 helicase (ZIKVHel, PDB: 5K8L), and NS5 methyltransferase (ZIKVMTase, PDB: 5M5B). Next, the ligand structures were generated using PubChem Sketcher V2.4 [48] and optimized using UCSF Chimera 1.17.3 [49] through default parameters. Molecular docking simulations were performed at Dockthor “https://dockthor.lncc.br/v2/ (accessed on 25 February 2024)” [50]. The grid size did not exceed 24 Å, and the discretization used was 0.25. The search precision was defined as virtual screening, with 500,000 evaluations, and a population size of 750 over 12 runs in soft docking mode. Finally, the intermolecular interactions were described using Discovery Studio Visualizer v21.1.0.20298 software.
2.8. Statistical Analysis
The statistical analyses were conducted using GraphPad Prism® 7.00.288 software. The experiments and biological assays were performed in triplicate. The results are represented as the mean ± standard deviation. Values were considered significantly different for p ≤ 0.05.
3. Results
3.1. Determination of Cytotoxicity by the MTT Assay
Cell viability could be determined for all samples. In general, the results showed a dose-dependent response on treated cells, where higher concentrations of the CHE and fractions resulted in high cytotoxicity compared to the untreated controls (Figure 1). The HE fraction showed the lowest cytotoxicity, with CC50 = 191.10 ± 4.15 µg/mL, while the DCM fraction was the most cytotoxic (CC50 = 92.40 ± 3.12 µg/mL). No cytotoxic effect was observed in control cells, i.e., cells treated with 0.5% DMSO (v/v), indicating that the vehicle used in the assays did not interfere with cell viability, as expected.
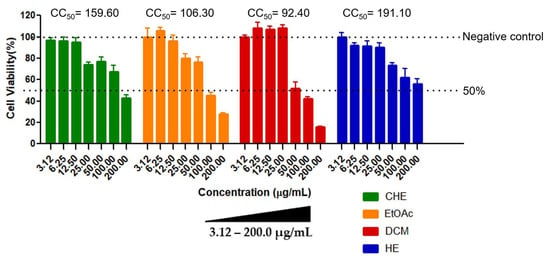
Figure 1.
Cytotoxicity of B. holophylla extract and fractions in Vero cells. The cells were treated with different concentrations of the samples (3.12–200 µg/mL). Negative control: Vero cells + 0.5% DMSO. Results were expressed as the mean ± standard deviation of three independent experiments. CC50 values (µg/mL) were calculated by nonlinear regression analysis using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA). The values were considered significantly different when p ≤ 0.05. CHE: crude hydroethanolic extract; EtOAc: ethyl acetate fraction; DCM: dichloromethane fraction; HE: hydroethanolic fraction.
3.2. Global Antiviral Assay
In general, CHE and fractions of B. holophylla showed a dose-dependent effect on cell viability, with greater effectiveness at the highest concentrations tested for MOI 1.0 and 5.0 (Figure 2). These results suggest that the protective effects of the samples increase with higher concentrations. CHE showed greater global antiviral activity against ZIKV, with EC50 = 11.93 ± 4.74 µg/mL and SI = 13.38 at MOI = 1.0. The EtOAc, DCM and HE fractions presented selectivity indices between 2.94 and 5.36. At MOI = 5.0, CHE showed a SI < 10, and the fractions were ineffective (Table 1).
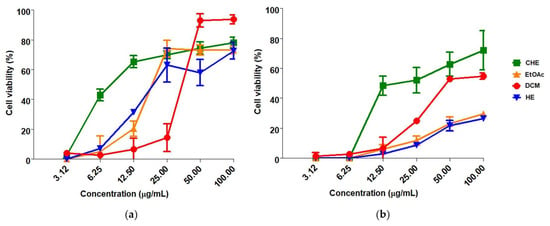
Figure 2.
Anti-ZIKV activity of Bauhinia holophylla extract and fractions. (a) MOI = 1.0; (b) MOI = 5.0. ZIKV and Vero cells were pretreated with the samples for 30 min at 37 °C and incubated for 48 h at 37 °C. The cell viability was evaluated using the MTT method, at 492 nm. Results were expressed as the mean ± standard deviation of three independent experiments. EC50 values were calculated by nonlinear regression analysis using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA). The values were considered significantly different when p ≤ 0.05. CHE: crude hydroethanolic extract; EtOAc: ethyl acetate fraction; DCM: dichloromethane fraction; HE: hydroethanolic fraction.

Table 1.
Antiviral activity of Bauhinia holophylla extract and fractions using MOI = 1.0 and MOI = 5.0. Results were expressed as the mean ± standard deviation of three independent experiments. CC50 and EC50 values were calculated by nonlinear regression analysis using GraphPad Prism 7.0 (GraphPad Software Inc., La Jolla, CA, USA). The values were considered significantly different when p ≤ 0.05.
The results observed for the antiviral activity of CHE were supported by the reduction in the cytopathic effects on Vero cells infected with ZIKV and treated with CHE at different concentrations (50.0–12.5 μg/mL) (Figure 3), 48 h after infection, compared to cell and viral controls (Figure 3a–c). The results showed that CHE was effective in protecting cells infected by ZIKV, reducing viral infection and ensuring the maintenance of cell monolayer integrity despite the increase in the number of viral particles (MOI 1.0 to 5.0), especially at concentrations of 25.00 and 50.00 µg/mL (Figure 3d,e,h,i). These results were corroborated by the EC50 values observed for CHE (Table 1).
