Abstract
This study investigated the development of aptamer-based molecular probes to detect Methicillin-Resistant Staphylococcus aureus (MRSA) and evaluated the antibacterial activity. Early detection of MRSA infection will improve patients’ recovery and reduce the cost for treating patients. S. aureus can become resistant to methicillin and other β-lactam antibiotics through the expression of PBP2A protein, which is resistant to the action of methicillin. We have developed two aptamer molecular probes against PBP2A protein and whole bacterial cell (MRSA) under optimized in vitro conditions using SELEX approach. Target aptamer sequences were identified, and chemically synthesized aptamer probes were evaluated using fluorescently-labelled aptamer probes using flow cytometry and confocal imaging. Antibacterial activities of those aptamers were also evaluated using a bacterial killing assay. The results showed that high specific aptamers were developed against purified PBP2A protein. However, these aptamers showed less specificity to detect MRSA under in vitro condition. These aptamers showed no cytotoxic effect on 3T3 cells and no antibacterial activity against MRSA. The results suggested that the specific aptamer development and the in vitro selection methodology require further refinement to improve the diagnostic and therapeutic utility of these aptamers.
1. Introduction
Aptamers are short, single-stranded oligonucleotides (DNA or RNA) discovered in the early 1990s. They have found diverse applications in biotechnology [1,2]. Aptamers are isolated through a process called Systematic Evolution of Ligands by Exponential Enrichment (SELEX) [3,4] in which a starting library of oligonucleotides are screened for sequences that can bind specifically and with high affinity to a target of choice. The process itself is very versatile and the choice of targets to obtain aptamers can vary from small molecules (nucleotides, vitamins, amino acids, carbohydrates, dyes, antibiotics) [5,6], peptides, and proteins to whole cells [2,7]. Aptamers are also described as chemical versions of antibodies and can inhibit their targets through specific and strong interactions that are superior to those of biologics and small molecule therapeutics. They also avoid the toxicity and immunogenicity concerns of these traditional agents due to their nucleic acid compositions [2].
The library contains aptamers that have a variable or randomised region in the middle flanked by two constant regions called primer-binding regions [3,4]. The number of rounds required to drive a selection to completion depends on both the target and how much stringency is increased in each round; selection can be completed in 8–18 rounds [8]. The counter-SELEX method was introduced to increase the efficiency of aptamer selection by traditional SELEX [9]. Compared to traditional SELEX, counter-SELEX has a pre-clearing step using closely related structural analogs of the target to effectively discard a non-specific aptamer. This allows improvement in aptamer selection and can also be applied to other modified SELEX methods. Aptamers have great potential in optical imaging as a cost-effective imaging method that typically uses fluorescent or bioluminescent molecules. Aptamer-based optical imaging can be divided into direct targeting and activatable probes [10]. Direct conjugation with an aptamer and a fluorescent molecule has been widely studied [11,12,13,14,15,16].
The simplest way to image via a visualized aptamer is by using fluorescence probes. Aptamer probes have been used in therapy [17], diagnosis [18], in vitro bioanalysis [19], and in vivo imaging [10,20]. The combination of aptamers with nanotechnology is also an actively studied field [21]. Aptamers can bind to their cognate protein and efficiently inhibit the function of target as an antagonist. Many therapeutic studies using aptamers have been conducted to treat certain diseases by inhibiting therapeutic target activity and decreasing the partner binding property [22,23]. The first FDA-approved aptamer drug for age-related macular degeneration is a typical antagonistic aptamer [24]. Macugen (pegaptanib) is a 28-base RNA oligonucleotide with two branched 20 kDa PEG moieties [25]. It selectively binds to the vascular endothelial growth factor (VEGF)165 isoform when introduced intravitreally.
Staphylococcus (from the Greek; grape-berry) were first described and classified in 1882 by the Scottish surgeon Sir Alexander Ogston [26]. S. aureus is a commonly isolated human microorganism that can colonise the anterior nares and other skin regions of healthy individuals [27]. It is one of the main causes of hospital- and community-acquired infections, and can result in serious consequences. S. aureus is an important cause of skin and soft tissue infections, endovascular infections, pneumonia, septic arthritis, endocarditis, osteomyelitis, foreign body infections, and sepsis [28,29,30,31]. In addition, S. aureus causes infections of surgical wounds and prosthetic implants [32].
Methicillin was introduced by Beecham in 1959. However, one year later, methicillin-resistant S. aureus (MRSA) was detected in the United Kingdom [33]. Unlike resistance to penicillin, the mechanism underlying methicillin resistance protects the bacteria from the entire class of β-lactam antibiotics, including penicillin, cephalosporins, and carbapenems. The mortality rate from severe MRSA infections is about 20% and it has been estimated that MRSA infections are the leading cause of death by a single infectious agent in the US, exceeding deaths caused by HIV/AIDS [34,35]. MRSA carries the mecA gene that codes for an alternative penicillin binding protein, PBP2A, with low binding affinity to all β-lactams [36].
PBP2A belongs to the group of high molecular mass (78 kDa) family of PBPs and consists of a transpeptidase domain and a non-penicillin binding domain of unknown function [37]. PBP2A is known to possess a low affinity for β-lactams that allows MRSA strains to grow in antibiotic concentrations that inactivate all native PBPs [38]. PBP2A is a poorly active enzyme when compared to other native PBPs that synthesize highly cross-linked peptidoglycan [39]. Even when the transpeptidase activity of all native PBPs is inhibited by the presence of methicillin, PBP2A has been shown to rely on the transglycosylase, β-lactam-insensitive domain of the native PBP2 to confer resistance in MRSA [40].
