Characterization of Carbapenem-Resistant Gram-Negative Bacilli Isolates in Multispecialty Private Hospitals in Lagos, Nigeria
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Identification of Gram-Negative Bacilli
2.3. Detection of Carbapenem-Resistant Gram-Negative Bacilli
2.4. Antibacterial Susceptibility Test of the Presumptive Carbapenemase-Producing Gram-Negative Bacilli
2.5. Molecular Detection of Carbapenemase Genes
2.6. Data Analysis
3. Results
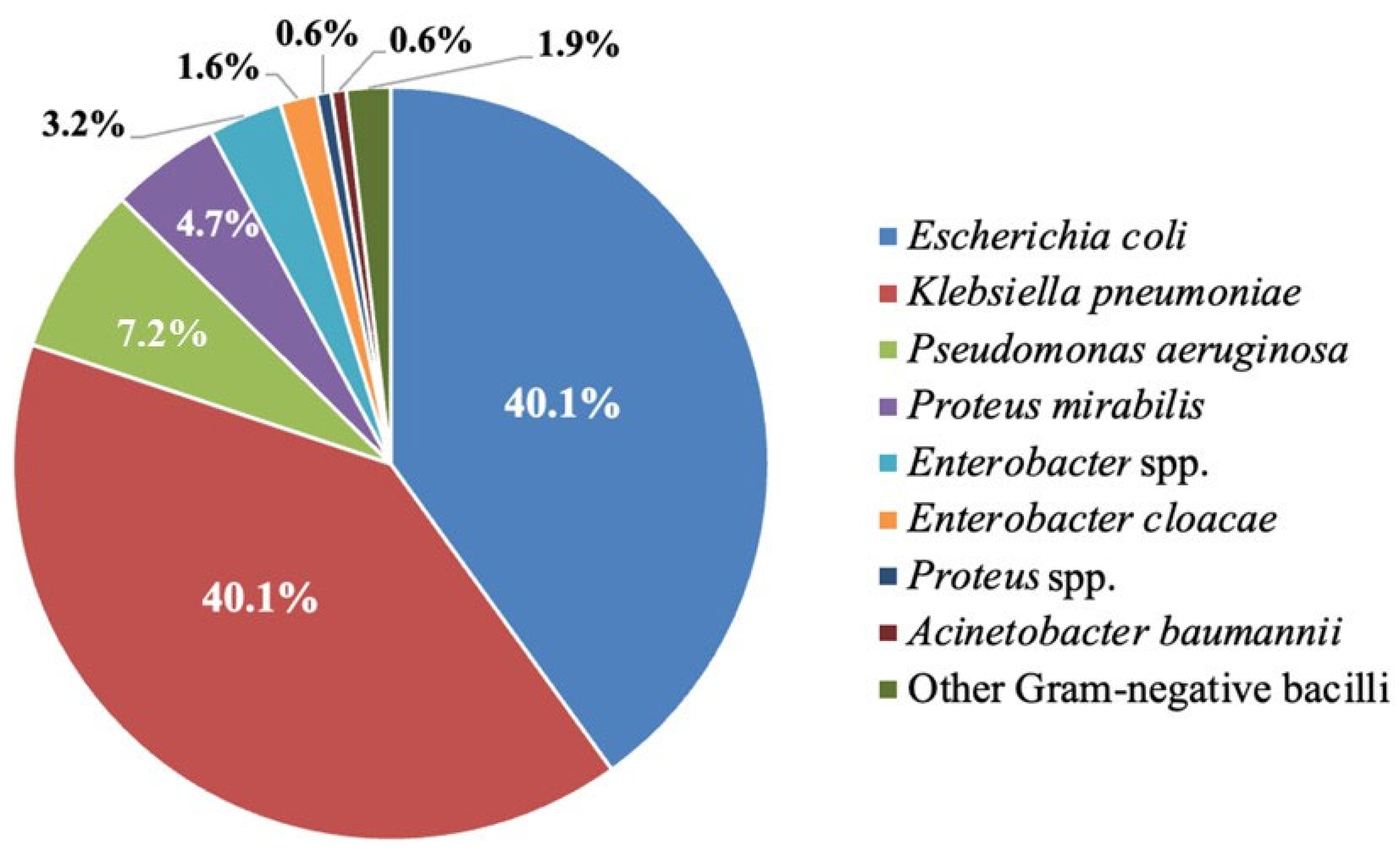
3.1. The Identified Gram-Negative Bacilli Isolate
3.2. Demographic Distribution of Carbapenem-Resistant Gram-Negative Bacilli
3.3. Prevalence and Proportion of Carbapenem-Resistant Gram-Negative Bacilli
3.4. Antimicrobial Resistance Profiles and Carbapenemase-Encoding Genes of Presumptive Carbapenemase-Producing Gram-Negative Bacilli
3.5. Demographic Data and Clinical Outcome of Presumptive Carbapenemase-Producing Gram-Negative Bacilli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Genava, Switzerland, 2024. [Google Scholar]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The current burden of carbapenemases: Review of significant properties and dissemination among Gram-negative bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.A.; Matsumura, Y. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019, 33, e00102-19. [Google Scholar] [CrossRef]
- Olalekan, A.; Bader, B.K.; Iwalokun, B.; Wolf, S.; Lalremruata, A.; Dike, A.; Mannie-Udoh, M.; Lo Presti, L.; Liese, J.; Guther, J.; et al. High incidence of carbapenemase-producing Pseudomonas aeruginosa clinical isolates from Lagos, Nigeria. JAC Antimicrob. Resist. 2023, 5, dlad038. [Google Scholar] [CrossRef]
- Shaker, O.A.; Gomaa, H.E.; ElMasry, S.A.; Halim, R.M.A.; Abdelrahman, A.H.; Kamal, J.S. Evaluation of Combined Use of Temocillin Disk and Mastdisks Inhibitor Combination Set Against Polymerase Chain Reaction for Detection of Carbapenem-Resistant Enterobacteriaceae. Open Access Maced. J. Med. Sci. 2018, 6, 242–247. [Google Scholar] [CrossRef]
- Shettima, S.A.; Tickler, I.A.; Dela Cruz, C.M.; Tenover, F.C. Characterisation of carbapenem-resistant Gram-negative organisms from clinical specimens in Yola, Nigeria. J. Glob. Antimicrob. Resist. 2020, 21, 42–45. [Google Scholar] [CrossRef]
- Ghanbarinasab, F.; Haeili, M.; Ghanati, S.; Moghimi, M. High prevalence of OXA-48-like and NDM carbapenemases among carbapenem resistant Klebsiella pneumoniae of clinical origin from Iran. Iran. J. Microbiol. 2023, 15, 609–615. [Google Scholar] [CrossRef]
