Abstract
The Unified Medical Language System (UMLS) is an internationally recognized medical vocabulary that enables semantic interoperability across various biomedical terminologies. To use its knowledge, the users must understand its complex knowledge structure, a structure that is not interoperable or is not compliant with any known biomedical and healthcare standard. Further, the users also need to have good technical skills to understand its inner working and interact with UMLS in general. These barriers might cause UMLS usage concerns among inter-disciplinary users in biomedical and healthcare informatics. Currently, there exists no terminology service that normalizes UMLS’s complex knowledge structure to a widely accepted interoperable healthcare standard and allows easy access to its knowledge, thus hiding its workings. The objective of this research is to design and implement a light-weight terminology service that allows easy access to UMLS knowledge structured using the fast health interoperability resources (FHIR) standard, a widely accepted interoperability healthcare standard. The developed terminology service, named UMLS FHIR, leverages FHIR resources and features, and can easily be integrated into any application to consume UMLS knowledge in the FHIR format without the need to understand UMLS’s native knowledge structure and its internal working.
1. Introduction
Experts have developed multiple semantic standards, ranging from databases (e.g., protein, organism, etc.) to controlled terminologies (e.g., International Classification of Diseases (ICD), Systematized Nomenclature of Medicine (SNOMED), etc.) to ontologies (e.g., Gene Ontology, Diseases Ontology [1], etc.), with varying complexity to capture biomedical and healthcare semantics. In general, these standards express the same term in diverse ways. For example, the term atrial fibrillation is referred to as Atrial Fibrillations in MeSH (Medical Subject Heading), Auricular Fibrillation in PSY (Psychological Index) terms, Auricular Fibrillations in MeSH, Fibrillation—atrial in SNOMED, AFib in NCI (National Cancer Institute), etc. Furthermore, the position of the term in the hierarchical structure varies across the standards.
The National Library of Medicine (NLM) has undertaken the Unified Medical Language System (UMLS) [2,3,4] to bring different semantic standards together by integrating the similar medical terms expressed in diverse ways into a single entity, called a concept, and it also capture the relationships between the terms defined in the integrating sources. The current version of UMLS has integrated 211 semantic standards, generating 4.26 million concepts from 15.2 million medical terms, and more than 10 million relationships. The UMLS has three modules—Lexical tools, Metathesaurus, and Semantic Network [5]. Metathesaurus holds the concepts, mapping between concepts to terms from the source standard, and concept attributes, synonyms, relationships, etc. Semantic Network is an ontological resource that categorizes the Metathesaurus concepts into a higher-level category. For the rest of the article, UMLS knowledge refers to both UMLS Metathesaurus and Semantic Network.
Currently, users have two ways to access UMLS knowledge. First, obtain UMLS knowledge as raw files (.nlm) and execute MetamorphoSys, an installation wizard, to generate a set of RRF (Rich Release Format) files (e.g., MRCONSO.RRF, MRSAB.RRF, MRREL.RRF, MRSTY.RRF, etc.) and SQL scripts. Executing the SQL scripts will create SQL tables (with the same name as the RRF file) and copy the data from the RRF files into the respective tables. Each RRF file has a corresponding SQL table, and the table structure reflects the RRF file structure. Individual users write SQL queries to obtain the desired UMLS knowledge, and also a custom software to parse the results to use in an application. Second, users query the UMLS API endpoints [6] developed by NLM and the results are communicated in JSON format. With the former approach, users must first understand the complex UMLS knowledge structure to query and use it, a complex and slow process. The UMLS knowledge structural complexity is due to knowledge distribution across multiple SQL tables (or RRF files), each holding attributes, relationships, and other characteristics related to a concept. The users must tackle the perplexing task of understanding what each table (or RRF file) holds, the columns of the table, and how to join the tables (or parse RRF files) to obtain the desired UMLS knowledge. The user must also have good SQL knowledge to write efficient SQL queries, and installing UMLS from scratch or updating it is another dreaded process. This approach is a convoluted process and daunting for non-technical users, who are a majority in the domain of biomedical and healthcare informatics.
Figure 1 presents a query output, using the latter approach, to find medical terms equivalent to atrial fibrillation in the JSON format. There are 26 terms from multiple source standards equivalent to the query term atrial fibrillation. The figure only displays the JSON structure of one term due to space constraints. To use this knowledge, users need to refer to the documentation of one or more RRF files (or SQL tables) to understand the keys (ui, obsolete, rootSource, etc.) and associated value in the output JSON structure. For example, the key “obsolete” indicates if the term is outdated in the source. The user needs to refer to the MRCONSO RRF file (or SQL table) documentation to get this information. Similarly, “rootSource” indicates the original semantic source of the term. The user must refer to the MRSAB RRF file (or SQL table) documentation to obtain information on the abbreviation. For keys (e.g., “relation”) with a link (another endpoint) as its value, the users need to query the endpoint and understand the output’s JSON structure by referring to one or more RRF files. For some keys (e.g., parents, ancestors, etc.), the user needs to refer to the UMLS API documentation.

