Lysinibacillus sphaericus III(3)7 and Plasmid Vector pMK4: New Challenges in Cloning Platforms
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis
2.2. Cell-Free Extract (CFE) Preparation
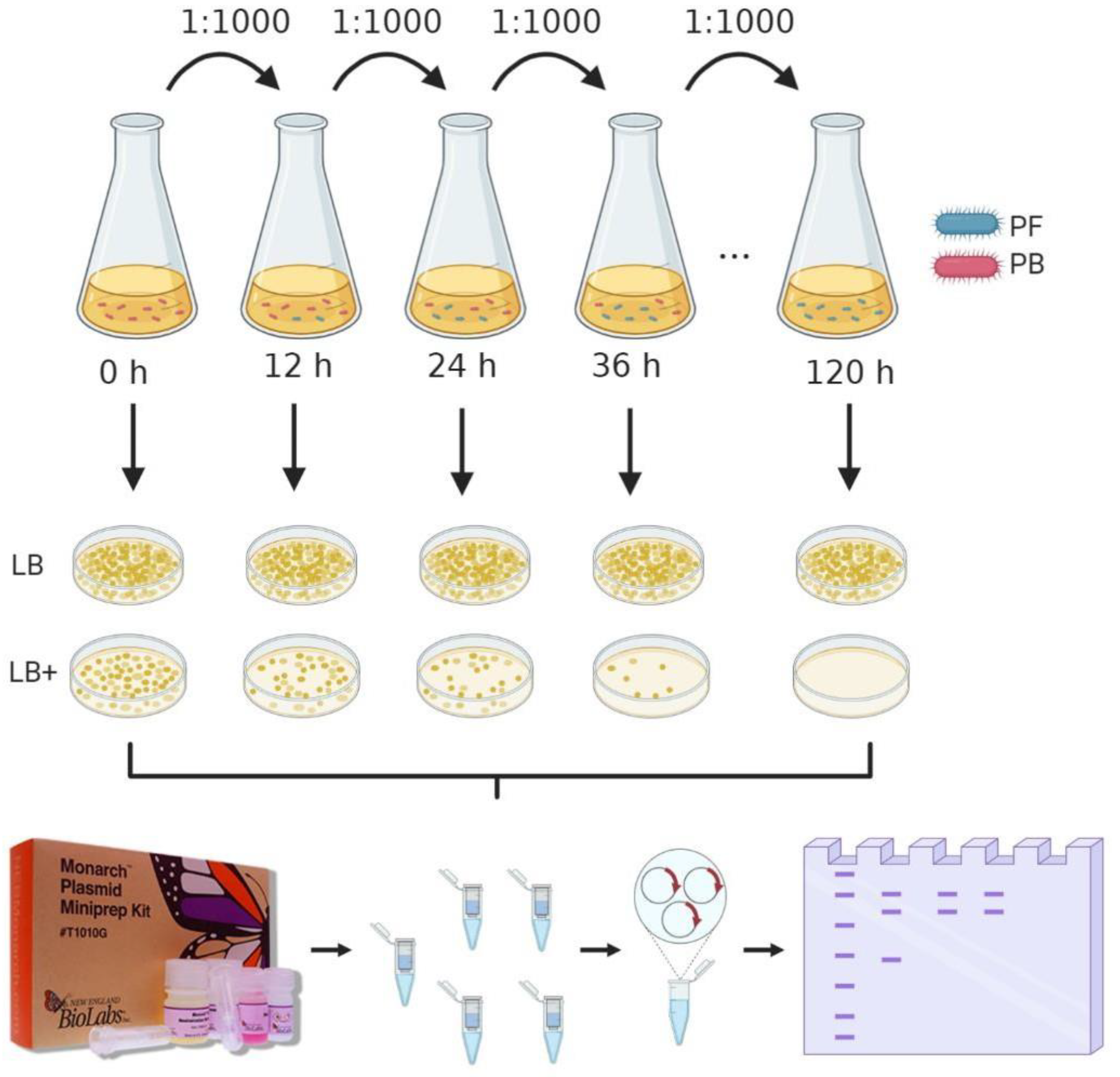
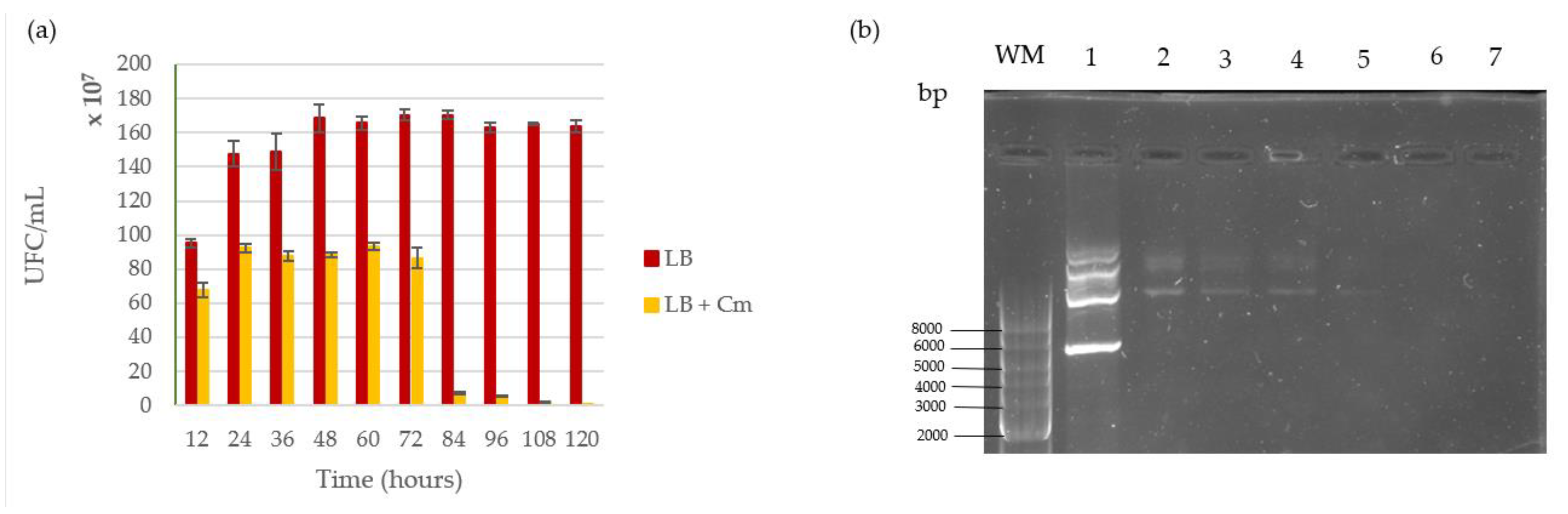
2.3. Stability Assay of Vector pMK4 within L. sphaericus III(3)7 Cells
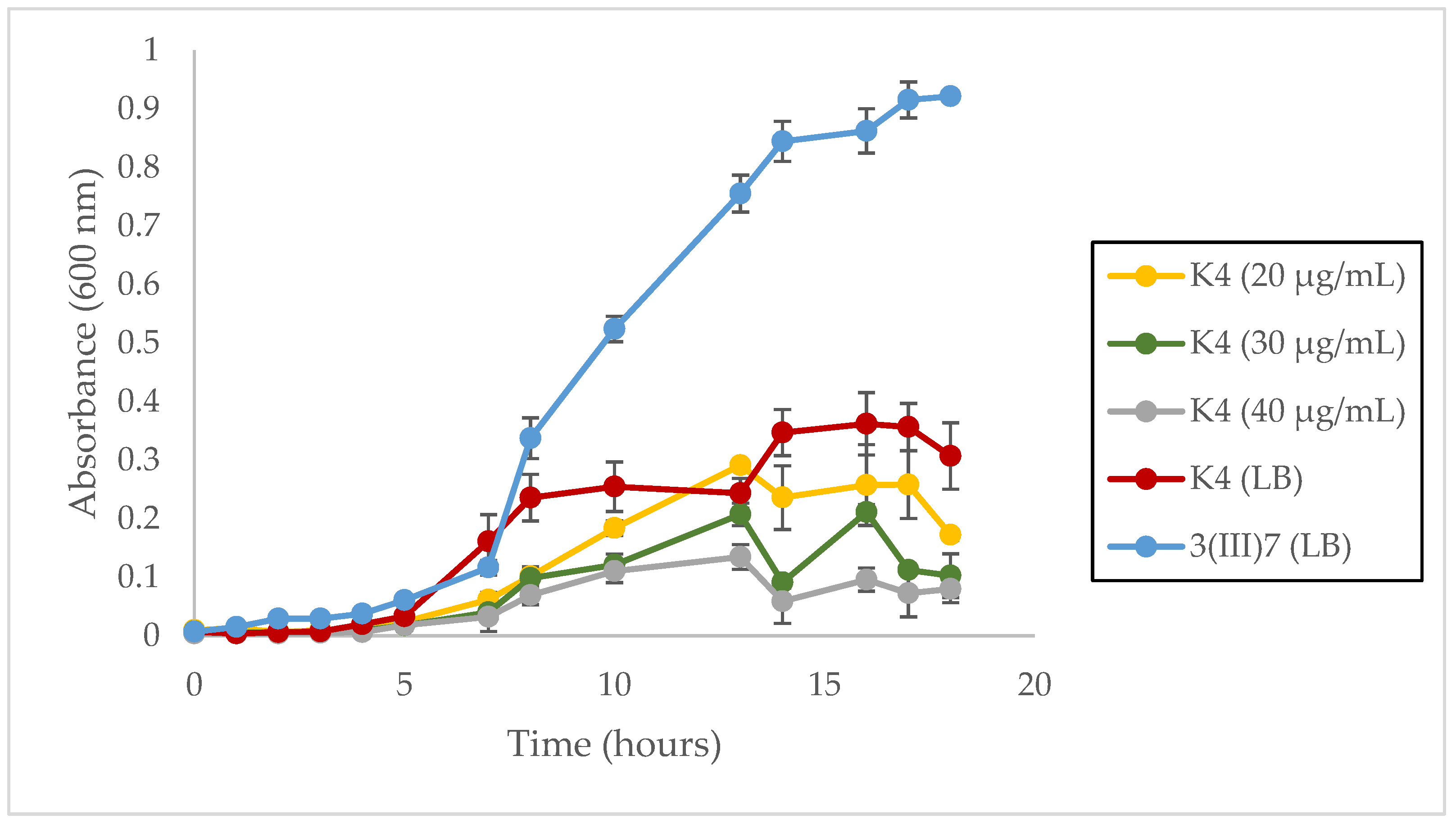
2.4. Growth Rate of Plasmid-Bearing and Plasmid-Free Cells
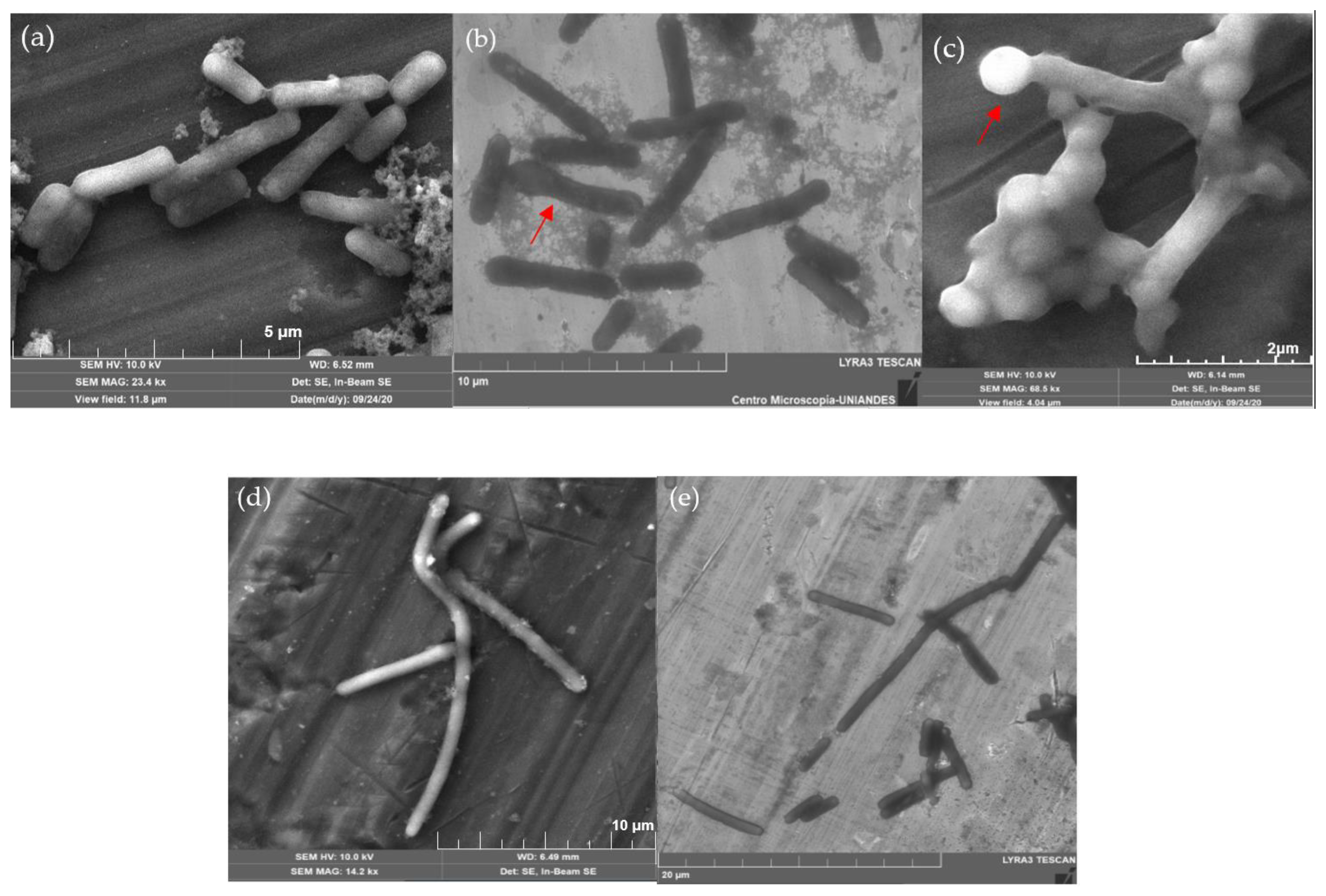
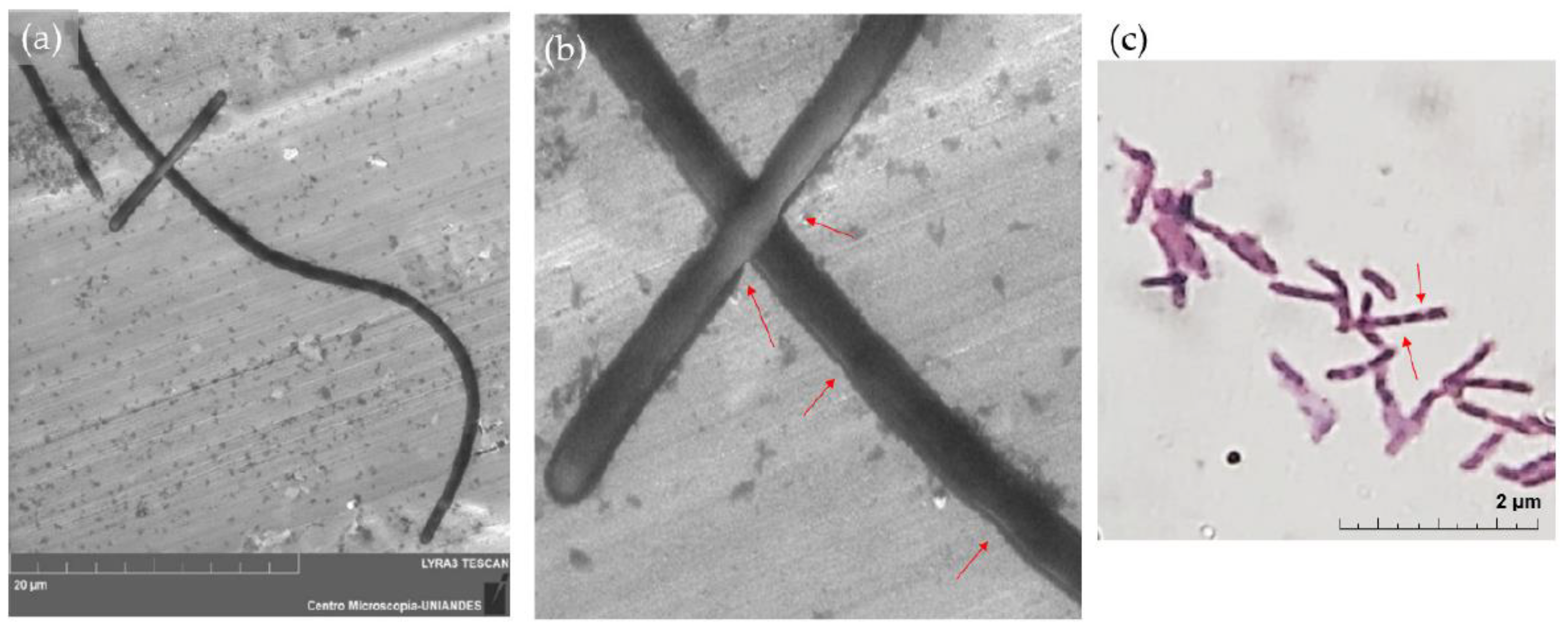
2.5. Morphological Changes
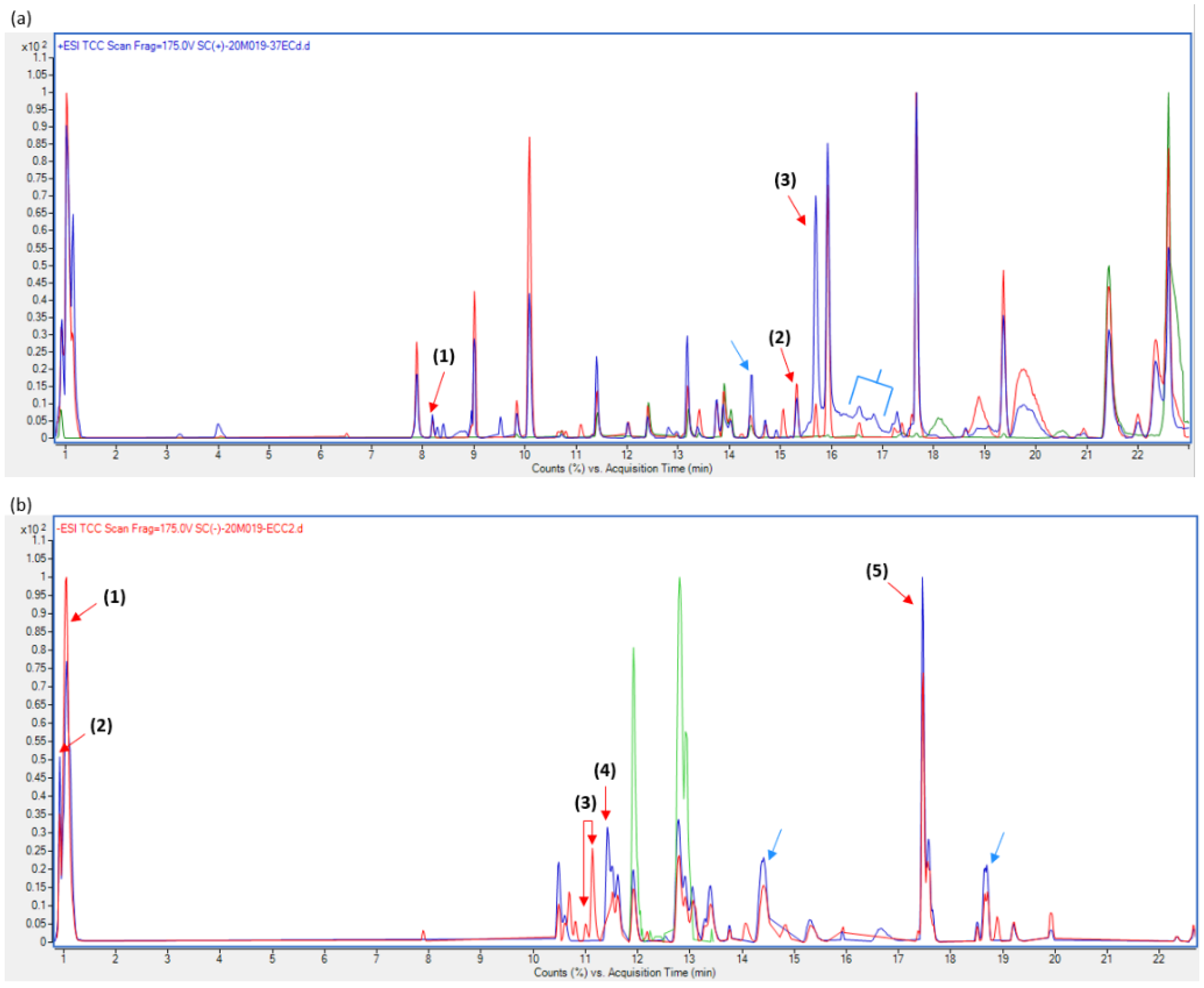
2.6. Metabolite Characterization—Total Compound Chromatogram (TCC)
2.7. Larvicidal Activity
2.8. Statistical Analyses
3. Results
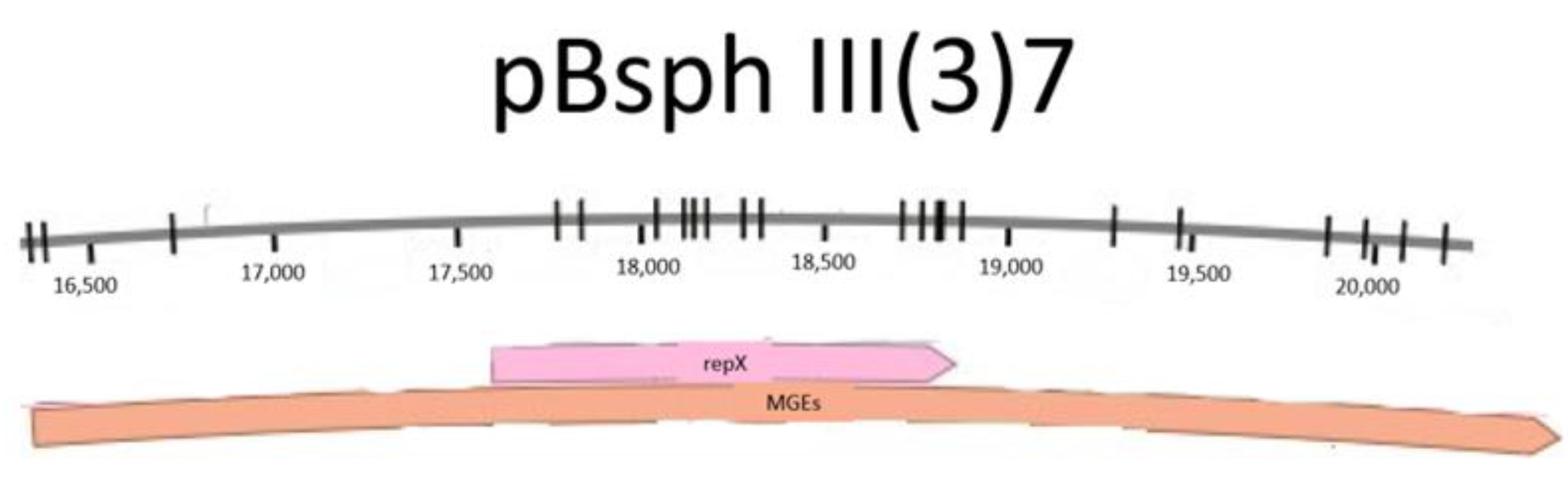
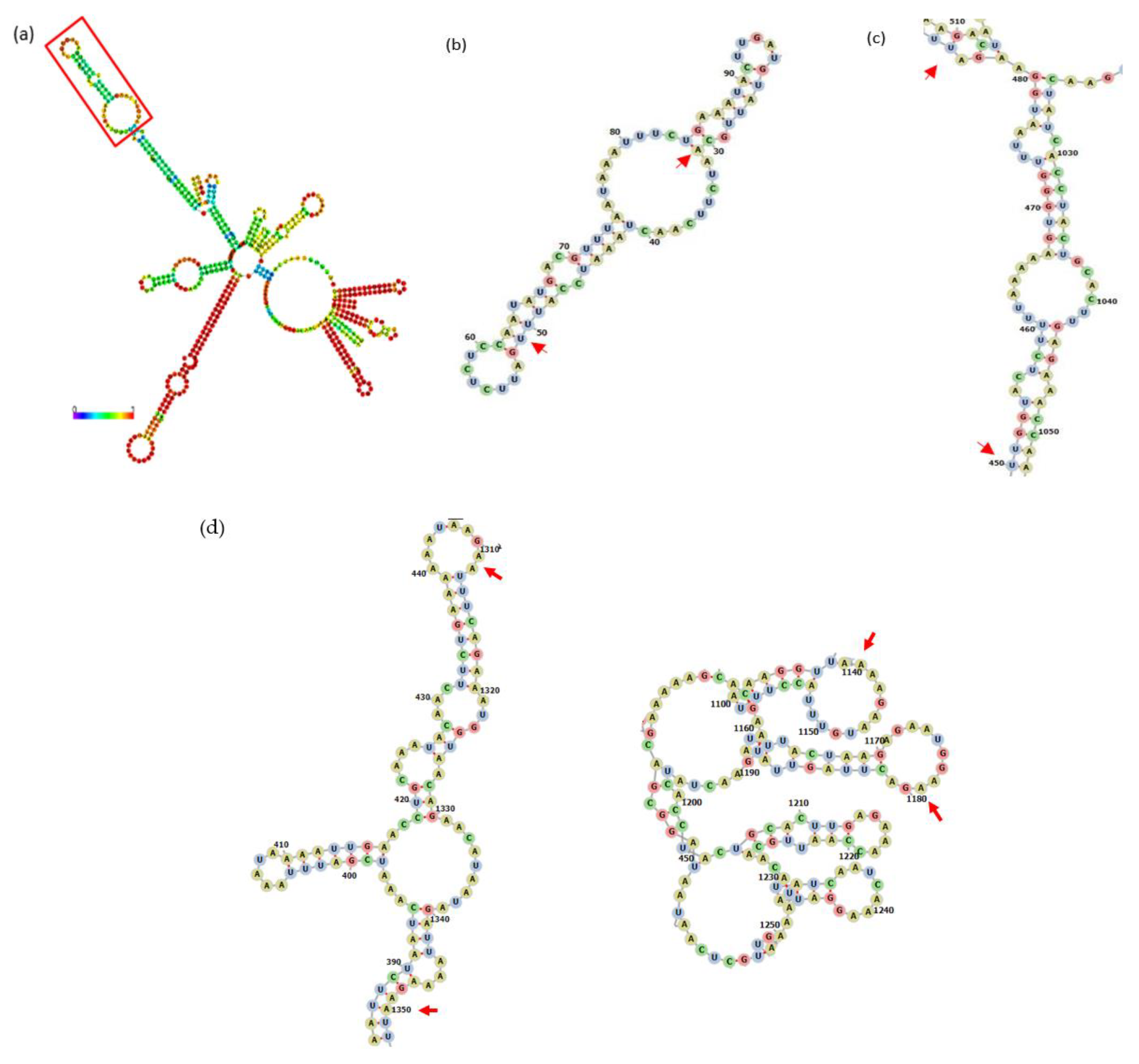
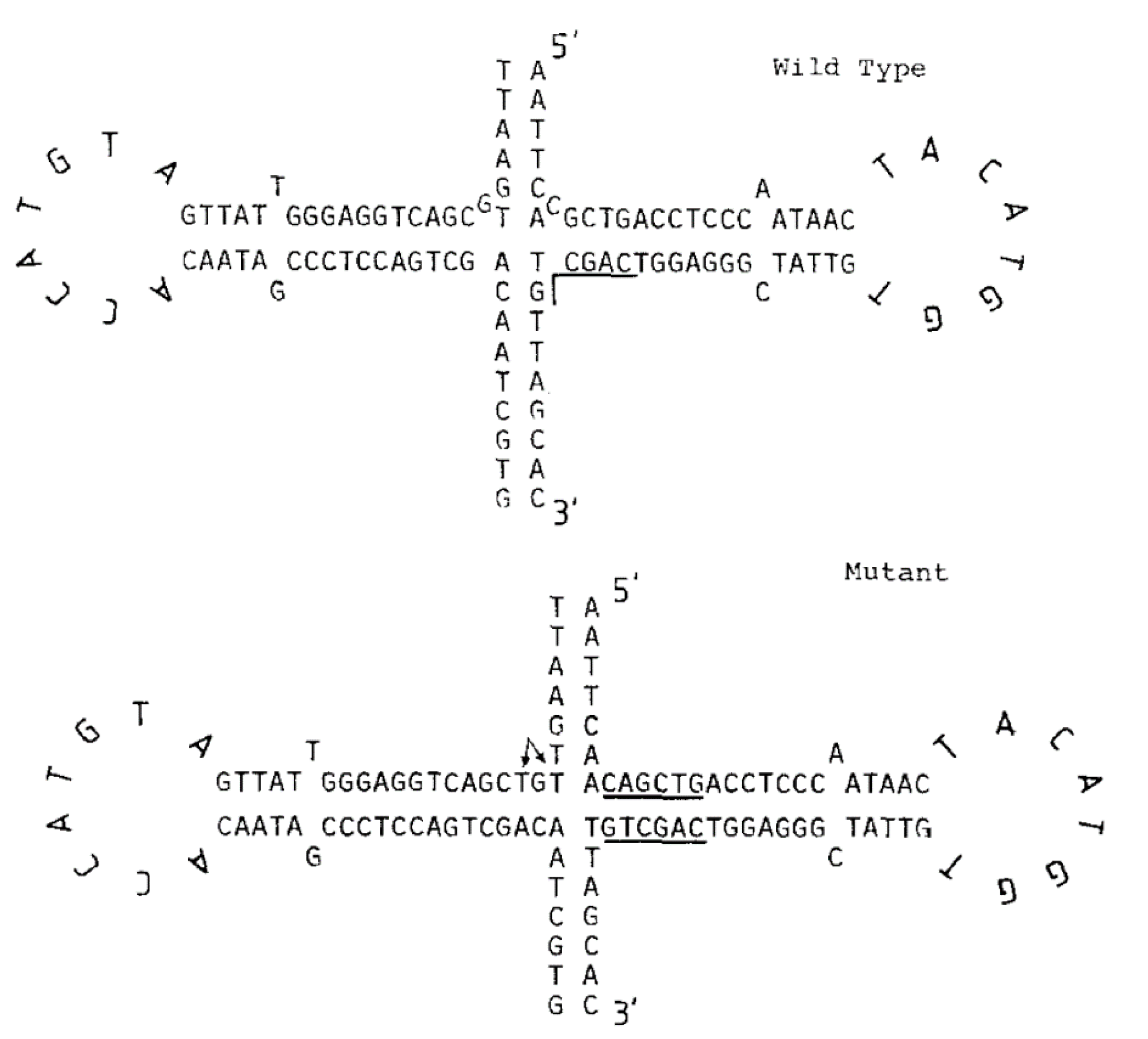
3.1. Bioinformatical Approach to Incompatibility
3.2. Plasmid Stability
3.3. Growth Rate
3.4. Morphological Changes
3.5. Metabolite Characterization
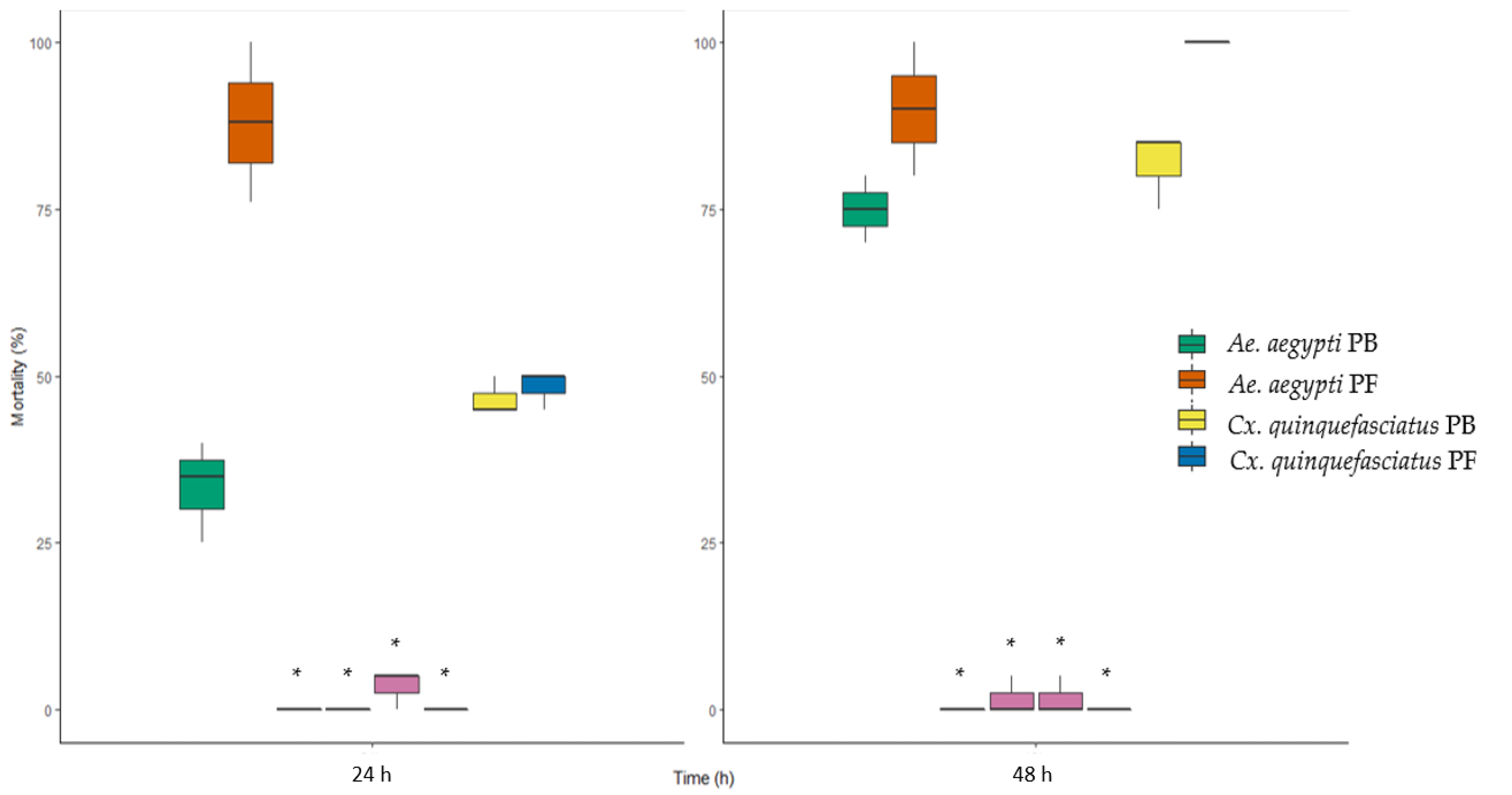
3.6. Larvicidal Activity against Cx. quinquefasciatus and Ae. aegypti Larvae
4. Discussion
4.1. Bioinformatical Approach to Incompatibility
4.2. Shuttle Vector pMK4 Stability Inside L. sphaericus III(3)7 Cells
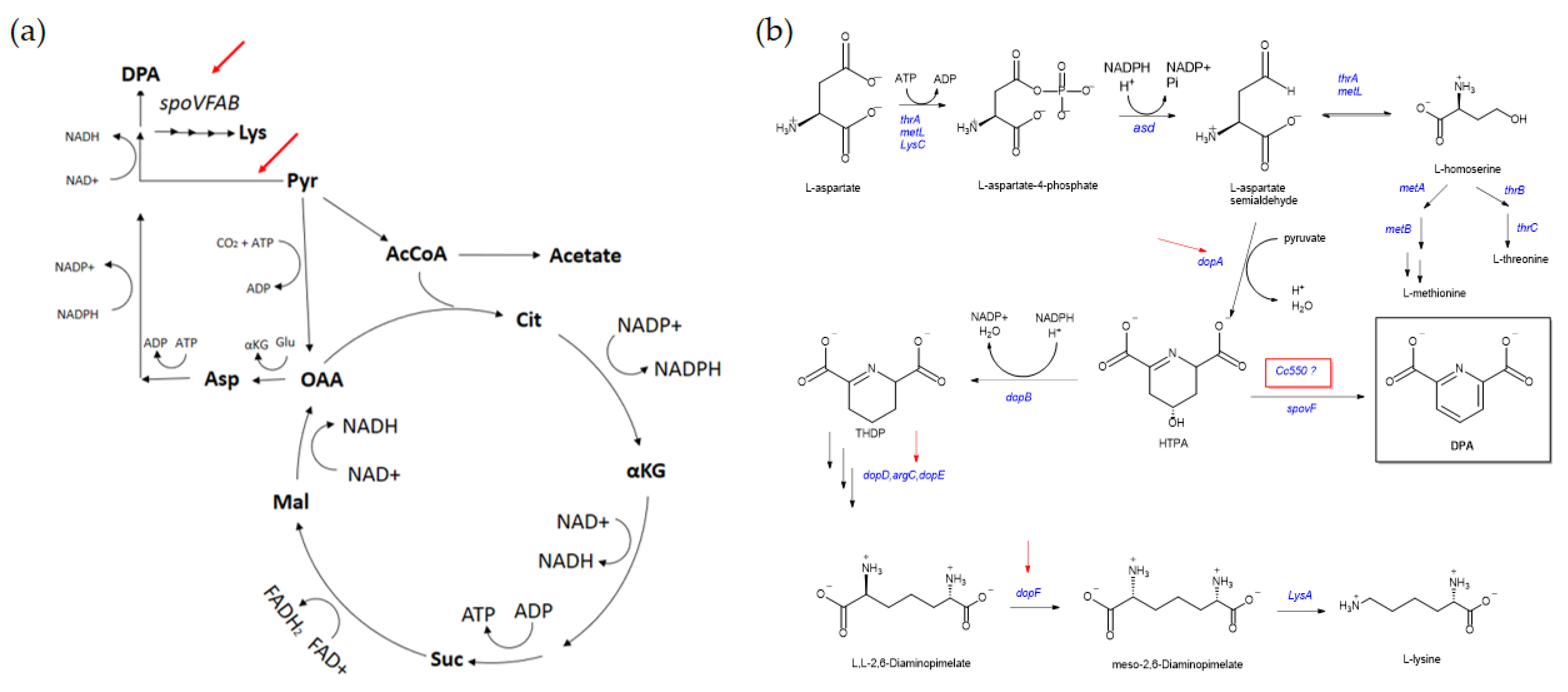
4.3. Metabolic Burden in L. sphaericus III(3)7
4.4. Metabolite Production
4.5. Changes in Larvicidal Activity against Cx. quinquefasciatus and Ae. aegypti Larvae
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gómez-Garzón, C.; Hernández-Santana, A.; Dussán, J. Comparative Genomics Reveals Lysinibacillus sphaericus Group Comprises a Novel Species. BMC Genom. 2016, 17, 709. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Ge, Y.; Wu, Y.; Zhao, N.; Yuan, Z.; Hu, X. The LspC3–41I Restriction-Modification System Is the Major Determinant for Genetic Manipulations of Lysinibacillus sphaericus C3–41. BMC Microbiol. 2017, 17, 116. [Google Scholar] [CrossRef] [PubMed]