- Tula, M.Y.; Enabulele, O.I.; Ophori, E.A.; Aziegbemhin, A.S.; Iyoha, O.; Filgona, J. A systematic review of the current status of carbapenem resistance in Nigeria: Its public health implication for national intervention. Niger. Postgrad. Med. J. 2023, 30, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Adesanya, O.A.; Igwe, H.A. Carbapenem-resistant Enterobacteriaceae (CRE) and gram-negative bacterial infections in south-west Nigeria: A retrospective epidemiological surveillance study. AIMS Public Health 2020, 7, 804–815. [Google Scholar] [CrossRef]
- Ugah, U.I.; Udeani, T.K. Prevalence of Phenotypic Carbapenem-Resistant Enterobacterales Isolates and Their Distribution by Sex, Age Groups, State and Species in South-East Nigeria. Gomal J. Med. Sci. 2022, 20, 89–96. [Google Scholar] [CrossRef]
- Anibijuwon, I.I.; Gbala, I.D.; Adebisi, O.O. Carbapenem-Resistant Enterobacteriaceae among In-Patients of Tertiary Hospitals in Southwest, Nigeria. Not. Sci. Biol. 2018, 10, 310–317. [Google Scholar] [CrossRef]
- Reyes, J.; Komarow, L.; Chen, L.; Ge, L.; Hanson, B.M.; Cober, E.; Herc, E.; Alenazi, T.; Kaye, K.S.; Garcia-Diaz, J.; et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): A prospective cohort study. Lancet Microbe 2023, 4, e159–e170. [Google Scholar] [CrossRef]
- Ling, W.; Furuya-Kanamori, L.; Ezure, Y.; Harris, P.N.A.; Paterson, D.L. Adverse clinical outcomes associated with carbapenem-resistant Acinetobacter (CRA) infections: A systematic review and meta-analysis. JAC Antimicrob. Resist. 2021, 3, dlab157. [Google Scholar] [CrossRef]
- Wilson, G.M.; Suda, K.J.; Fitzpatrick, M.A.; Bartle, B.; Pfeiffer, C.D.; Jones, M.; Rubin, M.A.; Perencevich, E.; Evans, M.; Evans, C.T.; et al. Risk factors associated with carbapenemase-producing carbapenem-resistant Enterobacteriaceae positive cultures in a cohort of US Veterans. Clin. Infect. Dis. 2021, 73, 1370–1378. [Google Scholar] [CrossRef]
- Kedisaletse, M.; Phumuzile, D.; Angela, D.; Andrew, W.; Mae, N.F. Epidemiology, risk factors, and clinical outcomes of carbapenem-resistant Enterobacterales in Africa: A systematic review. J. Glob. Antimicrob. Resist. 2023, 35, 297–306. [Google Scholar] [CrossRef]
- Odewale, G.; Adefioye, O.J.; Ojo, J.; Adewumi, F.A.; Olowe, O.A. Multidrug resistance of Acinetobacter baumannii in Ladoke Akintola university teaching hospital, Osogbo, Nigeria. Eur. J. Microbiol. Immunol. 2016, 6, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Ogbolu, D.O.; Alli, O.A.T.; Oluremi, A.S.; Ogunjimi, Y.T.; Ojebode, D.I.; Dada, V.; Alaka, O.O.; Foster-Nyarko, E.; Webber, M.A. Contribution of NDM and OXA-type carbapenemases to carbapenem resistance in clinical Acinetobacter baumannii from Nigeria. Infect. Dis. 2020, 52, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Health Facilities Monitoring and Accreditation Agency. An Overview of Healthcare in Lagos. Available online: https://hefamaa.lagosstate.gov.ng/ (accessed on 7 July 2025).