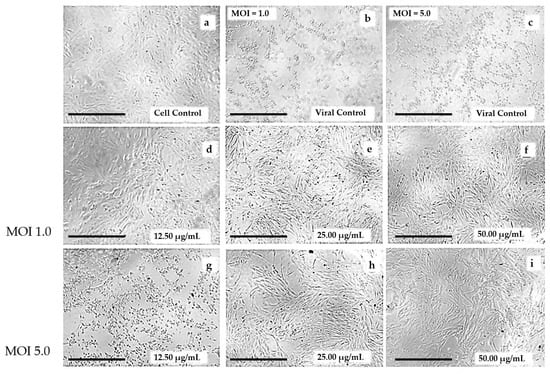
Figure 3.
Crude hydroethanolic extract (CHE) of Bauhinia holophylla leaves reduced cytopathic effect in Vero cells induced by ZIKV infection. (a) Control cells (non-infected Vero cells, only with the addition of DMSO at 0.5% v/v); (b,c) viral controls (Vero cells infected with ZIKV at MOI 1.0 and 5, 0 respectively, only with the addition of DMSO at 0.5% v/v, respectively); (d–f) Vero cells infected with ZIKV at MOI = 1.0 and treated with CHE at 12.50; 25.00 and 50.00 µg/mL, respectively, after 48 h of incubation at 37 °C; (g–i) Vero cells infected with ZIKV at MOI = 5.0 and treated with CHE at 12.50; 25.00 and 50.00 µg/mL, respectively, after 48 h of incubation at 37 °C. Scale bar = 400 µm.
3.3. LC-HRMS/MS and Molecular Networking Analyses
In the molecular network formed by the FBMN workflow, some classes of O-glycosylated flavonoids were annotated and considered promising candidates for antiviral activity, particularly, quercetin-O-deoxyhexoside (Figure 4), found in CHE.
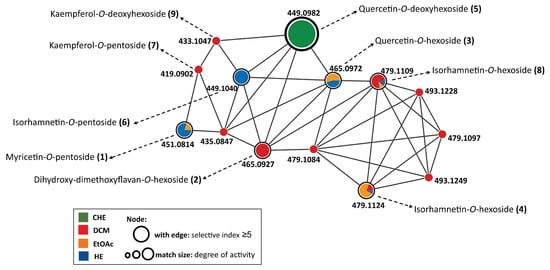
Figure 4.
Molecular network obtained from FBMN workflow and annotated compounds from Bauhinia holophylla leaves based on spectral matches within the public spectral libraries in the GNPS platform.
In Figure 4, the colors correspond to the samples tested, the node sizes refer to activity (a large node is comparable to an ion present in active samples), and the presence of a border is equivalent to ions present in samples with an SI greater than 2.0 (MOI = 1.0).
O-glycosylated flavonoids show a fragmentation pattern with a neutral loss of sugar units, i.e., 162 to hexoses, 146 to deoxyhexoses, and 132 to pentoses. All these fragments were observed in the annotated compounds (Table 2).

Table 2.
Metabolites annotated by LC-HRMS/MS and molecular network in the Bauhinia holophylla extracts and fractions (positive ionization mode).
In addition, it was possible to differentiate some isomers by analyzing the fragmentation pattern. For example, compounds 2 and 3, m/z 465 [M + H]+, have the same molecular formula, although compound 2 shows typical flavans fragments (m/z 153, 125, 121), while compound 3 shows typical fragments of flavonols (m/z 53, 165, 137), such as quercetin derivatives [51]. Compound 2 has a slightly lower polarity than compound 3 and was found mainly in the DCM, while compound 3 was present in the EtOAc and in the HE due to a slightly higher polarity.
For compounds 5 and 6, with m/z 449 [M + H]+, differentiation was also achieved through the presence of aglycone fragments: m/z 303 for quercetin in compound 5, and m/z 317 for isorhamnetin in compound 6.
3.4. Molecular Docking
Quercetin-O-deoxyhexoside showed promising antiviral activity against ZIKV, observed by molecular network analysis. This finding motivated us to study the putative receptor of these natural compounds using molecular docking tools. Thus, all possible isomers of quercetin-O-deoxyhexoside were generated considering three different positions of the O-deoxyhexoside portion in relation to the aglycone. Quercetin (aglycone) was also used to verify the influence of glycosylation on interactions with ZIKV receptors. The three-dimensional molecular structures of ligands are represented in Figure 5. As can be seen, the geometry of the aglycone structure remains invariable independent of the O-deoxyhexoside moiety position, due to its aromatic rigid structure.
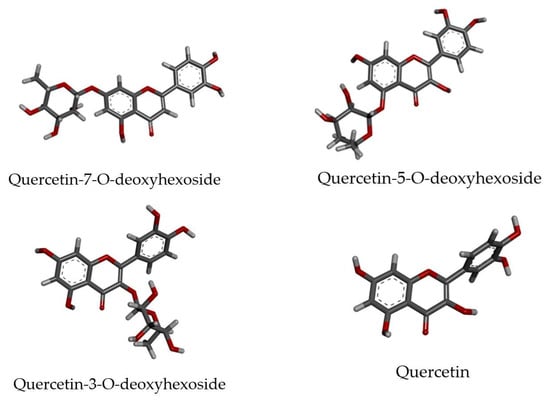
Figure 5.
Quercetin and selected derivatives structures submitted to molecular docking studies.