- Edo, J.; Dussán, V.; Dussán, J. Adsorption of Toxic Metals and Control of Mosquitos-Borne Disease by Lysinibacillus sphaericus: Dual Benefits for Health and Environment. Biomed. Environ. Sci. 2016, 29, 187–196. [Google Scholar] [CrossRef]
- Lozano, L.C.; Dussán, J. Synergistic Activity Between S-Layer Protein and Spore-Crystal Preparations from Lysinibacillus sphaericus Against Culex Quinquefasciatus Larvae. Curr. Microbiol. 2017, 74, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Rey, A.; Silva-Quintero, L.; Dussán, J. Complete Genome Sequencing and Comparative Genomic Analysis of Functionally Diverse Lysinibacillus sphaericus III(3)7. Genom. Data 2016, 9, 78–86. [Google Scholar] [CrossRef][Green Version]
- Lozano, L.C.; Dussán, J. Metal Tolerance and Larvicidal Activity of Lysinibacillus sphaericus. World J. Microbiol. Biotechnol. 2013, 29, 1383–1389. [Google Scholar] [CrossRef]
- Radeck, J.; Kraft, K.; Bartels, J.; Cikovic, T.; Dürr, F.; Emenegger, J.; Kelterborn, S.; Sauer, C.; Fritz, G.; Gebhard, S.; et al. The Bacillus BioBrick Box: Generation and Evaluation of Essential Genetic Building Blocks for Standardized Work with Bacillus Subtilis. J. Biol. Eng. 2013, 7, 29. [Google Scholar] [CrossRef]
- Silva, F.; Queiroz, J.A.; Domingues, F.C. Evaluating Metabolic Stress and Plasmid Stability in Plasmid DNA Production by Escherichia coli. Biotechnol. Adv. 2012, 30, 691–708. [Google Scholar] [CrossRef]
- Friehs, K. Plasmid Copy Number and Plasmid Stability. Adv. Biochem. Eng. Biotechnol. 2004, 86, 47–82. [Google Scholar] [CrossRef]
- Juan, C.A.; Tolmasky, M.E. Plasmids: Biology and Impact in Biotechnology and Discovery; American Society of Microbiology: Washington, DC, USA, 2015; ISBN 978-1-55581-897-5. [Google Scholar]