- Smith, A.; Hussey, M. Gram Stain Protocols; American Society for Microbiology (ASM): Washington, DC, USA, 2005. [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing. Breakpoint Table for Interpretation of MICs and Zone Diameters; Version 14.0; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2024. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Pasteran, F.; Lucero, C.; Soloaga, R.; Rapoport, M.; Corso, A. Can we use imipenem and meropenem Vitek 2 MICs for detection of suspected KPC and other-carbapenemase producers among species of Enterobacteriaceae? J. Clin. Microbiol. 2011, 49, 697–701. [Google Scholar] [CrossRef]
- Koehne, W.J.; Peritz, T.; Privette, K.; Gould, J. 1441. Using Carbapenem Resistance Levels to Discriminate Between Carbapenemase Producing and Non-Carbapenemase Producing Carbapenem Resistant Enterobacteriaceae. Open Forum Infect. Dis. 2020, 7, S724. [Google Scholar] [CrossRef]
- Rafailidis, P.I.; Kofteridis, D. Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev. Anti Infect. Ther. 2022, 20, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Manchanda, V.; Rai, S.; Gupta, S.; Rautela, R.S.; Chopra, R.; Rawat, D.S.; Verma, N.; Singh, N.P.; Kaur, I.R.; Bhalla, P. Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J. Med. Microbiol. 2011, 29, 249–253. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, X.; Yang, R.; Shen, X.; Li, G.; Zhang, C.; Li, P.; Li, S.; Xie, J.; Yang, Y. Carbapenem-resistant Gram-negative bacteria (CR-GNB) in ICUs: Resistance genes, therapeutics, and prevention—A comprehensive review. Front. Public Health 2024, 12, 1376513. [Google Scholar] [CrossRef]
- Mohammed, M.U.; Manisha, D.; Nagamani, K. Clinical, phenotypic and genotypic profile of carbapenem resistant Gram negative infections in intensive care units. Indian J. Microbiol. Res. 2021, 8, 28–34. [Google Scholar] [CrossRef]
- Ismail, H.; Zwane, T.B.C.; Du Toit, E.; da Costa, R.M.A.; Franceschi, F.; Perovic, O. Carbapenem-resistant Enterobacterales among patients with bloodstream infections in South Africa: Consolidated surveillance data, 2015–2021. PLoS ONE 2025, 20, e0324262. [Google Scholar] [CrossRef] [PubMed]
- Maina, J.W.; Mutua, J.M.; Musyoki, A.M. Carbapenem-resistant gram-negative bacterial infections and risk factors for acquisition in a Kenyan intensive care unit. BMC Infect. Dis. 2024, 24, 522. [Google Scholar] [CrossRef]