Figure 1.
The output JSON structure as a result of a UMLS API query to find terms equivalent to term atrial fibrillation.
In either approach, users face the same challenges—the need to understand the UMLS’s native knowledge structure so as to use UMLS knowledge. These challenges—the complex UMLS knowledge structure that does not align with a healthcare standard or any standard in general, the need to write complex SQL queries and custom software by the user to parse the query results—make it difficult to interact with the UMLS and use its knowledge. However, when a widely accepted standard is used to represent the UMLS knowledge, the standard normalizes the UMLS knowledge structure, hiding the discussed issues. Further, the user can write a reusable software program or use an existing library to parse the standard’s structure to use the UMLS knowledge. As UMLS is recognized internationally and used in a wide range of applications, there is a need for a terminology service—a light-weight solution, when compared to the current UMLS installation process, which expresses its knowledge in an interoperable format using mature standard(s).
The emerging interoperability standard that can represent healthcare data and also has the ability to express a semantic standard’s metadata and its content is Fast Interoperability Health Resources (FHIR) [7,8,9]. FHIR is an emerging new member of the HL7 family, developed to meet healthcare data interoperability requirements, and it has the great benefit of it being relatively easy to implement and rapidly deploy FHIR compliant applications when compared to other existing standards. Some of the salient features of FHIR are as follows: data are captured and shared as resource-modular data units; integrated support to REST with well-defined guidelines [10] allowing developers to manage and manipulate the resources using an API; and FHIR extensions allow experts to extend the FHIR resource to meet new requirements. Furthermore, when compared with existing standards (e.g., HL7 Clinical Document Architecture (CDA) [11], HL7 V2 messaging [11], openEHR [12], etc.), FHIR by design has the ability to represent semantic knowledge using FHIR terminology resources; other standards do not have any foundational support. FHIR’s ability to represent semantic knowledge along with FHIR features, and the availability of open-source implementation of the FHIR standard, the HAPI library [13], makes the FHIR standard a prime candidate to be evaluated and used to represent UMLS knowledge. Other healthcare standards lack an open-source implementation of the standard’s specification.
The overall objective of this research is to implement a terminology service, which allows users to query and easily access the UMLS knowledge structured using the FHIR standard. An interoperable representation of UMLS knowledge allows any FHIR-compliant application to consume UMLS knowledge without the need to understand its native knowledge structure and inner workings. Towards this objective, this research presents the following: A detailed analysis of similarities and differences between UMLS’s knowledge structure and FHIR ConceptMap, an FHIR terminology resource; A design of FHIR extensions to profile ConceptMap to align it with UMLS’s knowledge structure, which then allows us to express UMLS knowledge as ConceptMap and implement the extensions using the HAPI library to demonstrate the validity of the research; Finally, it allows us to design and implement the proposed REST-based terminology service, named UMLS FHIR, using the HAPI library that allows users to query the desired UMLS knowledge and expresses it in an interoperable format using the profiled FHIR ConceptMap. The UMLS FHIR terminology service is available at http://umls.it.ilstu.edu/umlsfhir/, whereas the API which drives the service is available at http://umls.it.ilstu.edu/umlsfhir/fhir. As like any other API, UMLS FHIR API endpoints can be called from any FHIR compliant application to consume the FHIR formatted knowledge. The terminology service presents a simple web interface, on top of UMLS FHIR API, to query UMLS, returning the results in the FHIR format. The service acts as a simple interoperable facade hiding the discussed UMLS complexities.