S. aureus specific ssDNA aptamer panels are developed by a whole bacterium-based SELEX procedure. After several rounds of selection with S. aureus as the target and Streptococcus pneumoniae and S. epidermidis as counter targets, a panel of aptamers to detect S. aureus was developed using cell-based SELEX by [41]. They demonstrated that the simultaneous probing of several targets by a set of specific aptamers proved to be more efficient than probing by an individual aptamer in detection of S. aureus. Unlike single target SELEX, which generates an enriched pool of aptamers to the single target, the complex SELEX ends up with an enriched pool of aptamers to various targets on the cells [42,43]. In another study, a novel and sensitive fluorescence bioassay for the simultaneous detection of S. typhimurium and S. aureus was developed. This technique used aptamer-conjugated magnetic nanoparticles for both recognition and concentration elements and using nanoparticles as highly sensitive dual-colour labels [44]. The luminescent signal was effectively amplified with the help of both magnetic separation and concentration. This method demonstrates the first use of aptamer-conjugated magnetic nanoparticles as the capture and concentration element and the use of up-conversion nanoparticles as the fluorescence element for the simultaneous detection of two types of pathogenic bacteria. According to published literature, several studies have focused on developing S. aureus specific aptamers in in vitro.
However, MRSA-specific aptamer molecules have not yet been developed and currently we do not have reliable and sensitive techniques to detect in vivo MRSA over other Staphylococcus species. Hence, this study focused on developing bacterial cell MRSA aptamer and PBP2A aptamer molecular probes to detect MRSA and generate therapeutic applications.
2. Materials and Methods
2.1. Bacterial Culture and Reagents
All bacterial strains used in this study were log-phase cultures of bacteria grown in Miller’s Luria–Bertani (LB) broth, and bacterial titers were estimated by optical density. The following bacterial strains were used in this study: Staphylococcus aureus (MRSA-ATCC 252), Staphylococcus aureus (MSSA-ATCC 25923), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (PAO1), Staphylococcus epidermidis-clinical strain, Klebsiella pneumoniae (ATCC BAA 1706), Burkholderia cepacia (ATCC 25416), and E. coli BL21 (DE3).
2.2. PBPA2A Protein Overexpression
mecA gene was cloned and PBP2A protein was over-expressed and purified according to Haghighat et al. [45].
2.3. SDS-PAGE Protein Gel
Purified protein PBP2A was run on pre-cast 4–12% Bis-Tris polyacrylamide gels NuPAGE-Novex from Life technologies (Cat# NP0321BOX, Invitrogen, Waltham, MA, USA) in order to visualize the protein purity and to determine the size of the protein.
2.4. Latex Agglutination Test for Penicillin Binding Protein (PBP2A)
This test is a rapid latex agglutination assay to detect PBP2A protein in MRSA. The Oxoid PBP2A Latex Test (Cat# DR0900) was performed according to the manufacturer’s instructions in order to detect purified PBP2A protein.
2.5. Designing of Aptamer Libraries
Aptamer libraries were designed according to Sefah [46] and ordered from Integrated DNA Technologies (IDT). The ‘antisense’ strand was functionalised at the 5′ end with biotin. All oligonucleotide sequences were purified by high-performance liquid chromatography (HPLC) in 1 μM scale. Fluorescein isothiocyanate (FITC) labelled forward primers were used to monitor the progress of selection by flow cytometry.
2.6. PCR Amplification of Aptamer Libraries
Aptamer libraries were amplified by 10 PCR cycles using the following amplification conditions: hot start at 95 °C for 2 min, denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 30 s, followed by final extension for 4 min at 72 °C.
2.7. Agarose Gel Electrophoresis
A 4% agarose gel was prepared with Tris-borate-EDTA (TBE) buffer (Promega-Cat No.: V4251, Madison, WI, USA) and GelRed (Cambridge Bioscience-Cat No.: BT41003, Cambridge, UK) (1:10,000) added to visualize the ssDNA. After making the gel, 5 μL of DNA sample was loaded with gel loading dye (1:6) and electrophoresis was performed at 100 V for 40 min in TBE buffer. The bands were observed under UV light and images of the gels captured.
2.8. Experimental Design and SELEX Procedure
The ssDNA library pool was prepared by adding 50 μL of 100 μM (5 nmol) DNA libraries to 500 μL of PBS, mixed and heated at 95 °C for 5 min. The resulting mixture was snap-cooled on ice and kept on ice until ready to use later the same day.
The aptamer library and the target molecules (bacteria and PBP2A) were incubated in 1 mL PBS for 30 min at 37 °C on a rotary shaker. After incubation, unbound aptamer was removed by washing three times in PBS and centrifuged at 13,000× g for 1 min at 4 °C. Then we discarded the supernatants. The pellet mixture was mixed in DNase-free water and heated at 95 °C for 10 min, centrifuged at 13,000× g for 5 min, and supernatant containing eluted DNA was collected for PCR amplification.
MRSA-aptamer:
The SELEX procedure was carried out independently against MRSA (ATCC252), MSSA (ATCC 25923), PAO1-Pseudomonas aeruginosa, Kebsiella pneumoniae (ATCC BAA1706), E. coli (ATCC 25922), and alveolar epithelial cell line (A549) for five rounds without counter selection. These ssDNA aptamers were sequenced using an Ion Torrent sequencing method. We attempted to eliminate non-specific aptamers at the sequence level; the sequenced data were carefully analysed and common sequences shared with the other bacteria were eliminated.
PBP2A-aptamer:
SELEX was carried out against PBP2A protein (0.1 mg/mL) coated Ni-NTA Agarose beads (Qiagen-Cat No.: 30210, Hilden, Germany) and 15 SELEX rounds were carried out before Ion Torrent sequencing with counter selection against BSA and FBS.
2.9. Preparation of ssDNA from PCR Products
Next, 200 μL of Streptavidin Sepharose beads (GE Healthcare Life Sciences-Cat No.: 17-5113-01, Buckinghamshire; UK) were mixed with 1 mL of PCR products in a 1.5 mL Eppendorf tube and incubated for 5 min at room temperature on a rotary shaker and washed three times with water. Single stranded aptamers (non-biotinylated strand) were separated from the immobilized complementary strand with a 5 min incubation of 50 μL of fresh 200 mM NaOH. Supernatant was taken after centrifugation at 13,000× g for 1 min (ssDNA aptamer). The supernatant was diluted into 1 mL of PBS-T, containing 10 μL of 200 mM monobasic phosphate buffer to adjust the pH to 7.5. Finally, the material was heated to 95 °C for 10 min then immediately placed at 4 °C until the next round of SELEX.