- Venne, D.M.; Hartley, D.M.; Malchione, M.D.; Koch, M.; Britto, A.Y.; Goodman, J.L. Review and analysis of the overlapping threats of carbapenem and polymyxin resistant E. coli and Klebsiella in Africa. Antimicrob. Resist. Infect. Control 2023, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Omoregbe, F.; Fagade, O. Carbapenemase Producers among Gram Negative Bacteria from Environmental and Clinical Samples in Makurdi, Nigeria. J. Adv. Microbiol. 2020, 20, 11–20. [Google Scholar] [CrossRef]
- Sabour, S.; Harrington, K.R.V.; Martinson, E.; Bhatnagar, A.S.; Huang, J.Y.; Duffy, D.; Bantle, K.; Lutgring, J.D.; Karlsson, M.; Brown, A.C. Characterization of carbapenem-resistant Enterobacterales and Pseudomonas aeruginosa carrying multiple carbapenemase genes-Antimicrobial Resistance Laboratory Network, 2018–2022. J. Clin. Microbiol. 2024, 62, e0122024. [Google Scholar] [CrossRef]
- Strich, J.R.; Lawandi, A.; Warner, S.; Demirkale, C.Y.; Sarzynski, S.; Babiker, A.; Dekker, J.P.; Kadri, S.S. Association between piperacillin/tazobactam MIC and survival among hospitalized patients with Enterobacterales infections: Retrospective cohort analysis of electronic health records from 161 US hospitals. JAC Antimicrob. Resist. 2023, 5, dlad041. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Lob, S.H.; Bauer, K.A.; Esterly, J.; Siddiqui, F.; Young, K.; Motyl, M.R.; Sahm, D.F. Activity of ceftolozane/tazobactam, imipenem/relebactam and ceftazidime/avibactam against clinical Gram-negative isolates-SMART United States 2019–21. JAC Antimicrob. Resist. 2024, 6, dlad152. [Google Scholar] [CrossRef]
- Leong, Q.; Chew, K.L. Drug resistance rates of difficult to treat Pseudomonas aeruginosa isolates to ceftolozane-tazobactam and ceftazidime-avibactam from a tertiary hospital, Singapore. Pathology 2022, 54, 966–968. [Google Scholar] [CrossRef]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, genetics and worldwide spread of New Delhi Metallo-beta-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Mohammed, Y.; Zailani, S.B.; Onipede, A.O. Characterization of KPC, NDM and VIM type carbapenem resistance Enterobacteriaceae from North Eastern, Nigeria. J. Biosci. Med. 2015, 3, 100–107. [Google Scholar] [CrossRef]
- Stoesser, N.; Phan, H.T.T.; Seale, A.C.; Aiken, Z.; Thomas, S.; Smith, M.; Wyllie, D.; George, R.; Sebra, R.; Mathers, A.J.; et al. Genomic Epidemiology of Complex, Multispecies, Plasmid-Borne bla(KPC) Carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014. Antimicrob. Agents Chemother. 2020, 64, e02244-19. [Google Scholar] [CrossRef]