- Muschiol, S.; Balaban, M.; Normark, S.; Henriques-Normark, B. Uptake of Extracellular DNA: Competence Induced Pili in Natural Transformation of Streptococcus Pneumoniae. Bioessays 2015, 37, 426–435. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans Gen. Nov. Sp. Nov., and Transfer of Bacillus fusiformis to Lysinibacillus fusiformis Comb. Nov. and Bacillus sphaericus to Lysinibacillus sphaericus Comb. Nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Velappan, N.; Sblattero, D.; Chasteen, L.; Pavlik, P.; Bradbury, A.R.M. Plasmid Incompatibility: More Compatible than Previously Thought? Protein Eng. Des. Sel. 2007, 20, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.A.; Yasbin, R.E.; Young, F.E. New Shuttle Vectors for Bacillus subtilis and Escherichia coli Which Allow Rapid Detection of Inserted Fragments. Gene 1984, 29, 21–26. [Google Scholar] [CrossRef]
- Alonso, J.C.; Trautner, T.A. A Gene Controlling Segregation of the Bacillus subtilis Plasmid PC194. Mol. Gen. Genet. 1985, 198, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Alvarado, A.; de la Cruz, F. Identification of Bacterial Plasmids Based on Mobility and Plasmid Population Biology. FEMS Microbiol. Rev. 2011, 35, 936–956. [Google Scholar] [CrossRef]
- Alonso, J.C.; Tailor, R.H. Initiation of Plasmid PC194 Replication and Its Control in Bacillus Subtilis. Mol. Gen. Genet. 1987, 210, 476–484. [Google Scholar] [CrossRef]
- Santana-Martinez, J.C.; Silva, J.J.; Dussan, J. Efficacy of Lysinibacillus sphaericus against Mixed-Cultures of Field-Collected and Laboratory Larvae of Aedes aegypti and Culex quinquefasciatus. Bull. Entomol. Res. 2019, 109, 111–118. [Google Scholar] [CrossRef]
- San Millan, A.; MacLean, R.C. Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Carloni, E.; Andreoni, F.; Omiccioli, E.; Villa, L.; Magnani, M.; Carattoli, A. Comparative Analysis of the Standard PCR-Based Replicon Typing (PBRT) with the Commercial PBRT-KIT. Plasmid 2017, 90, 10–14. [Google Scholar] [CrossRef]