2.10. Ion Torrent Sequencing and Data Analysis
Seven aptamer DNA samples were sequenced at The Institute of Genetics and Molecular Medicine, Western General Hospital, Edinburgh, UK. The sequenced data was analysed using TextWrangler, a text editor software, which generated two probable aptamer probes called PBP2A specific and MRSA specific. The hairpin structures of MRSA and PBP2A aptamer probes were determined using the website at Integrated DNA Technologies (IDT), the OligoAnalyzer 3.1. To evaluate the binding efficacy of the aptamer probes, they were re-synthesised with 5′ FITC modification by SIGMA-ALDRICH, UK.
2.11. Evaluation of PBP2A-Aptamer Probe Binding In Vitro; Ni-NTA Beads Assay
Ni-NTA agarose beads were incubated with 100 μg/mL of PBP2A, BSA, FBS, and trypsin separately at room temperature for 1 h on a rotary shaker, and the unbound proteins were washed. These protein coated beads were used as target substrates for the FITC labelled PBP2A-aptamer probe to evaluate the efficacy of binding by using plate reader, confocal microscopy, and fibre-based confocal microendoscopy (FCM-Cellvizio) after incubation for 30 min at 37 °C. Before analysis, the beads were washed three times with PBS in order to remove excess aptamer probes. For the plate reader analysis, 100 μL of labelled beads were loaded into 96-well plates (three wells) and fluorescence-measured at 488/520 nm. The beads were also visualised with live confocal microscopy (Zeiss LSM510meta) and FCM-Cellvizio).
2.12. Evaluation of MRSA and PBP2A-Aptamer Probe Binding In Vitro Bacterial Binding Assay
A panel of bacteria were grown overnight and washed three times in PBS. A concentration of 108 cfu/mL bacteria and the FITC labelled aptamer probes at 100 nM was co-incubated in 1 mL PBS for 30 min at 37 °C on rotatory shaker. Excess unbound aptamer was washed off and analysed using flow cytometry.
2.13. Study of Aptamer as a Therapeutic Agent for Bacterial Infection
The assay was carried out in 96-well plates with aptamers at 1 μM concentration in 100 μL of Mueller-Hinton broth (MHB). In test wells, 50 μL of bacterial inoculum (2 × 106 cfu/mL) in MHB was added to 50 μL of aptamer solution in MHB. Controls contained 50 μL of inoculum and 50 μL of MHB without aptamer. Bacterial growth was measured by recording the optical density (O.D.) at 600 nm over 16 h of incubation at 35 °C. The O.D. was measured every 15 min using a spectrophotometer. Growth inhibition was determined as the concentration of aptamer that completely inhibited bacterial growth after incubation. Experiments were conducted in triplicate on three separate occasions [47,48].
2.14. Aptamer Cytotoxicity Studies on 3T3 Cells
To evaluate the potential cytotoxicity of the aptamers, the CellTiter-Glo® Luminescent Cell Viability Assay (Promega—Cat No.: G7570, Madison, WI, USA) was carried out according to the manufacturer instructions.
2.15. Statistics
All statistical analyses were performed on GraphPad Prism (version 5). The results were represented as the mean ± SEM. Data sets were compared using a two-tailed unpaired Student’s t-test. Statistical significance was set at p < 0.05.
3. Results and Discussion
3.1. Designing of Aptamer Libraries
In this study two DNA aptamer libraries have been developed to test, as shown in the Table 1 below, and we used the SELEX protocol to develop specific aptamer probes. The table below shows that each library has different lengths of variable regions of 30 and 40 nucleotides.

Table 1.
Two aptamer libraries used in this study.
3.2. Agarose Gel Electrophoresis to Visualize Naïve Aptamer Libraries
An agarose gel was run to determine the length of the naïve aptamer libraries as a quality control of the naïve aptamer library (Figure 1). The libraries were PCR-amplified using specific primers to determine the optimum PCR conditions for the full amplification of the libraries.
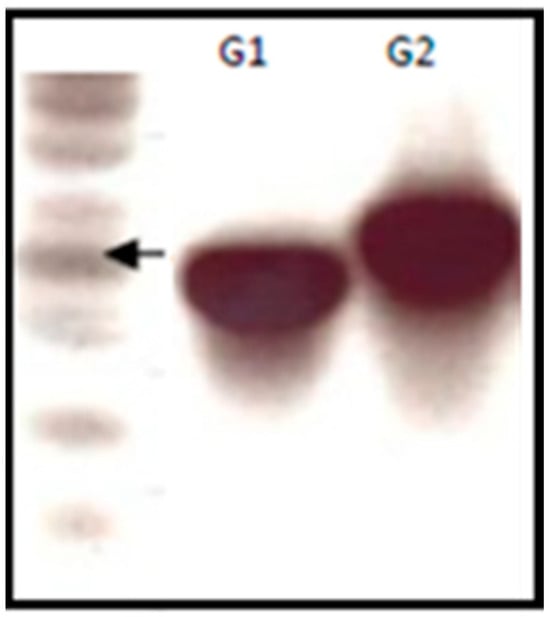
Figure 1.
Agarose (4% TBE) gel electrophoresis image showing the products of the naïve aptamer libraries (1 μL/lane). G1 and G2 contained 30 and 40 nucleotides in the variable region respectively.
3.3. Over-Expression and Purification of PBP2A Protein
PBP2A protein was over-expressed and purified according to [45]. Purified proteins were visualized by running an SDS-polyacrylamide protein gel and staining with coomassie brilliant blue (Figure 2). The BCA assay was performed to quantify the over-expressed protein, and the PBP2A protein concentrations was achieved above 1 mg/mL of 10 mL.
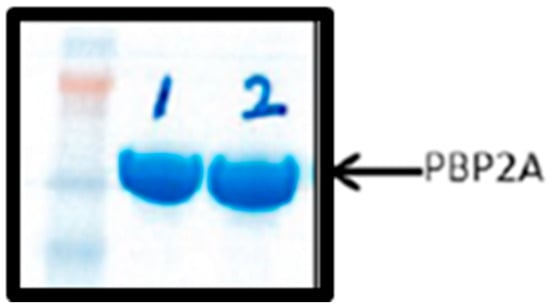
Figure 2.