- Odih, E.E.; Oaikhena, A.O.; Underwood, A.; Hounmanou, Y.M.G.; Oduyebo, O.O.; Fadeyi, A.; Aboderin, A.O.; Ogunleye, V.O.; Argimon, S.; Akpunonu, V.N.; et al. Correction for Odih et al., “High genetic diversity of carbapenem-resistant Acinetobacter baumannii isolates recovered in Nigerian hospitals in 2016 to 2020”. mSphere 2024, 9, e0065923. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Song, C.; Zhang, J.; Diao, S.; Heinrichs, T.M.; Martins, F.S.; Lv, Z.; Zhu, Y.; Yu, M.; Sy, S.K.B. Effects of amikacin, polymyxin-B, and sulbactam combination on the pharmacodynamic indices of mutant selection against multi-drug resistant Acinetobacter baumannii. Front. Microbiol. 2022, 13, 1013939. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Harnod, D.; Hsueh, P.R. Global threat of carbapenem-resistant Gram-negative bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Zhan, Q.; Xu, Y.; Wang, B.; Yu, J.; Shen, X.; Liu, L.; Cao, X.; Guo, Y.; Yu, F. Distribution of fluoroquinolone resistance determinants in Carbapenem-resistant Klebsiella pneumoniae clinical isolates associated with bloodstream infections in China. BMC Microbiol. 2021, 21, 164. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Biswas, I.; Veeraraghavan, B. Accurate identification of clinically important Acinetobacter spp.: An update. Future Sci. OA 2019, 5, FSO395. [Google Scholar] [CrossRef]
- Becker, B.; Weiss, C.; Holzapfel, W.H. An evaluation of the use of three phenotypic test-systems for biochemical identification of Enterobacteriaceae and Pseudomonadaceae. Food Control 2009, 20, 815–821. [Google Scholar] [CrossRef]
- Jeong, S.; Hong, J.S.; Kim, J.O.; Kim, K.H.; Lee, W.; Bae, I.K.; Lee, K.; Jeong, S.H. Identification of Acinetobacter Species Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. Ann. Lab. Med. 2016, 36, 325–334. [Google Scholar] [CrossRef]
- MarÝ-Almirall, M.; Cosgaya, C.; Higgins, P.G.; Van Assche, A.; Telli, M.; Huys, G.; Lievens, B.; Seifert, H.; Dijkshoorn, L.; Roca, I. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: Inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin. Microbiol. Infect. 2017, 23, 210.e1–210.e9. [Google Scholar] [CrossRef] [PubMed]
- De Florio, L.; Riva, E.; Giona, A.; Dedej, E.; Fogolari, M.; Cella, E.; Spoto, S.; Lai, A.; Zehender, G.; Ciccozzi, M. MALDI-TOF MS identification and clustering applied to Enterobacter species in nosocomial setting. Front. Microbiol. 2018, 9, 1885. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Pourhoseingholi, M.A.; Vahedi, M.; Rahimzadeh, M. Sample size calculation in medical studies. Gastroenterol. Hepatol. Bed Bench 2013, 6, 14. [Google Scholar] [PubMed]
| Primer | Sequence (5′–3′) | Amplicon Size (bp) | PCR Condition (35 Cycles) | Reference | ||
|---|---|---|---|---|---|---|
| Denature | Annealing | Extension | ||||
| BlaNDM-F | GGGCAGTCGCTTCCAACGGT | 475 | 95 °C | 58 °C | 72 °C | [25] |