- Hu, X.; Fan, W.; Han, B.; Liu, H.; Zheng, D.; Li, Q.; Dong, W.; Yan, J.; Gao, M.; Berry, C.; et al. Complete Genome Sequence of the Mosquitocidal Bacterium Bacillus sphaericus C3-41 and Comparison with Those of Closely Related Bacillus Species. J. Bacteriol. 2008, 190, 2892–2902. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, G.-Q.; She, Z.-S.; Zhu, H. Leaderless Genes in Bacteria: Clue to the Evolution of Translation Initiation Mechanisms in Prokaryotes. BMC Genom. 2011, 12, 361. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Weisblum, B. Nucleotide Sequence and Functional Map of PC194, a Plasmid That Specifies Inducible Chloramphenicol Resistance. J. Bacteriol. 1982, 150, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Persson, C.; Wagner, E.G.; Nordström, K. Control of Replication of Plasmid R1: Structures and Sequences of the Antisense RNA, CopA, Required for Its Binding to the Target RNA, CopT. EMBO J. 1990, 9, 3767–3775. [Google Scholar] [CrossRef]
- Slagter-Jäger, J.G. CopA and CopT: The Perfect RNA Couple. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 2003. [Google Scholar]
- Herskovitz, M.A.; Bechhofer, D.H. Endoribonuclease RNase III Is Essential in Bacillus subtilis. Mol. Microbiol. 2000, 38, 1027–1033. [Google Scholar] [CrossRef]
- Commichau, F.M.; Stülke, J. A Mystery Unraveled: Essentiality of RNase III in Bacillus subtilis Is Caused by Resident Prophages. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef]
- Brantl, S. Plasmid Replication Control by Antisense RNAs. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Del Solar, G.; Espinosa, M. Plasmid Copy Number Control: An Ever-Growing Story. Mol. Microbiol. 2000, 37, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zhao, N.; Hu, X.; Shi, T.; Cai, Q.; Yuan, Z. A Novel Transcriptional Activator, TubX, Is Required for the Stability of Bacillus sphaericus Mosquitocidal Plasmid PBsph. J. Bacteriol. 2014, 196, 4304–4314. [Google Scholar] [CrossRef]
- Meijer, W.J.J.; Wisman, G.B.A.; Terpstra, P.; Thorsted, P.B.; Thomas, C.M.; Holsappel, S.; Venema, G.; Bron, S. Rolling-Circle Plasmids from Bacillus subtilis: Complete Nucleotide Sequences and Analyses of Genes of PTA1015, PTA1040, PTA1050 and PTA1060, and Comparisons with Related Plasmids from Gram-Positive Bacteria. FEMS Microbiol. Rev. 1998, 21, 337–368. [Google Scholar] [CrossRef] [PubMed]