The binding energies of these compounds against the ZIKV molecular targets were obtained through docking methodology. Thus, Table 3 shows the binding energy for each crystallographic ligand of the molecular target studied, obtained from the redocking process.

Table 3.
Interaction energy between ligands and selected targets (NS2B-NS3 protease, NS3 helicase, and NS5 methyltransferase).
The binding energies of benzimidazole-1ylmethanol (NS2B-NS3 protease), 5′-guanosine-diphosphate-monothiophosphate (NS3 helicase), and S-adenosylmethionine (NS5 methyltransferase) were −21.63, −60.42, and −71.34 Kcal/mol, respectively. Thus, among the molecular targets, the natural compounds showed higher affinity against NS2B-NS3 protease. For instance, quercetin-5-O-deoxyhexoside had the lowest interaction energy (−33.06 Kcal/mol) among flavonoids, even lower than that of the crystallographic ligand benzimidazole-1ylmethanol. In contrast, the crystallographic ligands of NS3 helicase and NS5 methyltransferase had higher affinity for their respective receptors compared to flavonoid compounds. In summary, these findings suggest that NS2B-NS3 protease is the main molecular target of flavonoids due to their higher affinity for this receptor.
Hence, the molecular interactions between quercetin-5-O-deoxyhexoside and NS2B-NS3 protease are highlighted in Figure 6. As can be seen, this flavonoid can carry out van der Waals interactions with Tyr114, Pro115, Ser119, Asn136, and Gly137; hydrogen bonds with Lys38, Asp113, and Gly135; Pi-interactions with His35, Ala116, and Tyr145 residues. It is noteworthy that the catalytic residues triad of NS2B-NS3 protease, consisting of His35, Ser119, and Asp113, are close to the flavonoids, suggesting a competitive inhibition process.
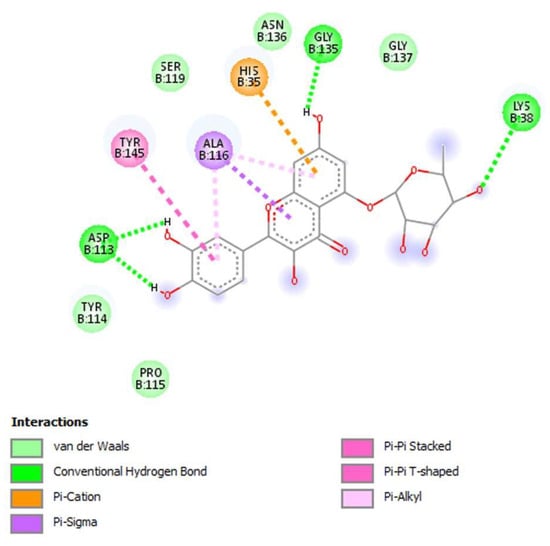
Figure 6.
Intermolecular interactions between the amino acid residues of NS2B-NS3 protease and quercetin-5-O-deoxyhexoside.
4. Discussion
Flavonoids correspond to an essential and diversified group of plant secondary metabolites, widely distributed in the plant kingdom, often in a glycoside form, and associated with various physiological roles, such as antioxidants, protection against UV radiation, and chemical defense of plants [52,53,54,55]. In addition to this ecological role, several flavonoids stand out as potent plant bioactive compounds with antimicrobial, anti-inflammatory, antioxidant, cardioprotective, neuroprotective, and antitumoral effects [16,38,55,56,57]. O-glycosylated flavonoids are widely distributed in Bauhinia spp., and previous studies describe these compounds as one of the main contributors to the antimicrobial effect against bacteria, fungi, and viruses in this genus [14,58,59]. The low toxicity, low cost, and wide distribution in the plant kingdom make flavonoids good candidates for application in antimicrobial therapy [60]. Extracts and fractions of B. holophylla leaves predominantly contain flavonoids of the flavonol-3-O-glycoside type, such as myricetin, quercetin, kaempferol, and isorhamnetin O-glycosylated [14,15,38,61]. A significant antiviral effect of O-glycosylated flavonoids from B. holophylla leaves against some Dengue virus strains was reported by Dos Santos et al. [14]. Dengue virus is genetically and serologically related to ZIKV and other flaviviruses [1].
In the current study, we observed that CHE obtained from B. holophylla leaves showed a significant global viral activity against ZIKV, especially at MOI = 1.0 (EC50 = 11.93 ± 4.74 µg/mL and SI = 13.38). However, the same activity was not observed for DCM, EtOAc, and HE, with EC50 > 25 µg/mL and SI < 10 (Table 1). All samples showed moderate cytotoxicity according to Niksic et al. [62], as they presented CC50 between 21–200 µg/mL. Samples with an SI greater than 10.0 are safe, effective and, therefore, more active [44,63]. Despite moderate toxicity, CHE from B. holophylla leaves has a protective effect attributable to the presence of flavonoids, which are described as antioxidants, inhibitors of DNA adduct formation, and activators of detoxifying enzymes [15]. CHE was also able to reduce structural changes on Vero cells caused by ZIKV infection, minimizing cytopathic effects, such as cell individualization, rounding, detachment of cells, and cytolysis, maintaining the cell monolayer integrity even though the MOI increased from 1.0 to 5.0 (Figure 3). There is limited understanding regarding the mechanisms through which ZIKV causes these conditions or which viral proteins are responsible for the associated ZIKV-induced cytopathic effects, including cell hypertrophy, growth restriction, cell-cycle dysregulation, and cell death [64]. According to Bernardo-Menezes et al. [65], ZIKV proteins are major virulence factors that modulate the host cell machinery by disrupting and/or blocking specific cellular systems and organelle functions, such as endoplasmic reticulum stress and mitochondrial dysfunction.