SDS-polyacrylamide protein gel showing the purified protein Lanes 1 (1-sample) and 2 (2-sample) represent the purified PBP2A proteins from two different clones.
3.4. Penicillin Binding Protein (PBP2A) Latex Agglutination Test
To confirm the activity of the over-expressed PBP2A protein, a Latex Agglutination test was carried out. The PBP2A latex agglutination test is a 20-min phenotypic test that detects PBP2A in isolated colonies [49]. The test results show that the purified PBP2A protein is biochemically similar to the active proteins in the MRSA but not in MSSA (Figure 3).
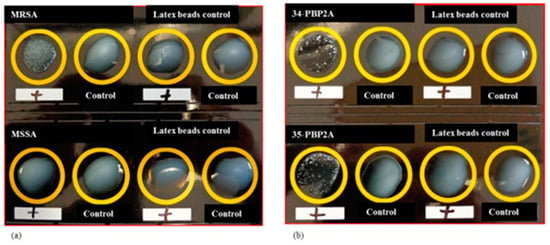
Figure 3.
Latex beads agglutination assay for purified PBP2A protein (a) MRSA and MSSA bacterial strains showing the positive and negative results respectively for the latex beads agglutination test. (b) Two samples of PBP2A over-expressed protein (34 and 35) respond to the test as MRSA. This indicates that the over-expressed PBP2A protein is showing the same functional properties to its native protein present in MRSA but not in MSSA.
3.5. Experimental Design of the SELEX Procedure
SELEX assays were performed on bacterial cells and PBP2A protein using ssDNA aptamer libraries using PBS as a buffer. We used 10 PCR cycles to amplify the bound ssDNA from the target molecules. An agarose gel was run to quantify and visualise the seven samples of dsDNA before sending for sequencing (Figure 4).
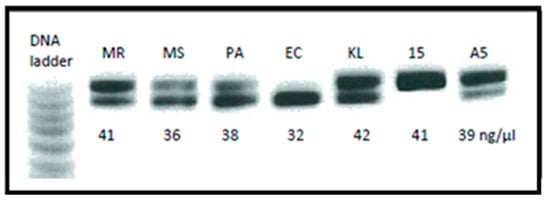
Figure 4.
Agarose (4% TBE) gel electrophoresis image showing the final PCR products before sending for Ion Torrent sequencing. Two bands of DNA represent the combination of 74 bp and 84 bp aptamer G-libraries also indicate the concentrations of DNA present in the sample. MR-MRSA; MS-MSSA; PA-PAO1 Pseudomonas; EC—E.coli; KL—Kebsiella pneumoniae; 15-PBP2A 15 rounds of SELEX; A5-A549 eukaryotic cells.
3.6. Sequence Alignment between Two Aptamer Probes
Sequences of two aptamer probes are shown below indicating that they have 50% identity. Underlined sequences are the primer binding sites of the aptamer probes. Red color indicates the common nucleotides shared between two aptamer probes.
PBP2A-aptamer:
5′CTACACGACGCTCTTCCGATCTCGCGGTGTGGAATGGAAAGGCAGAGGGGGTAGACGGAGAAGATCGGAAGAGCGGTTCAGCA
MRSA-aptamer:
5′CTACACGACGCTCTTCCGATCTGGCGGCGGGGGATGGTGGCGGAATGGTGGTGGTGAGCTGGAGATCGGAAGAGCGGTTCAGCA
Red color indicates the common nucleotides shared between two aptamer probes.
3.7. The Hairpin Structures of MRSA and PBP2A Aptamer Probes
Figure 5 shows the hairpin structures of the aptamer probes. Their minus Gibbs free energy (ΔG) indicates higher stability and higher melting temperatures, and also indicates that they are very stable aptamer probes at room temperature.
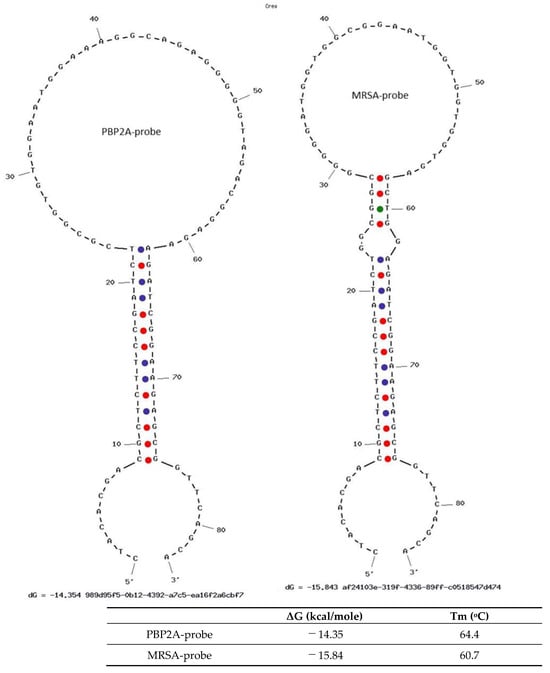
Figure 5.
The hairpin structures of MRSA and PBP2A aptamer probes. The minus Gibbs free energy (ΔG) indicates that the hairpin structures of aptamer probes are stable, and the melting temperatures are well above room temperature.
3.8. Evaluation of PBP2A-Aptamer Probe Binding In Vitro; Ni-NTA Beads Assay
The two lead aptamer probes were re-synthesised with 5′ FITC modification from SIGMA-ALDRICH. For the assays, an FITC modified PBP2A-aptamer probe was used to evaluate the specificity of the aptamer candidate on different proteins including PBP2A protein. These protein coated beads were used as target molecules for the PBP2A-aptamer probe and the efficacy of binding was evaluated by using a plate reader, confocal microscopy, and confocal laser endomicroscopy (Figure 6). The images clearly indicate that only the PBP2A coated Ni beads are fluorescently labelled with the PBP2A-aptamer probe showing the expected protein specificity of the probe.