| BlaNDM-R | GTAGTGCTCAGTGTCGGCAT | 30 s | 30 s | 1.30 s | ||
| BlaOXA-F | TTGGTGGCATCGATTATCGG | 438 | 95 °C | 55 °C | 72 °C | [26] |
| BlaOXA-R | GAGCACTTCTTTTGTGATGGC | 30 s | 30 s | 1.30 s | ||
| BlaKPC-F | CATTCAAGGGCTTTCTTGCTGC | 538 | 95 °C | 55 °C | 72 °C | [27] |
| BlaKPC-R | ACGACGGCATAGTCATTTGC | 30 s | 30 s | 1.30 s | ||
| BlaIMP-F | GGAATAGAGTGGCTTAAYTC | 232 | 95 °C | 55 °C | 72 °C | [28] |
| BlaIMP-R | TCGGTTTAAYAAAACAACCACC | 30 s | 30 s | 1.30 s | ||
| BlaVIM-F | GATGGTGTTTGGTCGCATA | 390 | 95 °C | 55 °C | 72 °C | [28] |
| BlaVIM-R | CGAATGCGCAGCACCAG | 30 s | 30 s | 1.30 s | ||
| Non-CR-GNB (n = 225) | CR-GNB (n = 92) | p-Value | |
|---|---|---|---|
| Age, year (IQR) | 36 (29–45) | 38.5 (29.8–49.0) | 0.050 c |
| Sex, n (%) | 0.758 a | ||
| Female | 181 (80.4) | 72 (78.3) | |
| Male | 44 (19.6) | 20 (21.7) | |
| Patient Type, n (%) | |||
| Outpatient | 181 (80.4) | 80 (87.0) | 0.196 a |
| Inpatient | 44 (19.6) | 12 (13.0) | |
| Setting, n (%) | |||
| Outpatient Departments | |||
| General medicine | 154 (85.1) | 51 (63.7) | <0.001 b,* |
| Obstetrics and gynecology | 13 (7.2) | 14 (17.5) | 0.150 a |
| Medicine | 4 (2.2) | 3 (3.8) | 0.441 b |
| Pediatrics | 3 (1.7) | 0 (0.0) | 0.555 b |
| Ear nose and throat | 3 (1.7) | 4 (5.0) | 0.206 b |
| Surgery | 1 (0.6) | 3 (3.8) | 0.087 b |
| Nephrology and dialysis | 1 (0.6) | 0 (0.0) | 1.000 b |
| Neurology | 1 (0.6) | 0 (0.0) | 1.000 b |
| Urology | 1 (0.6) | 2 (2.5) | 0.223 b |
| Oncology | 0 (0.0) | 3 (3.8) | 0.028 b,* |
| Inpatient Departments | |||
| General medicine | 26 (59.1) | 3 (25.0) | 0.052 a |
| Obstetrics and gynecology | 5 (11.4) | 0 (0.0) | 0.574 b |
| Medicine | 3 (6.8) | 0 (0.0) | 1.000 b |
| Pediatrics | 2 (4.5) | 1 (8.3) | 0.552 b |
| Surgery | 2 (4.5) | 0 (0.0) | 1.000 b |
| Intensive care unit | 2 (4.5) | 6 (50.0) | 0.001 b,* |
| Nephrology and dialysis | 1 (2.3) | 0 (0.0) | 1.000 b |
| Neurology | 1 (2.3) | 0 (0.0) | 1.000 b |
| Oncology | 1 (2.3) | 2 (16.7) | 0.113 b |
| Cardiology | 1 (2.3) | 0 (0.0) | 1.000 b |
| Specimen type, n (%) | |||
| Urine | 178 (79.1) | 73 (79.3) | 1.000 a |
| High vagina swab | 13 (5.8) | 6 (6.5) | 1.000 a |
| Stool | 10 (4.4) | 1 (1.1) | 0.186 b |
| Wound swab | 10 (4.4) | 1 (1.1) | 0.186 b |
| Ear swab | 6 (2.7) | 4 (4.3) | 0.484 b |
| Throat swab | 3 (1.3) | 1 (1.1) | 1.000 b |
| Sputum | 2 (0.9) | 1 (1.1) | 1.000 b |
| Catheter | 2 (0.9) | 2 (2.2) | 0.583 b |
| Semen | 1 (0.4) | 0 (0.0) | 1.000 b |
| Trachea aspirate | 0 (0.0) | 2 (2.2) | 0.084 b |
| Blood | 0 (0.0) | 1 (1.1) | 0.290 b |
| Non-carbapenem Resistant Gram-Negative Bacilli (n = 225), n (%) | Carbapenem-Resistant Gram-Negative Bacilli (n = 92), n (%) | |
|---|---|---|