Molecular networks built using the FBMN workflow in the GNPS platform and molecular docking indicated O-glycosylated flavonoids as promising candidates for the antiviral activity of B. holophylla leaves. Quercetin-O-deoxyhexoside was the main molecule associated with the significant global antiviral activity against ZIKV, demonstrated particularly by CHE at MOI = 1.0 (Figure 4), with inhibitory effects on NS2B-NS3 protease (Table 3 and Figure 6). Currently, molecular networking has become an essential bioinformatics tool to visualize and annotate the chemical space in non-targeted mass spectrometry data [28,66]. The goal of this platform is to provide as much chemical insight as possible in an untargeted MS dataset and connect this chemical insight to underlying biological questions [47]. In this tool, the clusters formed can be considered “molecular families”, since the compounds should share key chemical characteristics, thereby facilitating node connection. So, through this concept of molecular networks, unannotated compounds could belong to the same class of compounds, and they may be contributing to the observed activity through synergy. Due to chemical analysis by mass spectrometry, we cannot assert that the ion corresponding to the compound quercetin-O-deoxyhexoside is not present in the other extracts, since the process of ion suppression may be occurring, thus preventing its detection in the other samples. Molecular networks have been useful in the discovery of new bioactive molecules in complex matrices (natural products), correlated with various biological activities, such as antitumor, antimicrobial, and antiviral, among others [29,67,68,69,70,71]. Some studies indicate that the glycosylation of flavonoids increases the activity compared to isolated aglycone. Quercetin-3-β-O-d-glucoside was studied in in vivo models (knockout mice) infected with ZIKV and post-infection treatment showed a beneficial effect in reducing the viral load compared to the non-glycosylated form [72]. However, some glycosylated forms of quercetin with sugars such as rhamnose and arabinose have also been indicated as less active in studies with extracts of B. holophylla [14] and B. longifolia [73] against Dengue virus and Mayaro virus, respectively. It is known that the activity of these molecules depends on several factors, and that each type of virus has peculiarities, different molecular targets, and specific multiplication mechanisms, which can vary between species and strains [18,74,75].
The ability of flavonoids to interact with molecular targets of ZIKV has been assessed by using in silico molecular docking through the main molecular targets [76]. Molecular docking studies represent a strategy to clarify and improve the understanding of the mechanisms that contribute to the antiviral activity of flavonoids [75]. These studies describe the flavonoid’s affinity for molecular targets and the ligand–receptor interactions responsible for molecular recognition [77,78]. Myricetin and quercetin (in glycosylated or aglycone form), in addition to other flavonoids, can inhibit the penetration of ZIKV into the host cell and inhibit the NS2B-NS3 proteases, preventing the maturation of the virus [79]. Myricetin, quercetin, isorhamnetin, luteolin, and apigenin, along with curcumin, compounds commonly found in different vegetables and fruits, act as non-competitive allosteric inhibitors of the ZIKV NS2B-NS3 protease [80]. Virtual screening and docking assays identified three compounds from Pterogyne nitens (Fabaceae), a Brazilian medicinal plant, as potent NS2B-NS3 protease inhibitors. Among them, pedalitin proved to be a potent inhibitor of the ZIKV NS2B-NS3 protease, also acting as a non-competitive inhibitor in kinetic assays. However, this compound showed antiviral activity against ZIKV in Vero cell cultures at high concentrations, close to the cytotoxic concentration [81]. Two flavonoids, baicalein (glycosylated flavonoid) and baicalin (aglycone form), as well as quercetin-O-deoxyhexoside, demonstrated an affinity for ZIKV NS5, showing that flavonoids have in silico inhibitory activity on proteins essential for the ZIKV multiplication cycle [7].
5. Conclusions
The significant anti-ZIKV activity observed in B. holophylla leaves is particularly associated with quercetin-O-deoxyhexoside, which showed affinity with ZIKV target proteins, especially NS2B-NS3 protease, according to molecular docking assays. These results confirm the biotechnological potential of B. holophylla to produce bioactive compounds of economic and medicinal interest and suggest that other studies be carried out to confirm the activity and investigate the mechanism of action of quercetin-O-deoxyhexoside on ZIKV. In addition, stability and pharmacokinetic studies should be conducted, considering the reactivity of flavonoids and their physicochemical characteristics, which are critical in biological assays.