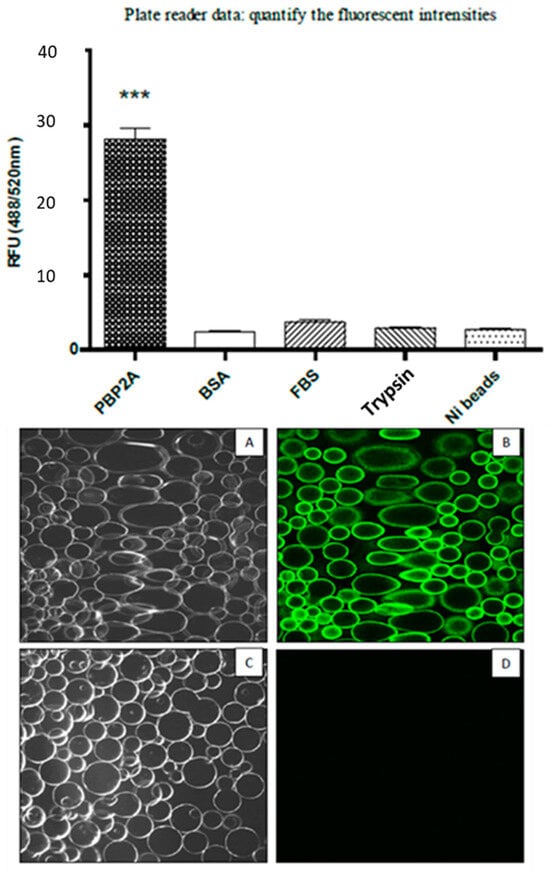
Figure 6.
PBP2A coated Ni-NTA protein binding assay shows that the PBP2A-aptamer is more specific to PBP2A protein compared with the BSA, FBS and Trypsin. (A). Transmitted image of PBP2A protein coated beads; (B). Fluorescent image of PBP2A coated beads; (C). Transmitted image of FBS coated beads; (D). Fluorescent image FBS coated beads. ***: p < 0.001.
The laser endomicroscopy (Cellvizio) experiments were carried out to prove that the FAM labelled aptamer can be used in parallel with the clinically approved Cellvizio imaging platform. To achieve this, Ni beads were visualised using the Cellvizio mini probe and data were analysed using proprietary image analysis tools (Cellvizio Viewer) (Figure 7).
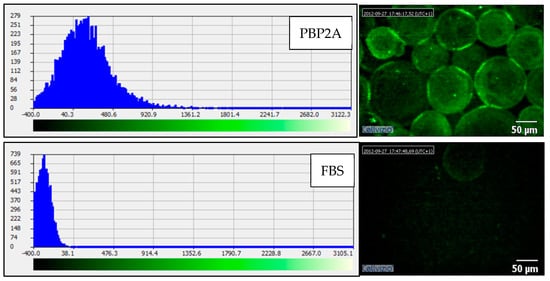
Figure 7.
PBP2A coated Ni-NTA protein binding assay; Cellvizio image analysis. Fluorescent image of the sample and the fluorescent intensity of the image. The reduced fluorescent shift to the right on the histogram indicates that the FBS beads are not labelled with the PBP2A-aptamer probe.
3.9. Evaluation of MRSA-Aptamer Probe Binding In Vitro Ni-NTA Bead Assay
The Ni-NTA beads assay was carried out to evaluate probe binding capacity to other proteins including PBP2A protein (Figure 8). The results indicated that the MRSA-aptamer probe also binds to PBP2A protein. This is not unexpected as the two aptamer probes share 50% sequence homology.
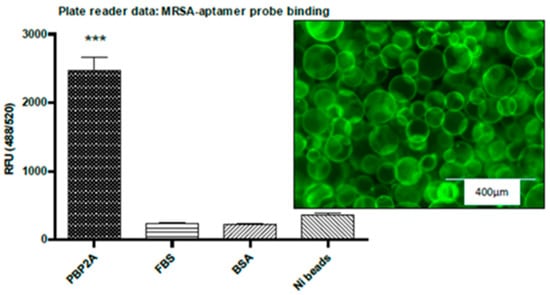
Figure 8.
PBP2A coated Ni-NTA protein binding assay shows that the MRSA-aptamer binds to PBP2A protein at a higher rate compared to FBS and BSA. The confocal image shows that the PBP2A coated Ni beads are labelled with MRSA-probe. ***: p < 0.001.
3.10. Evaluation of MRSA and PBP2A-Aptamer Probe Binding In Vitro Bacterial Binding Assay
The ultimate objective in developing these probes was to detect MRSA specifically over other bacterial species. To assess the specificity of these probes, bacterial binding assays were carried out and bacterial cells were analysed using flow cytometry (Figure 9). A panel of bacteria were used at 108 cfu/mL bacteria and probes at 100 nM in PBS. The PBP2A-probe showed no specificity towards MRSA, though it shows very high specificity to PBP2A protein in Ni-NTA beads binding assay. This may be due to the fact that the aptamer has been developed against the whole protein and it may share similar binding sites with the other bacterial proteins as well.
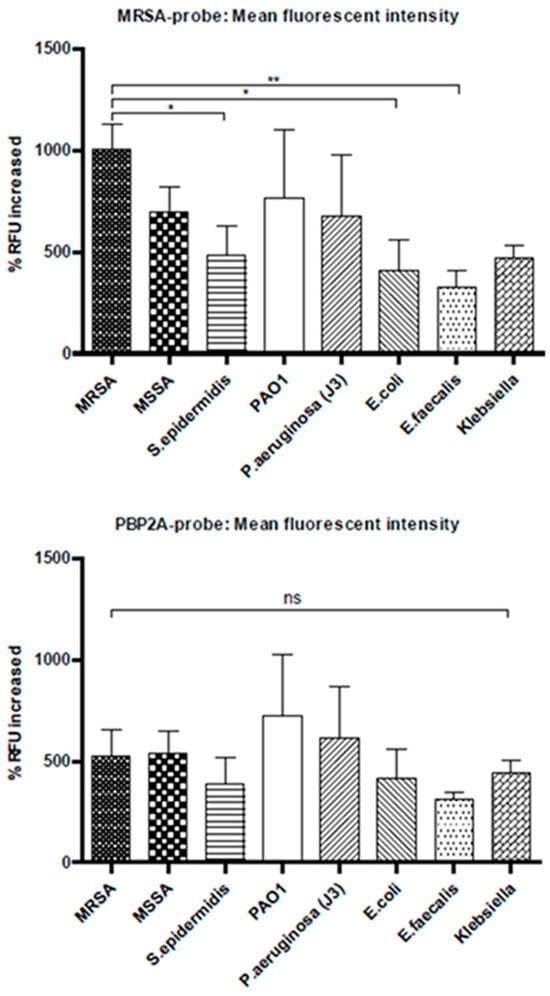
Figure 9.