| Klebsiella pneumoniae | 101 (44.9) | 26 (28.3) |
| Escherichia coli | 84 (37.3) | 43 (46.7) |
| Pseudomonas aeruginosa | 10 (4.4) | 13 (14.1) |
| Enterobacter species | 10 (4.4) | 0 (0.0) |
| Proteus mirabilis | 9 (4.0) | 6 (6.5) |
| Enterobacter cloacae | 5 (2.2) | 0 (0.0) |
| Proteus spp. | 2 (0.9) | 0 (0.0) |
| Aeromonas fergusonii | 1 (0.4) | 0 (0.0) |
| Burkholderia cepacia | 1 (0.4) | 0 (0.0) |
| Raoultella ornithinolytica | 1 (0.4) | 0 (0.0) |
| Salmonella enterica | 1 (0.4) | 0 (0.0) |
| Acinetobacter baumannii | 0 (0.0) | 2 (2.2) |
| Enterobacter asburiae | 0 (0.0) | 1 (1.1) |
| Escherichia fergusonii | 0 (0.0) | 1 (1.1) |
| Sample Code | Organisms | Carbapenemase Gene | SAM | TS | TZP | CTX | CTZ | CAZ | CZA | FEP | IMP | MEM | CIP | GEN | AMK | COL | Classify |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1a | P. aeruginosa | blaNDM | - | - | R | R | R | R | R | R | R | S | R | R | S | I | XDR |
| R123 | P. aeruginosa | Nd | - | - | R | R | R | R | R | R | R | S | R | R | S | S | XDR |
| L65 | P. aeruginosa | Nd | - | - | R | R | R | R | R | R | R | R | S | R | R | I | XDR |
| L73 | P. aeruginosa | Nd | - | - | R | R | S | R | R | R | R | S | R | S | S | S | MDR |
| L23 | P. aeruginosa | Nd | - | - | R | R | R | R | R | R | R | S | S | R | S | R | XDR |
| L74 | K. pneumoniae | blaKPC | R | R | R | R | R | R | R | R | R | R | R | S | I | S | XDR |
| L29 | K. pneumoniae | blaKPC | R | R | R | R | R | S | S | S | I | I | S | S | - | I | MDR |
| L26 | K. pneumoniae | blaKPC | R | R | R | R | R | S | S | S | I | I | S | S | - | I | MDR |
| R102 | K. pneumoniae | blaNDM | R | R | R | R | R | R | R | R | R | R | R | R | I | S | XDR |
| L19 | E. coli | blaNDM | R | R | R | R | R | R | R | R | R | R | R | R | R | R | PDR |
| R135 | E. coli | blaNDM | R | R | R | R | R | R | R | R | R | R | R | R | R | S | XDR |
| H26 | E. coli | blaNDM | R | R | R | R | R | R | R | R | R | R | R | R | S | S | XDR |
| R140 | E. coli | Nd | R | R | R | R | R | R | R | R | R | R | R | R | S | S | XDR |
| R104 | A. baumannii | blaNDM | R | R | R | R | R | R | R | R | R | R | R | R | S | S | XDR |
| R120 | A. baumannii | blaNDM | R | R | R | R | R | R | R | R | R | R | R | S | I | S | XDR |
| R92 | E. fergusonii | Nd | R | R | R | R | R | R | S | S | S | R | S | S | S | S | MDR |
| L61 | P. mirabilis | Nd | R | R | S | S | S | S | S | S | R | S | S | I | S | S | MDR |
| Sample Code | Organisms | Hospital Unit/Department | Specimen | Diagnosis | 30-Day Mortality Outcome |
|---|---|---|---|---|---|
| L23 | P. aeruginosa | ICU | Wound Swab | Surgical wound infection | * No |
| L65 | P. aeruginosa | ENT | Ear Swab | Otitis media | * No |