2. Literature Review
Structural and Semantic standards have played a crucial role in digitalizing healthcare data. A structural standard such as HL7 FHIR provides a structure to represent healthcare data, while a semantic standard such as LOINC complements the data with semantics added to it. Separately, UMLS, a semantic terminology, and FHIR, an interoperability standard, are well-known and widely used for various healthcare research and industrial applications. For example, UMLS is extensively used in clinical NLP [14,15,16,17,18], where UMLS is used as a reference terminology to identify medical terms in clinical notes. This improves UMLS, where UMLS is again used as a reference terminology to identify new medical terms or relationship from medical corpora (e.g., journals) using existing medical terms and then adding the new knowledge to be included in UMLS. As new knowledge is frequently added to UMLS [19,20,21,22], there are multiple audits and quality checks to ensure the quality of UMLS knowledge [23]. Clinical decision systems [15,24,25] have used UMLS in the past to extract similar medical cases for clinical references, etc. FHIR is primarily used to achieve data interoperability [7] across diverse healthcare systems, design customized Electronic Health Records (EHRs) and Personal Health Records (PHRs) [26] to meet physician and patient requirements, enable the easy access to and adoption of medical vocabulary [27,28,29], create computable case forms to identify patients suitable to cancer trails [30], capture genomic data [31], etc. A detailed discussion of the potential uses of UMLS or FHIR, separately, is beyond the scope of this research article.
Currently, there are a few terminology services and servers that allow access to semantic standards to satisfy diverse healthcare needs, and only one service that uses FHIR to serve the semantic content. The NLM UMLS API, a REST-based service, serves UMLS knowledge to a registered user via multiple endpoints. As discussed (Figure 1), the barrier is that the knowledge structure is directly related to the UMLS native knowledge structure, a non-interoperable format. Before UMLS API, NLM developed the UMLS Knowledge Source Server [32]. The authors used the innovative technology available at that time to service UMLS knowledge via the web. The server was implemented as a collection of Java servlets. The servlets queried UMLS using the RMI API. The query results, in XML format, are returned to the servlets that then apply the XSLT stylesheets to transform those results into HTML for display purposes. The server put UMLS on the web. Metke-Jimenez et al. have developed OntoServer [28]. The Ontoserver is a terminology server that uses FHIR terminology resources to expresses the semantic knowledge encapsulated by SNOMED CT terminology, LOINC codes, and select OWL ontologies, such as the Human Phenotype Ontology (HPO). Similarly, the LONIC FHIR Terminology Server [29] serves LONIC terminology codes using FHIR terminology resources. This research is developing a terminology service using FHIR to express UMLS knowledge, which encompasses SNOMED CT and LONIC along with 209 other internationally known terminologies. Jiang et al. proposed to develop a vocabulary mapping service that maps Observational Health Data Sciences and Informatics (OHDSI) vocabularies to FHIR resources [21]. However, the author did not take the proposal beyond the conceptual idea. Currently, there are no efforts, based on the authors’ knowledge, that leverage the FHIR standard and its terminology resources for representing UMLS knowledge.
3. Background
3.1. UMLS Knowledge Structure
In the UMLS Metathesaurus, terms (e.g., malaria, fever, etc.) from diverse standards (e.g., SNOMED, MeSH, etc.), referred to as atoms in UMLS, are mapped to a single concept—a central entity used to structure the knowledge in Metathesaurus. An atom has an AUI (Atom Unique Identifier, a unique identifier given by UMLS), a code (identifier from the source), and a name. The source standard has a fully qualified name, version, and abbreviation. A concept has a CUI (Concept Unique Identifier, a unique and permanent identifier given by UMLS), a name (a preferred name chosen by the NLM), one or more semantic type, and other attributes. In practice, the concepts and atoms are in the MRCONSO table (or RRF file), a concept’s semantic type(s) are in the MRSTY table (or RRF file), a concept’s relationships are in the MRREL table, and version information is in the MRSAB table. The CUI is used to join multiple tables to obtain the required knowledge [33]. Figure 2 shows atoms from diverse sources mapped to a single concept named Atrial Fibrillation with CUI C0004238.
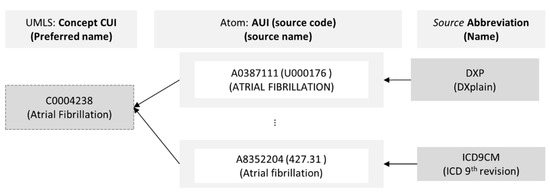
Figure 2.
Mapping of different atoms from diverse semantic standards equivalent to the unique UMLS concept named “Atrial Fibrillation”.
The Metathesaurus also encapsulates relationships, both hierarchical and associative, between concepts. The attribute’s type and label allow users to interpret the relationship. The type attribute categorizes a relationship, wherein the categories such as PAR (parent), CHD (child), RB (broader relationships), etc., are defined in a set by NLM, which will be referred to as the REL set in this article. The REL set has 15 values. The label attribute captures the name or label of the relationship. Figure 3 shows the UMLS knowledge structure as a UML class diagram, summarizing the above explanation. There are other attributes, not described here, that can also determine the status of a concept, atom, or relationship in UMLS. Refer to the UMLS reference manual for more details [5].