Author Contributions
Conceptualization, G.F.M.L., J.M.S.F. and A.H.F.C. Data curation, R.M.d.O.T., T.R.T., S.C.M., B.A.G., T.A.d.O. and S.T.D.d.F. Formal analysis, R.M.d.O.T., T.R.T., S.C.M., B.A.G. and T.A.d.O. Funding acquisition, S.G.L., A.G.T., J.M.S.F., L.A.R.d.S.L. and A.H.F.C. Investigation, R.M.d.O.T., G.F.M.L., S.C.M., B.A.G. and T.A.d.O. Methodology, R.M.d.O.T., T.R.T., G.F.M.L., S.C.M., B.A.G., T.A.d.O., S.T.D.d.F., A.G.T., L.A.R.d.S.L. and A.H.F.C. Project administration, A.H.F.C. Resources, S.G.L., A.G.T., J.M.S.F., L.A.R.d.S.L. and A.H.F.C. Supervision, A.H.F.C.; Validation, R.M.d.O.T., S.C.M. and T.A.d.O. Visualization, T.R.T., G.F.M.L., S.C.M., B.A.G., S.G.L., T.A.d.O., S.T.D.d.F., J.M.S.F., L.A.R.d.S.L. and A.H.F.C. Writing—original draft, R.M.d.O.T., S.C.M., B.A.G., A.G.T. and A.H.F.C. Writing—review and editing, R.M.d.O.T., T.R.T., G.F.M.L., S.C.M., B.A.G., S.G.L., T.A.d.O., S.T.D.d.F., A.G.T., J.M.S.F., L.A.R.d.S.L. and A.H.F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Processo 306581/2020-5), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) (APQ-02742-17, APQ-04559-22, CRA-APQ-00861-22). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES), Finance Code 001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
We thank Andreia Fonseca Silva for the botanical identification of this plant and Rafael Gonçalves Teixeira Neto for helping with the EVOS® FL Auto Imaging System microscope. The authors are grateful to the Universidade Federal de São João del-Rei (UFSJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for financial support. R.M.O.T. is grateful to Universidade Federal de São João del Rei (UFSJ) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for the M.Sc. fellowship. T.A.O is grateful for the PhD. fellowship, and A.G.T and A.H.F.C. are grateful for a productive fellowship from CNPq (310108/2020-9 and 308161/2023-8).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Faye, O.; Freire, C.C.M.; Iamarino, A.; Faye, O.; de Oliveira, J.V.C.; Diallo, M.; Zanotto, P.M.A.; Sall, A.A. Molecular evolution of Zika virus during its emergence in the 20th century. PLoS Negl. Trop. Dis. 2014, 8, e2636. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Polen, K.D.; Gilboa, S.M.; Hills, S.; Oduyebo, T.; Kohl, K.S.; Brooks, J.T.; Adamski, A.; Simeone, R.M.; Walker, A.T.; Kissin, D.M.; et al. Update: Interim guidance for preconception counseling and prevention of sexual transmission of Zika virus for men with possible Zika virus exposure—United States. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Pielnaa, P.; Al-Saadawe, M.; Saro, A.; Dama, M.F.; Zhou, M.; Huang, Y.; Huang, J.; Xia, Z. Zika virus-spread, epidemiology, genome, transmission cycle, clinical manifestation, associated challenges, vaccine, and antiviral drug development. Virology 2020, 543, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, M.; Fischer, M.; Staples, J.E. Zika virus spreads to new areas—Region of the Americas, May 2015–January 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.R.; Jamieson, D.J.; Powers, A.M.; Honein, M.A. Zika virus. N. Eng. J. Med. 2016, 374, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Oo, A.; Teoh, B.T.; Sam, S.S.; Bakar, S.A.; Zandi, K. Baicalein and baicalin as Zika virus inhibitors. Arch. Virol. 2018, 164, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Gubler, D.J. Zika virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed]
- Ventura, C.V.; Maia, M.; Bravo-Filho, V.; Góis, A.L.; Belfort, R., Jr. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 2016, 387, 228. [Google Scholar] [CrossRef]
- Meewan, I.; Shiryaev, S.A.; Kattoula, J.; Huang, C.T.; Lin, V.; Chuang, C.H.; Terskikh, A.V.; Abagyan, R. Allosteric inhibitors of Zika virus NS2B-NS3 protease targeting protease in “super-open” conformation. Viruses 2023, 15, 1106. [Google Scholar] [CrossRef]
- Silva, P.G.D.; Chaves, E.J.F.; Silva, T.M.S.; Rocha, G.B.; Dantas, W.M.; Oliveira, R.N.; Pena, L.J. Antiviral activity of flavonoids from geopropolis of the brazilian jandaira bee against Zika and Dengue viruses. Pharmaceutics 2023, 15, 2494. [Google Scholar] [CrossRef] [PubMed]
- da Vaz, A.M.S.; Bortoluzzi, R.L.C.; da Silva, L.A.E. Checklist of Bauhinia sensu stricto (Caesalpiniaceae) in Brazil. Plant Ecol. Evol. 2010, 143, 212–221. [Google Scholar] [CrossRef]
- Pinheiro, M.S.; Rodrigues, L.S.; Neto, L.S.; Moraes-Souza, R.Q.; Soares, T.S.; Americo, M.F. Effect of Bauhinia holophylla treatment in streptozotocin-induced diabetic rats. An. Acad. Bras. Cienc. 2017, 89, 263–272. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, M.; Teixeira, T.R.; Santos, F.R.S.; Lima, W.G.; Ferraz, A.C.; Silva, N.L.; Leite, F.J.; Siqueira, J.M.; Luyten, W.; Castro, A.H.F.; et al. Bauhinia holophylla (Bong.) Steud. leaves-derived extracts as potent anti-dengue serotype 2. Nat. Prod. Res. 2021, 35, 2804–2809. [Google Scholar] [CrossRef]