| L73 | P. aeruginosa | ENT | Ear swab | Otitis media | * No |
| R123 | P. aeruginosa | O&G | High Vaginal Swab | Pelvic Inflammatory Disease | * No |
| H1a | P. aeruginosa | Oncology | Throat Swab | Enlarged adenoid | * No |
| L19 | E. coli | ICU | Blood | Benign neoplasia/Pituitary adenoma | ** Died |
| H26 | E. coli | O&G | Urine | UTI/Preterm premature rupture of membrane | * No |
| R135 | E. coli | General Medicine/Surgery Outpatient | Urine | UTI | * No |
| R140 | E. coli | General Medicine/Surgery Outpatient | Urine | UTI | * No |
| L26 | K. pneumoniae | ICU | Catheter Tip | Prostate Enlargement/CVA | * No |
| L29 | K. pneumoniae | ICU | Trachea Aspirate | Adenocarcinoma of the prostate/COPD | * No |
| L74 | K. pneumoniae | General Medicine/Surgery Outpatient | Urine | UTI | * No |
| R10 | K. pneumoniae | Pediatrics | Stool | Neonatal sepsis | * No |
| R104 | A. baumannii | General Medicine/Surgery Outpatient | Urine | UTI | * No |
| R120 | A. baumannii | General Medicine/Surgery Outpatient | Urine | Pyelonephritis/UTI | * No |
| R92 | E. fergusonii | General Medicine/Surgery Outpatient | Urine | UTI | * No |
| L61 | P. mirabilis | General Medicine/Surgery Outpatient | Urine | UTI | * No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salau, M.; Kositanont, U.; Noisumdaeng, P.; Ogunsola, F.; Ettu, A.-W.O.-o.; Adewojo, D.; Ojimma, C.; Ojomaikre, O.; Changkaew, K. Characterization of Carbapenem-Resistant Gram-Negative Bacilli Isolates in Multispecialty Private Hospitals in Lagos, Nigeria. Infect. Dis. Rep. 2025, 17, 119. https://doi.org/10.3390/idr17050119
Salau M, Kositanont U, Noisumdaeng P, Ogunsola F, Ettu A-WO-o, Adewojo D, Ojimma C, Ojomaikre O, Changkaew K. Characterization of Carbapenem-Resistant Gram-Negative Bacilli Isolates in Multispecialty Private Hospitals in Lagos, Nigeria. Infectious Disease Reports. 2025; 17(5):119. https://doi.org/10.3390/idr17050119
Chicago/Turabian StyleSalau, Moruf, Uraiwan Kositanont, Pirom Noisumdaeng, Folasade Ogunsola, Abdul-Wahab Omo-ope Ettu, Damilola Adewojo, Chinonso Ojimma, Omamode Ojomaikre, and Kanjana Changkaew. 2025. "Characterization of Carbapenem-Resistant Gram-Negative Bacilli Isolates in Multispecialty Private Hospitals in Lagos, Nigeria" Infectious Disease Reports 17, no. 5: 119. https://doi.org/10.3390/idr17050119
APA StyleSalau, M., Kositanont, U., Noisumdaeng, P., Ogunsola, F., Ettu, A.-W. O.-o., Adewojo, D., Ojimma, C., Ojomaikre, O., & Changkaew, K. (2025). Characterization of Carbapenem-Resistant Gram-Negative Bacilli Isolates in Multispecialty Private Hospitals in Lagos, Nigeria. Infectious Disease Reports, 17(5), 119. https://doi.org/10.3390/idr17050119