- Azpiroz, M.F.; Laviña, M. Analysis of RecA-Independent Recombination Events between Short Direct Repeats Related to a Genomic Island and to a Plasmid in Escherichia coli K12. PeerJ 2017, 5. [Google Scholar] [CrossRef]
- Bzymek, M.; Lovett, S.T. Instability of Repetitive DNA Sequences: The Role of Replication in Multiple Mechanisms. Proc. Natl. Acad. Sci. USA 2001, 98, 8319–8325. [Google Scholar] [CrossRef] [PubMed]
- Michel, B.; Ehrlich, S.D. Illegitimate Recombination Occurs between the Replication Origin of the Plasmid PC194 and a Progressing Replication Fork. EMBO J. 1986, 5, 3691–3696. [Google Scholar] [CrossRef] [PubMed]
- Ballester, S.; Lopez, P.; Espinosa, M.; Alonso, J.C.; Lacks, S.A. Plasmid Structural Instability Associated with PC194 Replication Functions. J. Bacteriol. 1989, 171, 2271–2277. [Google Scholar] [CrossRef]
- Alonso, J.C.; Trautner, T.A. Generation of Deletions through a Cis-Acting Mutation in Plasmid PC194. Mol. Gen. Genet. 1985, 198, 432–436. [Google Scholar] [CrossRef]
- Baxter, J.; Funnell, B. Plasmid Partition Mechanisms. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Van Melderen, L. Bacterial Toxin-Antitoxin Systems. Mob. Genet. Elem. 2011, 1, 283–290. [Google Scholar] [CrossRef]
- Fu, P.; Ge, Y.; Hu, Y.; Yuan, Z.; Hu, X. A Toxin-Antitoxin System Is Essential for the Stability of Mosquitocidal Plasmid PBsph of Lysinibacillus Sphaericus. Microbiol. Res. 2018, 214, 114–122. [Google Scholar] [CrossRef]
- Chang, S.; Chang, S.Y.; Gray, O. Structural and Genetic Analyses of a Par Locus That Regulates Plasmid Partition in Bacillus subtilis. J. Bacteriol. 1987, 169, 3952–3962. [Google Scholar] [CrossRef]