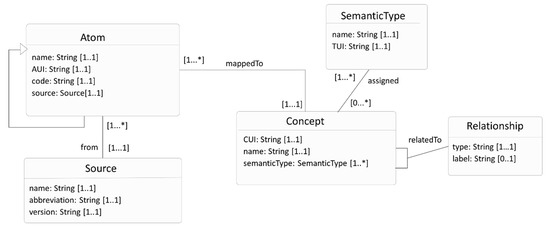
Figure 3.
UML class diagram representing UMLS knowledge structure where the Concept class represents the UMLS concept, Atom class represents atoms in UMLS, and other classes support the Concept class.
3.2. FHIR Standard
3.2.1. FHIR Resources
FHIR defines resources to describe and share healthcare data. For example, a patient resource captures demographics and administrative information about an individual, and observation supports the diagnosis, progress, baselines, etc. FHIR’s resources can capture diverse healthcare data and the necessary infrastructure and guidelines to handle resources. FHIR integrates REST into the standard, allowing standardized access to FHIR resources using API-style programming. Further, experts can profile FHIR resources without violating the fundamentals of FHIR specifications to meet any new healthcare requirements or application constraints. For example, a patient resource cannot capture an individual’s nationality. Experts can profile the patient resource by defining an FHIR extension for the patient resource to capture his/her nationality. FHIR has resources, categorized as terminology resources, to handle semantic standards and their knowledge, specifically, CodeSystem, ConceptMap, and ValueSet. This research profiles ConceptMap and uses CodeSystem and ValueSet to meet the research objective.
3.2.2. FHIR ConeptMap
The FHIR ConceptMap captures the mapping between a set of codes in a code system (e.g., SNOMED) to one or more codes in another coding system (e.g., MeSH, SNOMED, etc.). Figure 4 shows the ConceptMap as a UML class diagram from FHIR documentation [34]. The ConceptMap has a variety of attributes to describe the purpose of the mappings. A group (Group class) captures the mapping between the source code (SourceElement class) and the target code(s) (TargetElement class), and ConceptMap can have multiple groups. The equivalence attribute of the TargetElement class captures the exact nature of the mapping between the codes. Experts can specify more information about the mapping using the OtherElement class. For example, the SourceElement can be Atrial Fibrillation (code-C0004238, display—Atrial Fibrillation) with UMLS as its source, TargetElement is AFib (code 427.31, display—Atrial Fibrillation) from target ICD9CM and these codes are equal (equivalence in TargetElement).
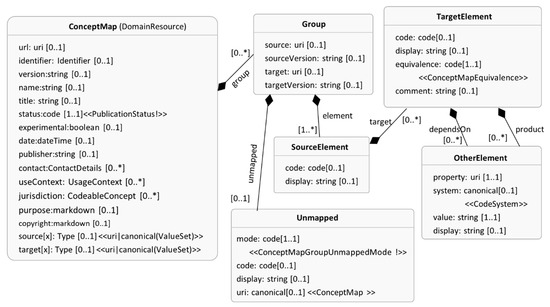
Figure 4.
Using the UML class diagram to represent FHIR ConceptMap. The classes and attributes represent entities in the FHIR ConceptMap.
3.2.3. FHIR CodeSystem
CodeSystem declares the existence of a code system, and its properties (e.g., URL, version, publication date, etc.). The CodeSystem has the ability to capture few codes from the code system, along with their basic properties (code, display, definition, etc.). The CodeSystem is not intended to support the process of representing the codes in the code systems, and is thus not an efficient way to represent the semantic knowledge captured by standards (e.g., SNOMED CT, LOINC, etc.).
3.2.4. FHIR ValueSet
A ValueSet encapsulates a set of codes drawn from one or more code systems, intended for use in a particular context. ValueSet is a bridge between CodeSystem and the usage of the codes across FHIR resources. For example, the equivalence attribute in the TargetElement class of ConceptMap can only accept values defined in the ConcetpMapEquivalence valueset. Please refer to the official documentation for more details and the structures of both ValueSet and CodeSystem.
3.3. Comparing UMLS Knowledge Structure and FHIR ConceptMap
Using the background knowledge on UMLS knowledge (Figure 3) and the ConceptMap (Figure 4), the following section explains the similarities between them and the shortcomings of ConceptMap.