Flow cytometry data showing percentage increased in mean fluorescence intensity. The top graph shows the results of MRSA-probe, and the bottom graph shows the PBP2A-probe. MRSA probe shows higher binding to MRSA bacteria compared to the rest of the bacteria. However, PBP2A-probe shows no specificity (Panels represent mean values ± SEM for n = 3). *: p < 0.05; **: p < 0.01; ns: Not significant.
These probes show no specificity towards MRSA in the bacterial binding assays described above. This is due to the non-specific binding of the probes towards the other bacteria tested. In addition, the in silico counter selection has not been successful. During ion torrent sequencing, far fewer short reads were observed and that may lead to lose some data for in silico counter selection. These sequences may have important and key potential aptamer sequences. Similarly, there are several other factors that may contribute to the unsuccessful SELEX and the development of aptamer probes [50,51].
Turek et al. [52] indicate that all MRSA clinical strains do not share similar surface properties. MRSA, S. aureus, and Enterococcus are all gram-positive bacteria. Based on their similarities, some common binding can be expected by the commonality of proteins among the strains, whereas others are more specific to each type of bacterium. However, they have not been tested with gram negative bacterial strains to study the efficacy of binding of these aptamers.
Modifying and optimising the selection buffer conditions may counteract or otherwise stabilise the interactions between the target and the unselected aptamer pool. With a negatively charged target, decreasing the pH below a target’s isoelectric point (pI) will neutralise the negative charge and encourage the binding of the negatively charged pool [53].
The SELEX buffer conditions may also vary with monovalent salt(s) identity, monovalent salt(s) concentration, and divalent salt identity, divalent salt concentration, buffer identity, buffer concentration, and pH. The optimised buffer conditions likely increase the probability of a successful selection and therefore promote higher ratios of successful aptamer selections against a variety of targets. Therefore, different buffer conditions were selected and tested on two different aptamer libraries with and without bacteria. The best buffer condition was selected as PBS [54,55], with Ca2+ and Mg2+ to eliminate the nonspecific binding and PCR amplification. Throughout the experimental protocol, this study used DNA low binding Eppendorf tubes to eliminate non-specific binding of DNA onto the wall of tubes. The BSA concentration in the selection buffer (PBS with Ca2+ and Mg2+) was increased to 1% to minimise the non-specific binding and increase the stringency of the selection.
3.11. Aptamer Cytotoxicity Studies on 3T3 Cells
This assay was carried out to investigate aptamer toxicity on mammalian cell lines in order to study the feasibility of using these probes in vivo in murine models. The 3T3 cells have been used as a standard cell line to study the cellular toxicity of potential drug targets. The aptamers demonstrated no cellular toxicity at the concentrations used (Figure 10). However more extensive studies are necessary before a conclusion could be made concerning the in vivo safety or toxicity of these aptamer.
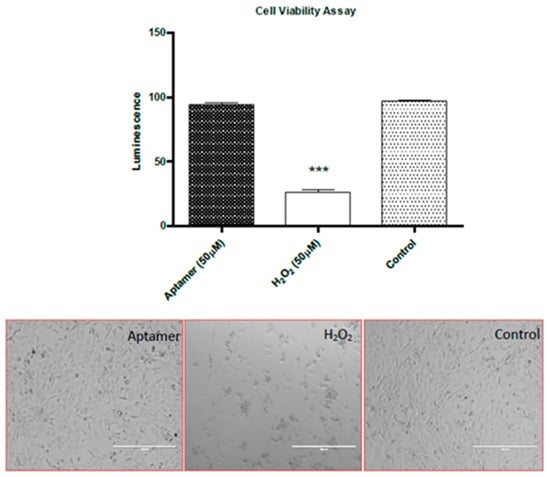
Figure 10.
Aptamer toxicity assay: The graph shows the ATP derived luminescence after incubation for 24 h. Below are the light microscopy images of healthy and non-viable 3T3 cells (Panels represent mean values ± SEM for n = 3). (Magnification ×40). ***: p < 0.001.
3.12. Study of Aptamer as a Therapeutic Agent for Bacterial Infection
A 242 bp fragment of mecA gene was amplified by PCR from S. aureus, expressed, purified, and used to stimulate humoral immune response in a murine model. This investigation revealed PBP2A as a potential to develop a vaccine against MRSA infection [45]. Therefore, a bacterial killing assay was carried to investigate them as a potential therapeutic agent if they bind to bacterial essential cell division proteins leading to killing of bacteria (Figure 11). The protein PBP2A is an essential cell division protein in MRSA, and it is a potential antibacterial drug target. Blocking its function will prevent bacterial cell division and leading to death.
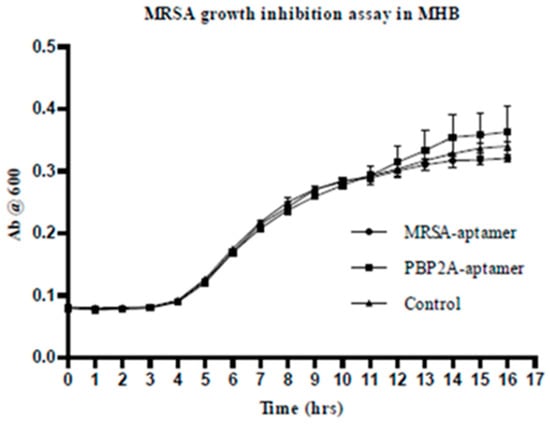
Figure 11.