- Ge, Y.; Hu, X.; Zhao, N.; Shi, T.; Cai, Q.; Yuan, Z. A New TubRZ Operon Involved in the Maintenance of the Bacillus sphaericus Mosquitocidal Plasmid PBsph. Microbiology 2014, 160, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Domínguez, Y.; Contreras-Ferrat, G.; Ramírez-Santos, J.; Membrillo-Hernández, J.; Gómez-Eichelmann, M.C. Plasmid DNA Supercoiling and Gyrase Activity in Escherichia coli Wild-Type and RpoS Stationary-Phase Cells. J. Bacteriol. 2003, 185, 1097–1100. [Google Scholar] [CrossRef]
- Butler, Y.X.; Abhayawardhane, Y.; Stewart, G.C. Amplification of the Bacillus subtilis Maf Gene Results in Arrested Septum Formation. J. Bacteriol. 1993, 175, 3139–3145. [Google Scholar] [CrossRef]
- Lenhart, J.S.; Schroeder, J.W.; Walsh, B.W.; Simmons, L.A. DNA Repair and Genome Maintenance in Bacillus Subtilis. Microbiol. Mol. Biol. Rev. 2012, 76, 530–564. [Google Scholar] [CrossRef]
- Jameson, K.H.; Wilkinson, A.J. Control of Initiation of DNA Replication in Bacillus subtilis and Escherichia coli. Genes 2017, 8, 22. [Google Scholar] [CrossRef]
- Jameson, K.H.; Rostami, N.; Fogg, M.J.; Turkenburg, J.P.; Grahl, A.; Murray, H.; Wilkinson, A.J. Structure and Interactions of the Bacillus subtilis Sporulation Inhibitor of DNA Replication, SirA, with Domain I of DnaA. Mol. Microbiol. 2014, 93, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Banse, A.V.; Hobbs, E.C.; Losick, R. Phosphorylation of Spo0A by the Histidine Kinase KinD Requires the Lipoprotein Med in Bacillus subtilis. J. Bacteriol. 2011, 193, 3949–3955. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mars, R.A.T.; Nicolas, P.; Denham, E.L.; van Dijl, J.M. Regulatory RNAs in Bacillus subtilis: A Gram-Positive Perspective on Bacterial RNA-Mediated Regulation of Gene Expression. Microbiol. Mol. Biol. Rev. 2016, 80, 1029–1057. [Google Scholar] [CrossRef]
- Kimura, T.; Kobayashi, K. Role of Glutamate Synthase in Biofilm Formation by Bacillus subtilis. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef] [PubMed]
- Mars, R. The Regulatory RNAs of Bacillus subtilis. Available online: https://research.rug.nl/en/publications/the-regulatory-rnas-of-bacillus-subtilis (accessed on 12 November 2020).