3.3.1. Similarities
- The equivalence attribute of TargetElement in ConceptMap is bound to ConceptMapEquivalence valueset, i.e., the equivalence attribute will only accept values defined in this ConceptMapEquivalence valueset. The ConceptMapEquivalence valueset defines ten values (e.g., equal, specializes, subsumes, wider, etc.), and these values describe the nature of the mapping between SourceElement and TargetElement. For example, when the equivalence attribute takes the value “equal”, the code captured by TargetElement is equal to the code captured by SourceElement;
3.3.2. Differences
- The ConceptMap cannot capture the label attribute of the relationship (Relationship class, Figure 3). For example, Atrial Fibrillation (source with CUI C0004238) is related to Digoxin (target with CUI C0012265), where the attribute’s type has the value related, and the label has the value may_be_treated_by.
- As mentioned previously, the equivalence attribute of TargetElement in ConceptMap is bound up with ConceptMapEquivalence valueset, and the values of the valueset describe the nature of the mapping between SourceElement and TargetElement. The equivalence attribute of the TargetElement class can be mapped to the type attribute of the UMLS relationship (Relationship class, Figure 3). A relationship type attribute can only take a value from the UMLS REL set. The mapping between the ConceptMapEquivalence valueset’s values and the REL set’s values is not exactly one to one. There is one to one mapping between a few values across the sets without loss of semantics, but for some values, there is no mapping. The lack of the mapping is either due to the unavailability of an equivalent value in the other set, or the fact that the official description of the value, particularly the values in the UMLS REL set, is vague, making it hard to determine a mapping. For example,
- ○
- The value PAR or CHD (representing hierarchy between UMLS concepts) from the REL set has an equivalent mapping to Subsumes or Specializes in ConceptMapEquivalence;
- ○
- The values AQ or QB or Empty from the REL set do not map to any value in the ConceptMapEquivalence valueset;
- ○
- The value RU in the UMLS REL set, described as “Related, unspecified”, is not clear, making it hard to determine a mapping to ConceptMapEquivalence values, and is hence unmapped.
- ConceptMap cannot capture the semantic type of a concept or atom.
Table 1 summarizes this discussion. The content of the table is formatted in *.attribute format, where * indicates the class name. For example, Concept.CUI (Figure 3) refers to the CUI attribute of the Concept class and ConceptMap.Group.target refers to the target attribute of the Group class in the class ConceptMap. Table 2 shows the values mapped (and unmapped) between REL set and ConceptMapEquivalence valueset, along with the value definition/description from the sources.

Table 1.
Mapping between UMLS Knowledge Structure and ConceptMap.

Table 2.
Mapping between REL set and ConceptMapEquivalence valueset values.
4. Method
This section describes the methodology followed to design and develop the UMLS FHIR terminology service, which represents UMLS knowledge, specifically, the mapping between the atoms and the UMLS concept and relationships, both hierarchical and associations between the UMLS concepts using FHIR. The workflow is represented in Figure 5.
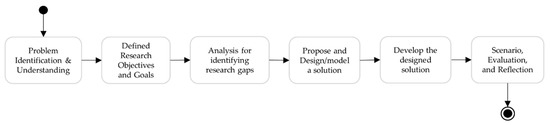
Figure 5.
Overview of the research methodology for designing and developing UMLS FHIR terminology service.
- Step 1: Problem identification and understanding, and defining research objectives and goals, discussed in Section 1.
- Step 2: An in-depth analysis to understand the fundamental similarities and differences between the UMLS knowledge structure and ConceptMap, discussed in Section 3.3.
- Step 4: Implement the designed FHIR extensions and profiled ConceptMap using the HAPI library. Develop the UMLS FHIR terminology service using HAPI library, profiled ConceptMap and other FHIR terminological resources.
- Step 5: Demonstrate the terminology server, evaluate the results, and reflect on the research.
5. UMLS FHIR Terminology Service: Design and Implementation
5.1. Design: FHIR Extensions to ConceptMap
To overcome the identified shortcoming in the analysis, the research designs three FHIR extensions to align the ConceptMap to the UMLS knowledge requirements for an interoperable representation of UMLS knowledge using ConceptMap.
- An extension to capture the semantic type(s) of the SourceElement (UMLS concept) and TargetElement (Atom).
- An extension to capture the label attribute associated with the UMLS relationship (Relationship class, Figure 3).
- An extension to capture the type attribute of the UMLS relationship (Relationship class, Figure 3). As discussed, the type attribute only takes values from the UMLS REL set. Thus, the REL set is translated into an FHIR valuleset, named UMLSRelationTypeCode. This extension is bound to the UMLSRelationTypeCode valueset. This design is similar to the equivalence attribute of the TargetElement class in ConceptMap that is bound to the ConceptMapEquivalence valueset.