- Ribeiro, D.L.; Cilião, H.L.; Specian, A.F.L.; Serpeloni, J.M.; de Oliveira, M.T.; Varanda, E.A.; Vilegas, W.; Saldanha, L.L.; MartínezLópez, W.; Dokkedal, A.L.; et al. Phytochemical study and evaluation of cytotoxicity, mutagenicity, cell cycle kinetics and gene expression of Bauhinia holophylla (Bong.) Steud. in HepG2 cells in vitro. Cytotechnology 2018, 70, 713–728. [Google Scholar] [CrossRef] [PubMed]
- Marena, G.D.; Saldanha, L.L.; Ramos, M.A.S.; de Grandis, R.A.; Dokkedal, A.L.; Bauab, T.M.; Pavan, F.R.; Resende, F.A. Antimicrobial, cytotoxic and mutagenic activities of Bauhinia holophylla hydroalcoholic extract. Am. J. Essent. Oil. Nat. Prod. 2021, 9, 1–6. [Google Scholar]
- Wu, K.X.; Chu, J.J.H. Antiviral screen identifies EV71 inhibitors and reveals camptothecin-target, DNA topoisomerase 1 as a novel EV71 host factor. Antivir. Res. 2017, 143, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jarenko, M. Antiviral activities of flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef] [PubMed]
- Di Petrillo, A.; Orrù, G.; Fais, A.; Fantini, M.C. Quercetin and its derivates as antiviral potentials: A comprehensive review. Phytother. Res. 2022, 36, 266–278. [Google Scholar] [CrossRef]
- Suebsaard, P.; Charerntantanakul, W. Rutin, α-tocopherol, and l-ascorbic acid up-regulate type I interferon-regulated gene and type I and II interferon expressions and reduce inflammatory cytokine expressions in monocyte-derived macrophages infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2021, 235, 110231. [Google Scholar] [CrossRef]
- Vissenaekens, H.; Criel, H.; Grootaert, C.; Raes, K.; Smagghe, G.; Van Camp, J. Flavonoids, and cellular stress: A complex interplay affecting human health. Crit. Rev. Food Sci. Nutr. 2022, 62, 8535–8566. [Google Scholar] [CrossRef] [PubMed]
- Chanu, N.R.; Gogoi, P.; Barbhuiya, P.A.; Dutta, P.P.; Pathak, M.P.; Sen, S. Natural flavonoids as potential therapeutics in the management of diabetic wound: A review. Curr. Top. Med. Chem. 2023, 23, 690–710. [Google Scholar] [CrossRef] [PubMed]
- Salatin, S.; Bazmani, A.; Shahi, S.; Naghili, B.; Memar, M.Y.; Dizaj, S.M. Antimicrobial benefits of flavonoids and their nanoformulations. Curr. Pharm. Des. 2022, 17, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.A.O.; Ponciano, C.S.; Caetano, A.H.D.; Wowk, P.F.; Bordignon, J.; Silva, H.; Almeida, M.V.; Ávila, E.P. The anti-Zika virus and anti-tumoral activity of the citrus flavanone lipophilic naringenin-based compounds. Chem. Biol. Interact. 2020, 331, 109218. [Google Scholar] [CrossRef]
- Eberle, R.J.; Olivier, D.S.; Pacca, C.C.; Avilla, C.M.S.; Nogueira, M.L.; Amaral, M.S.; Willbold, D.; Arni, R.K.; Coronado, M.A. In vitro study of Hesperetin and Hesperidin as inhibitors of Zika and Chikungunya virus proteases. PLoS ONE 2021, 16, e0246319. [Google Scholar] [CrossRef] [PubMed]
- Gaudry, A.; Bos, S.; Viranaicken, W.; Roche, M.; Krejbich-Trotot, P.; Gadea, G.; Desprès, P.; El-Kalamouni, C. The flavonoid isoquercitrin precludes initiation of Zika virus infection in human cells. Int. J. Mol. Sci. 2018, 19, 1093. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Nothias, L.F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends Pharmacol. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Vincenti, F.; Montesano, C.; Di Ottavio, F.; Gregori, A.; Compagnone, D.; Sergi, M.; Dorrestein, P. Molecular networking: A useful tool for the identification of new psychoactive substances in seizures by LC-HRMS. Front. Chem. 2020, 8, 572952. [Google Scholar] [CrossRef]
- Schmid, R.; Petras, D.; Nothias, L.-F.; Wang, M.; Aron, A.T.; Jagels, A.; Tsugawa, H.; Rainer, J.; Garcia-Aloy, M.; Dührkop, K.; et al. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 2021, 12, 3832. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Xu, R.; Lee, J.; Chen, L.; Zhu, J. Enhanced detection, and annotation of small molecules in metabolomics using molecular-network oriented parameter optimization. Mol. Omics 2021, 17, 665–676. [Google Scholar] [CrossRef]
- Afoullouss, S.; Balsam, A.; Allcock, A.L.; Thomas, O.P. Optimization of LC-MS2 data acquisition parameters for molecular networking applied to marine natural products. Metabolites 2022, 12, 245. [Google Scholar] [CrossRef]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Wani, T.A.; Zargar, S.; Hussain, A. Spectroscopic, thermodynamic, and molecular docking studies on molecular mechanisms of drug binding to proteins. Molecules 2022, 27, 8405. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.L.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, S.T.M.; Teixeira, T.T.; Siqueira, J.M.F.; Lima, L.A.R.S.; Luyten, W.; Castro, A.H.F. Flavonoid-rich fractions of Bauhinia holophylla leaves inhibit Candida albicans biofilm formation and hyphae growth. Plants 2022, 11, 1796. [Google Scholar] [CrossRef]