- McClintock, M.K.; Fahnhorst, G.W.; Hoye, T.R.; Zhang, K. Engineering the Production of Dipicolinic Acid in E. coli. Metab. Eng. 2018, 48, 208–217. [Google Scholar] [CrossRef]
- Toya, Y.; Hirasawa, T.; Ishikawa, S.; Chumsakul, O.; Morimoto, T.; Liu, S.; Masuda, K.; Kageyama, Y.; Ozaki, K.; Ogasawara, N.; et al. Enhanced Dipicolinic Acid Production during the Stationary Phase in Bacillus subtilis by Blocking Acetoin Synthesis. Biosci. Biotechnol. Biochem. 2015. [Google Scholar] [CrossRef]
- Korza, G.; Abini-Agbomson, S.; Setlow, B.; Shen, A.; Setlow, P. Levels of L-Malate and Other Low Molecular Weight Metabolites in Spores of Bacillus Species and Clostridium difficile. PLoS ONE 2017, 12, e0182656. [Google Scholar] [CrossRef]
- Ohné, M.; Rutberg, B. Repression of Sporulation in Bacillus subtilis by L-Malate. J. Bacteriol. 1976, 125, 453–460. [Google Scholar] [CrossRef]
- Boguslawski, K.M.; Hill, P.A.; Griffith, K.L. Novel Mechanisms of Controlling the Activities of the Transcription Factors Spo0A and ComA by the Plasmid-Encoded Quorum Sensing Regulators Rap60-Phr60 in Bacillus subtilis. Mol. Microbiol. 2015, 96, 325–348. [Google Scholar] [CrossRef]
- Doi, R.H. Sporulation and Germination. In Bacillus; Harwood, C.R., Ed.; Biotechnology Handbooks; Springer US: Boston, MA, USA, 1989; pp. 169–215. ISBN 978-1-4899-3502-1. [Google Scholar]
- Wang, J.; Mei, H.; Zheng, C.; Qian, H.; Cui, C.; Fu, Y.; Su, J.; Liu, Z.; Yu, Z.; He, J. The Metabolic Regulation of Sporulation and Parasporal Crystal Formation in Bacillus thuringiensis Revealed by Transcriptomics and Proteomics. Mol. Cell. Proteom. 2013, 12, 1363–1376. [Google Scholar] [CrossRef]
- Aronson, J.N.; Borris, D.P.; Doerner, J.F.; Akers, E. Gamma-Aminobutyric Acid Pathway and Modified Tricarboxylic Acid Cycle Activity during Growth and Sporulation of Bacillus thuringiensis. Appl. Microbiol. 1975, 30, 489–492. [Google Scholar] [CrossRef]
- Meyer, F.M.; Stülke, J. Malate Metabolism in Bacillus Subtilis: Distinct Roles for Three Classes of Malate-Oxidizing Enzymes. FEMS Microbiol. Lett. 2013, 339, 17–22. [Google Scholar] [CrossRef]
- Freese, E.B.; Vasantha, N.; Freese, E. Induction of Sporulation in Developmental Mutants of Bacillus Subtilis. Molec. Gen. Genet. 1979, 170, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Shin, I.; Ryu, H.-B.; Yim, H.-S.; Kang, S.-O. Cytochrome C550 Is Related to Initiation of Sporulation in Bacillus Subtilis. J. Microbiol. 2005, 43, 244–250. [Google Scholar] [PubMed]
- Aubert, J.P.; Millet, J.; Pineau, E.; Milhaud, G. N-Succinyl-L-glutamic acid in Bacillus megaterium during sporulation. Biochimica et Biophysica Acta 1961, 51, 529–537. [Google Scholar] [CrossRef]
- Gao, H.; Jiang, X.; Pogliano, K.; Aronson, A.I. The E1β and E2 Subunits of the Bacillus subtilis Pyruvate Dehydrogenase Complex Are Involved in Regulation of Sporulation. J. Bacteriol. 2002, 184, 2780–2788. [Google Scholar] [CrossRef] [PubMed]
- Ogura, M.; Sato, T.; Abe, K. Bacillus subtilis YlxR, Which Is Involved in Glucose-Responsive Metabolic Changes, Regulates Expression of TsaD for Protein Quality Control of Pyruvate Dehydrogenase. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Walter, T.; Aronson, A. Specific Binding of the E2 Subunit of Pyruvate Dehydrogenase to the Upstream Region of Bacillus thuringiensis Protoxin Genes. J. Biol. Chem. 1999, 274, 7901–7906. [Google Scholar] [CrossRef] [PubMed]
- Farrand, S.K.; Taber, H.W. Changes in Menaquinone Concentration during Growth and Early Sporulation in Bacillus subtilis. J. Bacteriol. 1974, 117, 324–326. [Google Scholar] [CrossRef]