Figure 6 shows the three FHIR extensions in the FHIR structure, and Figure A1 (Appendix A) shows the defined UMLSRelationTypeCode valueset in the JSON format. In Figure 6, Figure 6a shows the FHIR extension to capture the semantic type as an FHIR Coding datatype, Figure 6b shows FHIR extension to capture the label attribute of a UMLS relationship as a string datatype, and Figure 6c shows the extension bound to UMLSRelationTypeCode valueset, to capture the type attribute of a UMLS relationship (Relationship class, Figure 3). Figure A2 shows the profiled ConceptMap with the designed extensions in the FHIR structure.

Figure 6.
Three FHIR extensions for ConceptMap to enable an interoperable representation of UMLS knowledge.
5.2. Implementation: FHIR Extensions and Profiled ConceptMap
The HAPI library [13], an open-source implementation of the FHIR specifications in Java, allows users to implement the extensions and profiled ConceptMap. The HAPI library defines Java classes for every resource, data types, and enumerations for the valuesets defined by the FHIR. For example, the ConceptMap Java class in the HAPI library corresponds to the FHIR ConceptMap resource. The elements of the resource are programmed as class attributes. HAPI also supports data validation for the modeled FHIR resources to ensure resource confirmation to the FHIR specification. Figure 7 shows the profiled ConceptMap as a UML class diagram. The semantic type extension (Figure 6a) is translated as a semanticType attribute, and is added to both SourceElement and TargetElement, as both concepts and atoms have an assigned semantic type(s). The attributes mappingLabel (mapping label extension, Figure 6b) and mappingType (mapping type extension, Figure 6c) are added to TargetElement to capture the mapping (UMLS relationship) type and label. The mappingType attribute is bound to the UMLSRelationTypeCode valueset. Following HAPI documentation for implementing extensions and profiles [35], the profiled ConceptMap (Figure 7) is represented by creating a new class called UMLSConceptMap, which extends the resource ConceptMap. The UMLSRelationTypeCode is a Java enumeration with the same name. The source code is available at https://github.com/rishikanths/umlsfhir.
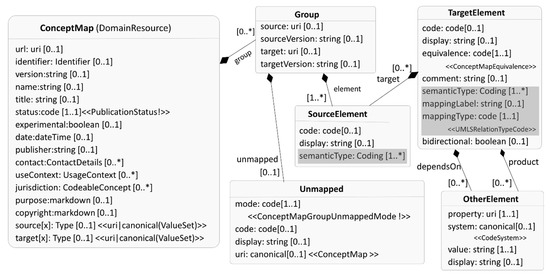
Figure 7.
Profiled FHIR ConceptMap as a UML class diagram with designed FHIR extensions (Figure 6) as attributes (highlighted).
5.3. Architecture and Impementation: UMLS FHIR Terminology Service
The FHIR specification integrates REST with the standard to manage resources and other related entities. The HAPI library has a set of predefined REST servers [36], and this terminology service architecture uses the HAPI plain REST server. As an implementation of the FHIR standard, the HAPI REST servers honor the FHIR Restful specifications [10]. The architecture, shown in Figure 8, is composed of three core entities:
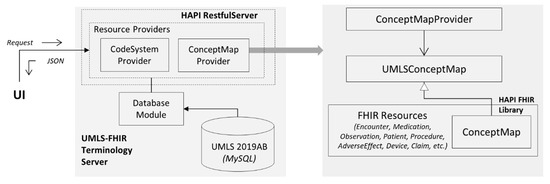
Figure 8.
Architecture of UMLS FHIR terminology service.
- A user-facing client application written in HTML and JavaScript;
- UMLS FHIR API developed using HAPI REST Server, running on the Apache Tomcat web server;
- A MySQL database for UMLS.
The terminology service can be accessed in two ways: a web application with a simple user interface, available at http://umls.it.ilstu.edu/umlsfhir/, or via an API, available at http://umls.it.ilstu.edu/umlsfhir/fhir. The web application uses the API. The web interface allows users to search for a concept using its name, where an autocomplete algorithm presents the search results based on the typed string. This search result is structured using the FHIR CodeSystem resource and returned to the user as an FHIR Bundle resource. From Section 3.2.3, CodeSystem is used to declare the existence of a code system, such as UMLS, and optionally define a part of its content. As the user search results are a subset of UMLS, the CodeSystem resource is used for an interoperable representation of the search results. When the user selects a term from the results, the selected term’s knowledge is returned as ConceptMap. Using the HAPI documentation, the REST operations on resources are performed through providers [37]. The server defines two providers: ConceptMapProvider, which allows read operation of profiled ConceptMap (Figure 7), and CodeSystemProvider allows the search operation on the CodeSystem. The Database module, a plug-in module, queries the locally installed MySQL UMLS database.