MRSA growth inhibition assay in MHB. 1 μM of aptamer has been used for this assay in MHB (100 μL) and 106 cfu/mL bacterial. The growth of bacteria was assessed by measuring the absorbance at 600 nm for 16 h. The data shows that there is no effect on MRSA growth from aptamer probes.
4. Conclusions
The PBP2A and MRSA aptamer molecular probes were able to show high specificity in in vitro Ni-NTA beads assay with 50% identical aptamer sequence. However, both PBP2A and MRSA aptamer probes were not specific enough to detect MRSA over other bacterial species in bacterial binding assays. These aptamers showed no toxicity towards 3T3 cells and had no antibacterial effects. The results suggested that the specific aptamer development and the in vitro selection methodology require further refinement to improve aptamers as diagnostic and therapeutic agents.
Funding
This study was funded by the University of Edinburgh, Scotland.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
I acknowledge the support and guidance from Kev Dhaliwal, Mark Bradley and Chris Haslett.
Conflicts of Interest
The author declares no conflict of interest.
References
- Gold, L.; Polisky, B.; Uhlenbeck, O.; Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995, 64, 763–797. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- He, F.; Wen, N.; Xiao, D.; Yan, J.; Xiong, H.; Cai, S.; Liu, Z.; Liu, Y. Aptamer-Based Targeted Drug Delivery Systems: Current Potential and Challenges. Curr. Med. Chem. 2020, 27, 2189–2219. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, F.; Rezaee, M.A.; Ebrahimzadeh, S.; Yousefi, L.; Nouri, R.; Kafil, H.S.; Gholizadeh, P. Novel Strategies to Combat Bacterial Biofilms. Mol. Biotechnol. 2021, 63, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Hu, J.; Peng, M.; Liu, J.; Liu, J.; Liu, H.; Zhao, X.; Tan, W. Generating Aptamers by Cell-SELEX for Applications in Molecular Medicine. Int. J. Mol. Sci. 2012, 13, 3341–3353. [Google Scholar] [CrossRef]
- Fitzwater, T.; Polisky, B. A SELEX primer. Methods Enzymol. 1996, 267, 275–301. [Google Scholar] [CrossRef]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-Resolution Molecular Discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef]
- Hong, H.; Goel, S.; Zhang, Y.; Cai, W. Molecular imaging with nucleic acid aptamers. Curr. Med. Chem. 2011, 18, 4195–4205. [Google Scholar] [CrossRef]
- Shi, H.; Tang, Z.; Kim, Y.; Nie, H.; Huang, Y.F.; He, X.; Deng, K.; Wang, K.; Tan, W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem.—Asian J. 2010, 5, 2209–2213. [Google Scholar] [CrossRef]
- Cui, Z.-Q.; Ren, Q.; Wei, H.-P.; Chen, Z.; Deng, J.-Y.; Zhang, Z.-P.; Zhang, X.-E. Quantum dot–aptamer nanoprobes for recognizing and labeling influenza A virus particles. Nanoscale 2011, 3, 2454–2457. [Google Scholar] [CrossRef] [PubMed]
- Talbot, L.J.; Mi, Z.; Bhattacharya, S.D.; Kim, V.; Guo, H.; Kuo, P.C. Pharmacokinetic characterization of an RNA aptamer against osteopontin and demonstration of in vivo efficacy in reversing growth of human breast cancer cells. Surgery 2011, 150, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-Z.; Yu, R.-N.; Chen, J.; Ma, Z.-Y.; Zhao, Y.-D. Targeted quantum dots fluorescence probes functionalized with aptamer and peptide for transferrin receptor on tumor cells. Nanotechnology 2012, 23, 485104. [Google Scholar] [CrossRef]
- Song, Y.; Zhu, Z.; An, Y.; Zhang, W.; Zhang, H.; Liu, D.; Yu, C.; Duan, W.; Yang, C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, X.; Zhang, Y.; Zhou, G.; Ke, X.; Wang, H.; Tinnefeld, P.; He, Z. One-pot synthesized aptamer-functionalized CdTe:Zn2+ quantum dots for tumor-targeted fluorescence imaging in vitro and in vivo. Anal. Chem. 2013, 85, 5843–5849. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Kulabhusan, P.K.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in bioanalytical applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef]
- López-Colón, D.; Jiménez, E.; You, M.; Gulbakan, B.; Tan, W. Aptamers: Turning the spotlight on cells. WIREs Nanomed. Nanobiotechnol. 2011, 3, 328–340. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Wang, K.; Li, W.; Cui, L.; Li, X. Angiogenin-mediated photosensitizer-aptamer conjugate for photodynamic therapy. Chem. Med. Chem. 2011, 6, 1778–1780. [Google Scholar] [CrossRef] [PubMed]
- Siller-Matula, J.M.; Merhi, Y.; Tanguay, J.-F.; Duerschmied, D.; Wagner, D.D.; McGinness, K.E.; Pendergrast, P.S.; Chung, J.-K.; Tian, X.; Schaub, R.G.; et al. ARC15105 is a potent antagonist of von willebrand factor mediated platelet activation and adhesion. Arter. Thromb. Vasc. Biol. 2012, 32, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Sell, S.; Kaczmarek, P.; Maasch, C.; Buchner, K.; Pruszynska-Oszmalek, E.; Kolodziejski, P.; Purschke, W.G.; Nowak, K.W.; Strowski, M.Z.; et al. A mixed mirror-image DNA/RNA aptamer inhibits glucagon and acutely improves glucose tolerance in models of type 1 and type 2 diabetes. Perspect. Surg. 2013, 288, 21136–21147. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, B. Pegaptanib for the treatment of age-related macular degeneration. Exp. Eye Res. 2006, 83, 615–619. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef]
- Wilson, L.G. The early recognition of streptococci as causes of disease. Med. Hist. 1987, 31, 403–414. [Google Scholar] [CrossRef]