- Zandi, K.; Ramedani, E.; Mahammadi, K.; Tajbakhsh, S.; Deilami, I.; Rastian, Z.; Farshadpour, F. Evaluation of antiviral activities of curcumin derivatives against HSV-1 in Vero cell line. Nat. Prod. Commun. 2010, 5, 1935–1938. [Google Scholar] [CrossRef]
- Aguilar, M.G.D.; Sousa, G.F.D.; Evangelista, F.C.; Sabino, A.P.; Camargo, K.C.; Vieira Filho, S.A.; Nunes, Y.R.F.; Lucienir, P.; Duarte, L.P. Friedelane triterpenes with cytotoxic activity from the leaves of Maytenus quadrangulata (Celastraceae). Braz. Chem. Soc. 2022, 33, 1281–1290. [Google Scholar] [CrossRef]
- Dulbecco, R. Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proc. Nat. Acad. Sci. USA 1952, 38, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Osti, R.Z.; Serrano, F.A.; Paschoalin, T.; Massaoka, M.H.S.; Travassos, L.R. The in vitro and in vivo antitumour activities of nitrosyl ruthenium amine complexes. Aust. J. Chem. 2012, 65, 1333–1341. [Google Scholar] [CrossRef]
- Ferraz, A.C.; Moraes, T.D.F.S.; da Cruz Nizer, W.S.; Dos Santos, M.; Totola, A.H.; Ferreira, J.M.S.; de Magalhaes, J.C. Virucidal activity of proanthocyanidin against Mayaro virus. Antivir. Res. 2019, 168, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Brandão, G.C.; Kroon, E.G.; Santos, J.R.; Stehmann, J.R.; Lombardi, J.A.; Oliveira, A.B. Antiviral activities of plants occurring in the state of Minas Gerais, Brazil. Part 2. Screening Bignoniaceae species. Rev. Bras. Farmacogn. 2010, 20, 742–750. [Google Scholar] [CrossRef]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to convert raw mass spectrometry data. Curr. Protoc. Bioinform. 2014, 46, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Ihlenfeldt, W.D.; Bolton, E.E.; Bryant, S.H. The PubChem chemical structure sketcher. J. Cheminform. 2009, 1, 20. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Cough, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrini, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New machine learning and physics-based scoring functions for drug discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Waridel, P.; Ndjoko, K.; Hobby, K.R.; Major, H.J.; Hostettmann, K. Evaluation of Q-TOF-MS/MS and Multiple Stage IT-MSn for the Dereplication of Flavonoids and Related Compounds in Crude Plant Extracts. Analysis 2000, 28, 895–906. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Liu, Z.; Guo, Y.; Lu, S.; Du, H.; Cao, Y. Antioxidant capacity of flavonoids from Folium Artemisiae Argyi and the molecular mechanism in Caenorhabditis elegans. J. Ethnopharmacol. 2021, 279, 114398. [Google Scholar] [CrossRef] [PubMed]
- Aquino, A.J.; Pereira-Filho, E.R.; Oliveira, R.V.; Cass, Q.B. Chromatography conditions development by design of experiments for the chemotype differentiation of four Bauhinia species. Front. Chem. 2022, 10, 800729. [Google Scholar] [CrossRef] [PubMed]
- Chagas, M.D.S.S.; Behrens, M.D.; Moragas-Tellis, C.J.; Penedo, G.X.M.; Silva, A.R.; Gonçalves-de-Albuquerque, C.F. Flavonols and flavones as potential anti-inflammatory, antioxidant, and antibacterial compounds. Oxid. Med. Cell. Longev. 2022, 2022, 9966750. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Kumar, V.; Rahman, M.; Verma, A.; Patel, D.K. New insights into the medicinal importance, physiological functions, and bioanalytical aspects of an important bioactive compound of foods ‘Hyperin’: Health benefits of the past, the present, the future. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 31–42. [Google Scholar] [CrossRef]
- Zhao, L.; Yuan, X.; Wang, J.; Feng, Y.; Ji, F.; Li, Z.; Bian, J. A review on flavones targeting serine/threonine protein kinases for potential anticancer drugs. Bioorg. Med. Chem. 2019, 27, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Phonghanpot, S.; Jarintanan, F. Antiproliferative, antibacterial, and antioxidant activities of Bauhinia strychnifolia Craib aqueous extracts in gut and liver perspective. BMC Complement. Med. Ther. 2021, 21, 276. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, P.S.; Orué, S.L.; Leite, D.F.; de Toledo Espindola, P.P.; Cassemiro, N.S.; Da Silva, D.B.; Carollo, C.A.; Nunes-Souza, V.; Rabelo, L.A.; Campos, J.F.; et al. Beneficial effects of Bauhinia rufa leaves on oxidative stress, prevention, and treatment of obesity in high-fat diet-fed c57bl/6 mice. Oxid. Med. Cell. Longev. 2022, 2014, 8790810. [Google Scholar] [CrossRef]
- Ansari, M.A.; Fatima, Z.; Hameed, S. Sesamol: A natural phenolic compound with promising anticandidal potential. J. Pathog. 2014, 2014, 895193. [Google Scholar] [CrossRef]
- Saldanha, L.L.; Delgado, A.Q.; Marcourt, L.; Camaforte, N.A.P.; Vareda, P.M.P.; Ebrahimi, S.N.; Vilegas, W.; Dokkedal, A.L.; Queiroz, E.F.; Wolfender, J.; et al. Hypoglycemic active principles from the leaves of Bauhinia holophylla: Comprehensive phytochemical characterization and in vivo activity profile. PLoS ONE. 2021, 16, e0258016. [Google Scholar] [CrossRef]