6. Results
This section will discuss the UMLS FHIR API endpoints and the returned FHIR resources. The base URL for the UMLS FHIR API is http://umls.it.ilstu.edu/umlsfhir/fhir. There are two endpoints.
- GET/CodingSystem?search={searchString}
- ○
- Description: Searches for all UMLS concepts whose name (Figure 3) begins with the user input string, captured by the parameter “searchString” and returns the result as an FHIR Bundle resource. A Bundle is a container for the collection of resources, and a search operation on any FHIR resource returns a Bundle [10]. In this case, the bundle contains a collection of CodeSystem.
- ○
- Example: http://umls.it.ilstu.edu/umlsfhir/fhir/CodeSystem?search=malaria returns all the concepts in whose name starts with “malaria”.
- ○
- Responses: Table 3 lists the response codes, its description, and resources for this endpoint.
 Table 3. Response codes and resources for the search endpoint.
Table 3. Response codes and resources for the search endpoint.
For the user search string “malaria”, the service returns all concepts whose name starts with “malaria” structured as an FHIR CodeSystem and wrapped in an FHIR Bundle. Figure 9 shows the output Bundle with CodeSystem in JSON format. The web application parses this JSON response to display the search results. If there are no results, the service will still return a Bundle resource, where the concept attribute within the CodeSystem resource will be empty.

Figure 9.
Search results returned using the CodeSystem resource.
- GET/ConceptMap/{concept_cui}
- ○
- Description: Searches for the UMLS concept using the user-provided identifier, captured by the “concept_cui” parameter, and returns the concept knowledge as a ConceptMap (Figure 7).
- ○
- Example: http://umls.it.ilstu.edu/umlsfhir/fhir/ConceptMap/C0004238 returns knowledge related to the concept identified by CUI C0004238 as ConceptMap.
- ○
- Response: Table 4 lists the response codes, its description, and resources for this endpoint.
 Table 4. Response codes and resources for the endpoint that represents UMLS knowledge as a ConceptMap.
Table 4. Response codes and resources for the endpoint that represents UMLS knowledge as a ConceptMap.
In the above example link, the CUI C0004238 identifies the UMLS concept, Atrial Fibrillation, and the knowledge on this concept is returned as a ConceptMap, the profiled ConceptMap (Figure 7). For explanation purposes, the output ConceptMap, in the JSON format, is divided into two figures. Figure 10 shows all the atoms from diverse semantic sources equivalent to the CUI C0004238, and Figure 11 shows its relationships. The table (left) in Figure 10 shows all atoms (63 of them) equivalent to Atrial Fibrillation and the respective ConceptMap representation (right). All the atoms from the same source and version are represented as a group (Group class, Figure 7). Each group has one source (SourceElement class, Figure 7) representing the UMLS concept and multiple targets (TargetElement class, Figure 7), each target representing an atom. For example, a group captures the mapping of six atoms from CHV (Consumer Health Vocabulary, 2011_02) to the UMLS concept with CUI C0004238. The source standards without an official URL are encoded as a URN using their abbreviation and version information within the context of UMLS. As discussed in the gap analysis, an “equal” value for the equivalence attribute says that the atoms from diverse sources are equal to the UMLS concept. Figure 11 shows the relationships of the concept Atrial Fibrillation. Like Figure 10, a group aggregates the relationships from the same source and version (not shown in the table due to space constraints). For each group, the source is Atrial Fibrillation with CUI C0004238, and the target(s) are other UMLS concepts. The attributes mappingType (code from UMLSRelationTypeCode valueset) and mappingLabel (a string) capture the information about the relationship (Relationship class, Figure 3).
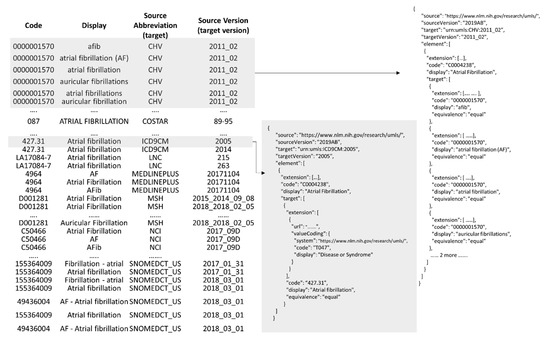
Figure 10.
Profiled ConceptMap representing atoms equivalent (Figure 3) to the UMLS concept Atrial Fibrillation with CUI C0004238.