- Winstedt, L.; von Wachenfeldt, C. Terminal Oxidases of Bacillus subtilis Strain 168: One Quinol Oxidase, Cytochrome Aa3 or Cytochrome Bd, Is Required for Aerobic Growth. J. Bacteriol. 2001, 182, 6557–6564. [Google Scholar] [CrossRef] [PubMed]
- Huyen, N.T.T.; Eiamphungporn, W.; Mäder, U.; Liebeke, M.; Lalk, M.; Hecker, M.; Helmann, J.D.; Antelmann, H. Genome-Wide Responses to Carbonyl Electrophiles in Bacillus subtilis: Control of the Thiol-Dependent Formaldehyde Dehydrogenase AdhA and Cysteine Proteinase YraA by the MerR-Family Regulator YraB (AdhR). Mol. Microbiol. 2009, 71, 876–894. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.A.; Huang, L.; Lee, D.-W.; Daldal, F.; Nagai, K.; Minagawa, N. Ascochlorin Is a Novel, Specific Inhibitor of the Mitochondrial Cytochrome Bc1 Complex. Biochim. et Biophys. Acta 2010, 1797, 360. [Google Scholar] [CrossRef]
- Fisher, N.; Meunier, B. Molecular Basis of Resistance to Cytochrome Bc1 Inhibitors. FEMS Yeast Res. 2008, 8, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Pinzón, P.A.; Dussán, J. Efficacy of the Vegetative Cells of Lysinibacillus sphaericus for Biological Control of Insecticide-Resistant Aedes aegypti. Parasites Vectors 2017, 10, 231. [Google Scholar] [CrossRef]

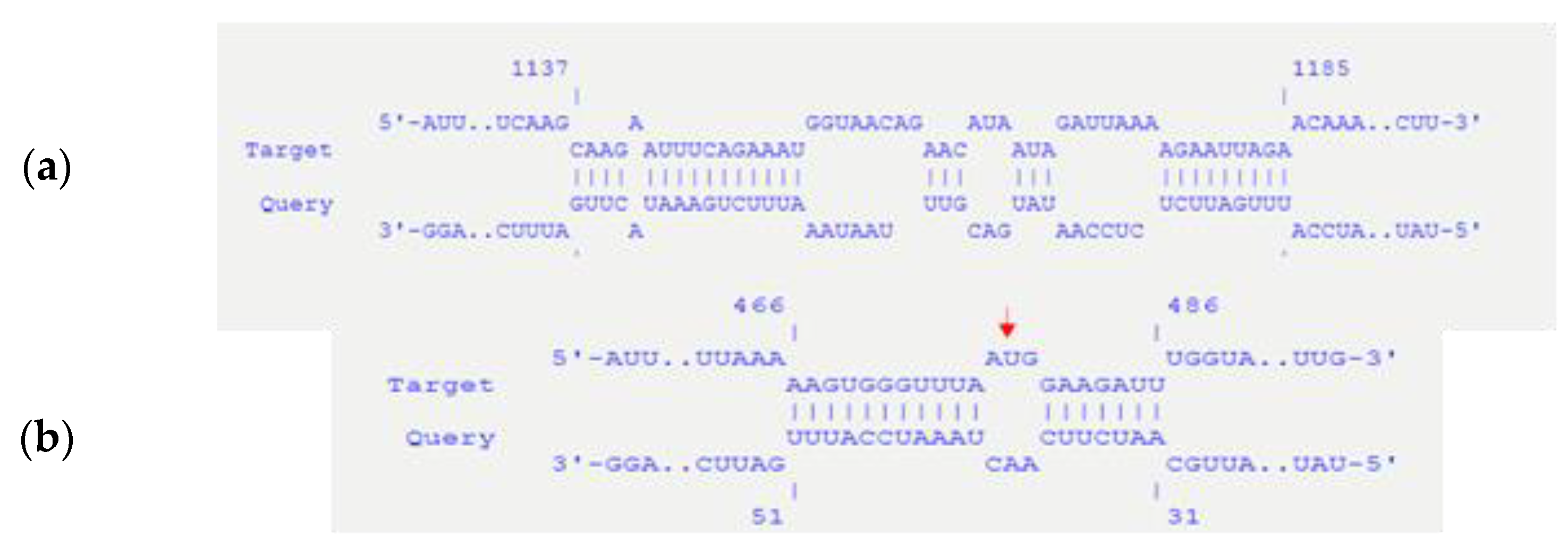
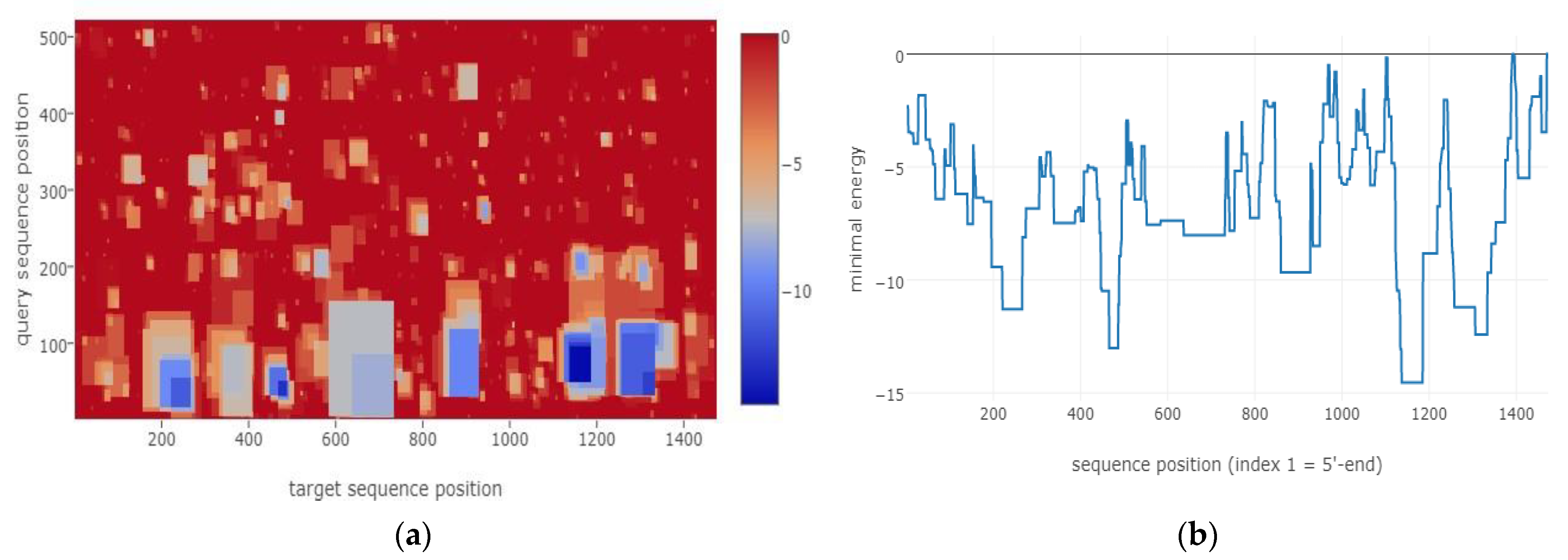


| Target | Start (T) | End (T) | Query | Start (Q) | End (Q) | Energy (kcal/mol) |
|---|---|---|---|---|---|---|
| pBsph-repX replicon | 1137 | 1185 | incA | 49 | 94 | −14.54 |
| pBsph-repX replicon | 466 | 486 | incA | 31 | 51 | −13.06 |
| MGE | Family | Identity Percentage of Tn Region with pBsph (MGEs Analysis Region) |
|---|---|---|
| Tn2 | Tn3 | 41.1% |
| Tn3 | Tn3 | 40.6% |
| Compound Name | Compound Structure | Found Predominantly in |
|---|---|---|
| L-malate | 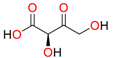 | Plasmid-bearing cells |
| Glycerol-1-phosphate | 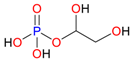 | Plasmid-free cells |
| Succynil glutamate |  | Plasmid-free cells |
| Acetyldihydrolipoamide | 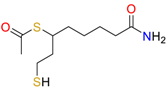 | Plasmid-free cells |
| Lysine | 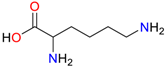 | Plasmid-free cells |
| Butylhydroquinone | 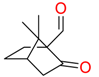 | Plasmid-free cells |
| Monoethanolamine | Plasmid-free cells | |
| Methyl lucidenate |  | Plasmid-bearing cells |
| Regulatory sRNA Name | Regulated Gene in B. subtilis | Percentage of Identity with pMK4 (%) |
|---|---|---|
| S1058 | spo0A | 84.62 |
| S1024 | sigA, sigK | 86.21 |
| S424 | spoVIF | 82.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
León, D.C.; Dussán, J. Lysinibacillus sphaericus III(3)7 and Plasmid Vector pMK4: New Challenges in Cloning Platforms. Microbiol. Res. 2021, 12, 455-479. https://doi.org/10.3390/microbiolres12020031
León DC, Dussán J. Lysinibacillus sphaericus III(3)7 and Plasmid Vector pMK4: New Challenges in Cloning Platforms. Microbiology Research. 2021; 12(2):455-479. https://doi.org/10.3390/microbiolres12020031
Chicago/Turabian StyleLeón, Diana C., and Jenny Dussán. 2021. "Lysinibacillus sphaericus III(3)7 and Plasmid Vector pMK4: New Challenges in Cloning Platforms" Microbiology Research 12, no. 2: 455-479. https://doi.org/10.3390/microbiolres12020031
APA StyleLeón, D. C., & Dussán, J. (2021). Lysinibacillus sphaericus III(3)7 and Plasmid Vector pMK4: New Challenges in Cloning Platforms. Microbiology Research, 12(2), 455-479. https://doi.org/10.3390/microbiolres12020031