- Wertheim, H.F.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Lowrey, J.A.; Savage, N.D.L.; Palliser, D.; Corsin-Jimenez, M.; Forsyth, L.M.G.; Hall, G.; Lindey, S.; Stewart, G.A.; Tan, K.A.L.; Hoyne, G.F.; et al. Induction of tolerance via the respiratory mucosa. Int. Arch. Allergy Immunol. 1998, 116, 93–102. [Google Scholar] [CrossRef]
- Biedenbach, D.J.; Moet, G.J.; Jones, R.N. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997–2002). Diagn. Microbiol. Infect. Dis. 2004, 50, 59–69. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial bloodstream infections in us hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004, 39, 309–317, Erratum in: Clin. Infect. Dis. 2004, 39, 93; Erratum in: Clin. Infect. Dis. 2005, 40, 1077. [Google Scholar] [CrossRef]
- Hoban, D.J.; Biedenbach, D.J.; Mutnick, A.H.; Jones, R.N. Pathogen of occurrence and susceptibility patterns associated with pneumonia in hospitalized patients in North America: Results of the SENTRY antimicrobial surveillance study (2000). Diagn. Microbiol. Infect. Dis. 2002, 45, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Pantosti, A.; Venditti, M. What is MRSA? Eur. Respir. J. 2009, 34, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Jevons, M. Methicillin resistance in staphylococci. Lancet 1963, 281, 904–907. [Google Scholar] [CrossRef] [PubMed]
- Klevens, R.M.; Edwards, J.R.; Richards, C.L., Jr.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef]
- Hanberger, H.; Walther, S.; Leone, M.; Barie, P.S.; Rello, J.; Lipman, J.; Marshall, J.C.; Anzueto, A.; Sakr, Y.; Pickkers, P.; et al. Increased mortality associated with meticillin-resistant Staphylococcus aureus (MRSA) infection in the Intensive Care Unit: Results from the EPIC II study. Int. J. Antimicrob. Agents 2011, 38, 331–335. [Google Scholar] [CrossRef]
- Ito, T.; Hiramatsu, K.; Tomasz, A.; de Lencastre, H.; Perreten, V.; Holden, M.T.G.; Coleman, D.C.; Goering, R.; Giffard, P.M.; Skov, R.L.; et al. Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 2012, 56, 4997–4999. [Google Scholar] [CrossRef]
- Goffin, C.; Ghuysen, J.-M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar] [CrossRef]
- Gaisford, W.C.; Reynolds, P.E. Methicillin resistance in Staphylococcus epidermidis. Relationship between the additional penicillin-binding protein and an attachment transpeptidase. JBIC J. Biol. Inorg. Chem. 1989, 185, 211–218. [Google Scholar] [CrossRef]
- de Jonge, B.L.M.; Tomasz, A. Abnormal Peptidoglycan Produced in a Methicillin-Resistant Strain of Staphylococcus aureus Grown in the Presence of Methicillin: Functional Role for Penicillin-Binding Protein 2A in Cell Wall Synthesis. Antimicrob. Agents Chemother. 1993, 37, 342–346. [Google Scholar] [CrossRef]
- Pinho, M.G.; Filipe, S.R.; de Lencastre, H.; Tomasz, A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, M.; Yang, G.; Zhang, D.; Ding, H.; Wang, H.; Fan, M.; Shen, B.; Shao, N. Single-stranded DNA aptamers that bind differentiated but not parental cells: Subtractive systematic evolution of ligands by exponential enrichment. J. Biotechnol. 2003, 102, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Cao, Z.C.; Li, Y.; Tan, W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007, 53, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Zhu, C.; Ma, X.; Wang, Z.; Yu, Y.; Jiang, Y. Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Anal. Chim. Acta 2012, 723, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, S.; Siadat, S.D.; Sorkhabadi, S.M.; Sepahi, A.A.; Mahdavi, M. Cloning, expression and purification of penicillin binding protein2a (pbp2a) from methicillin resistant Staphylococcus aureus: A study on immunoreactivity in Balb/C Mouse. Avicenna J. Med. Biotechnol. 2013, 5, 204–211. [Google Scholar]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Qazi, S.N.A.; Harrison, S.E.; Self, T.; Williams, P.; Hill, P.J. Real-time monitoring of intracellular Staphylococcus aureus replication. J. Bacteriol. 2004, 186, 1065–1077. [Google Scholar] [CrossRef]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef]
- Doern, G.V.; Vautour, R.; Gaudet, M.; Levy, B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 1994, 32, 1757–1762. [Google Scholar] [CrossRef]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Naturae. 2013, 5, 4–43. [Google Scholar] [CrossRef]
- Rudkin, J.K.; Laabei, M.; Edwards, A.M.; Joo, H.-S.; Otto, M.; Lennon, K.L.; O’Gara, J.P.; Waterfield, N.R.; Massey, R.C. Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014, 58, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Turek, D.; Van Simaeys, D.; Johnson, J.; Ocsoy, I.; Tan, W. Molecular recognition of live methicillin-resistant staphylococcus aureus cells using DNA aptamers. World J. Transl. Med. 2013, 2, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-S.; Tsai, Y.-C.; Hsu, K.-F.; Lee, G.-B. Optimization of aptamer selection on an automated microfluidic system with cancer tissues. Lab Chip 2021, 21, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kim, J.; Yim, G.; Ahn, H.J.; Lee, M.; Kim, T.-H.; Park, C.; Min, J.; Jang, H.; Lee, T. Fabrication of a surface-enhanced Raman spectroscopy-based analytical method consisting of multifunctional DNA three-way junction-conjugated porous gold nanoparticles and Au-Te nanoworm for C-reactive protein detection. Anal. Bioanal. Chem. 2022, 414, 3197–3204. [Google Scholar] [CrossRef] [PubMed]
- Sosic, A.; Meneghello, A.; Cretaio, E.; Gatto, B. Human thrombin detection through a sandwich aptamer microarray: Interac-tion analysis in solution and in solid phase. Sensors 2011, 11, 9426–9441. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).