- Niksic, H.; Becic, F.; Koric, E.; Gúsic, I.; Omeragic, E.; Muratovic, S.; Miladinovic, B.; Duric, K. Cytotoxicity screening of Thymus vulgaris L. essential oil in brine shrimp nauplii and cancer cell lines. Sci. Rep. 2021, 11, 13178. [Google Scholar] [CrossRef] [PubMed]
- Indrayanto, G.; Putra, G.S.; Suhud, F. Validation of In-Vitro Bioassay Methods: Application in Herbal Drug Research. Profiles Drug Subst. Excip. Relat. Methodol. 2021, 46, 273–307. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, D.; Li, C.; Zheng, Z.; Fu, M.; Ni, F.; Liu, Y.; Du, T.; Wang, H.; Griffin, G.E.; et al. Characterization of Zika virus endocytic pathways in human glioblastoma cells. Front. Microb. 2020, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Menezes, L.C.; Agrelli, A.; Oliveira, A.S.L.E.; Azevedo, E.A.N.; Morais, C.N.L. Zika virus: Critical crosstalk between pathogenesis, cytopathic effects, and macroautophagy. J. Cell. Biochem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.M.; Li, Y.Y.; Shi, Y.W.; Fang, Y.W.; Chao, R.; Gu, Y.C.; Wang, C.Y.; Shao, C.L. Integrating molecular networking and 1H NMR to target the isolation of Chrysogeamides from a library of marine derived Penicillium fungi. J. Org. Chem. 2019, 84, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Allard, P.M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.F.; Cachet, X.; et al. Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.F.; Zhang, X.; Zhu, F.; Huo, Z.Q.; Yao, Q.Q.; Feng, Q.; Liu, Z.; Zhang, G.M.; Yao, J.C.; Liang, H.B. MS/MS-Based molecular networking: An efficient approach for natural products dereplication. Molecules 2022, 28, 157. [Google Scholar] [CrossRef]
- Ramabulana, A.T.; Petras, D.; Madala, N.E.; Tugizimana, F. Metabolomics and molecular networking to characterize the chemical space of four Momordica plant species. Metabolites 2021, 11, 763. [Google Scholar] [CrossRef]
- Ramatapa, T.; Msobo, A.; Maphari, P.W.; Ncube, E.N.; Nogemane, N.; Mhlongo, M.I. Identification of plant-derived bioactive compounds using affinity mass spectrometry and molecular networking. Metabolites 2022, 12, 863. [Google Scholar] [CrossRef]
- Wong, G.; He, S.; Siragam, V.; Bi, Y.; Mbikay, M.; Chretien, M.; Qiu, X. Antiviral activity of quercetin-3-β-O-D-glucoside against Zika virus infection. Virol. Sin. 2017, 32, 545–547. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.E.; Kuster, R.M.; Yamamoto, K.A.; Salles, T.S.; Campos, R.; De Meneses, M.D.F.; Soares, M.R.; Ferreira, D. Quercetin and quercetin 3-O-glycosides from Bauhinia longifolia (Bong.) Steud. show anti-Mayaro virus activity. Parasit. Vectors 2014, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Loaiza-Cano, V.; Monsalve-Escudero, L.M.; Filho, C.S.B.; Martinez-Gutierrez, M.; Sousa, D.P. Antiviral role of phenolic compounds against Dengue virus: A review. Biomolecules 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Brognaro, H.; Prabhu, P.R.; de Souza, E.E.; Günther, S.; Reinke, P.Y.A.; Lane, T.J.; Ginn, H.; Han, H.; Ewert, W.; et al. Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease. Commun. Biol. 2022, 5, 805. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.S.; Santos, F.C.P.; Campana, P.R.V.; Costa, V.V.; de Pádua, R.M.; Souza, D.G.; Teixeira, M.M.; Braga, F.C. Natural products, and derivatives as potential Zika virus inhibitors: A comprehensive review. Viruses 2023, 15, 1211. [Google Scholar] [CrossRef]
- Santos, F.R.S.; Lima, W.G.; Maia, E.H.B.; Assis, L.C.; Davyt, D.; Taranto, A.G.; Ferreira, J.M.S. Identification of a potential Zika Virus inhibitor targeting NS5 methyltransferase using virtual screening and molecular dynamics simulations. J. Chem. Inf. Model. 2020, 24, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.R.S.; Nunes, D.A.F.; Lima, W.G.; Davyt, D.; Santos, L.; Taranto, A.G.; Ferreira, J.M.S. Identification of Zika virus NS2B-NS3 protease inhibitors by structure-based virtual screening and drug repurposing approaches. J. Chem. Inf. Model. 2019, 24, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Liu, H.; Li, J.; Yao, X.; Chen, Y.; Ke, C.; Liu, S. Structure-activity relationship of flavonoid bifunctional inhibitors against Zika virus infection. Biochem. Pharmacol. 2020, 177, 113962. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Choi, H.; Kim, Y.B. Therapeutic targets, and biological mechanisms of action of curcumin against Zika virus: In silico and in vitro analyses. Eur. J. Pharmacol. 2021, 904, 174144. [Google Scholar] [CrossRef]
- Lim, H.-J.; Nguyen, T.T.H.; Kim, N.M.; Park, J.S.; Jang, T.S.; Kim, D. Inhibitory effect of flavonoids against NS2B-NS3 protease of Zika virus and their structure activity relationship. Biotechnol. Lett. 2017, 39, 415. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).