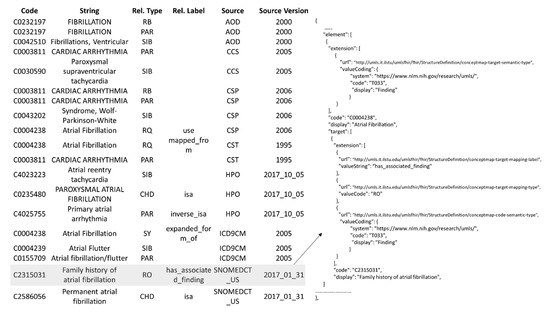
Figure 11.
Profiled ConceptMap representing relationships of Atrial Fibrillation (CUI C0004238).
When any other resources are accessed using the base URL, the FHIR server will generate a 404 error, which means the FHIR server does not support the REST operation on that resource [10]. The implemented valueset and extensions are available at the respective URLs, as shown in the figures.
7. Discussion
Applications employ semantic standards to annotate the data with semantics, which allows users (humans and machines) a uniform understanding of the data. There exists many semantic standards in the domain of biomedical and healthcare informatics that captured knowledge related to various health specializations. For example, ICD, a classification system, classifies diseases, a variety of signs, symptoms, abnormal findings, complaints, and external causes. SNOMED, a computer-processable collection of the medical terms (in both human and veterinary medicine), provides codes, terms, synonyms, and definitions for diseases, findings, procedures, substances, etc. LOINC provides codes for identifying laboratory and clinical observations, and Disease Ontology, which is a standard ontology that represents human diseases. Generally, there is an overlap of knowledge captured by these standards. The UMLS system has successfully encircled current and legacy semantics standards under one roof and integrated the collective knowledge to build a single semantic reference system. Integrating the standards into UMLS allows a unified format and the distribution of biomedical and healthcare knowledge.
However, the users are required to understand UMLS’s complex knowledge structure and its inner workings, and also need strong technical skills to interact with UMLS. Representing UMLS knowledge in an interoperable format would be easier to consume and understand for users—both humans and machines. Our research was motivated by the need to represent UMLS knowledge in an interoperable format using standards that are widely accepted and allow easy access to the knowledge without the need to understand its inner working. As such, the research developed a terminology server that allows easy access to UMLS knowledge structured using the FHIR standard. The UMLS FHIR terminology server, along with its API, acts as a semantic facade that hides the composite intricacies related to UMLS. As the UMLS knowledge is structured using the FHIR standard, a widely adopted standard in the industry, the users are immune to UMLS’s native knowledge structure and changes. The users need to know the FHIR standard, resources, and its features to use the terminology server. For any user in healthcare, being popular with the ability to represent a variety of healthcare data in an interoperable format, and applicable across the healthcare domain, the users gain much more learning from the FHIR than the UMLS native knowledge structure. Further, any FHIR-compliant application can easily consume the UMLS knowledge structured as FHIR resources, as the application is already capable of handling FHIR’s structure data. In succession, the application developers need not write custom software modules to query UMLS and parse its knowledge to be used in the application. Even with the availability of many different healthcare standards (HL7 CDA, openEHR, etc.), FHIR is the interoperability standard, and has gained incredible support from researchers, healthcare organizations, governments, and healthcare application (EHR, PHR, etc.) vendors. This supports our choice of the standard and the goal-representing UMLS knowledge using FHIR.
There exist different terminology servers and services, such as the VOSER vocabulary server [38], the UMLS Knowledge Source Server [32], UMLS API [6], GALEN terminology server [39] and OntoServer [28], to name a few. The UMLS API and OntoServer are REST APIs, while for the others, the user needs to install and configure the software on the server. As UMLS FHIR API is a REST-based service, the applications query the endpoints programmatically, as required, and use the knowledge. As the results are formatted using FHIR resources, users can use open-source FHIR libraries, such as HAPI, to get the knowledge from the structure. With other services, except for OntoServer, users need to understand the output structure and develop custom software modules to parse the output structure to get the knowledge. The FHIR standard integrates the REST specification into the standard. The FHIR specification presents detailed guidelines on how to manage FHIR resources via REST operation (read, update, delete, etc.), operation responses, naming style, operation return type, etc. Thus, any FHIR implementation (e.g., HAPI library, etc.) must follow the guidelines, and all the users must have a mutual understanding of how to access the resources. FHIR specification has standardized the REST API design. No other healthcare standard has this feature, and it is the responsibility of the developers to design the API specification for that terminology server and service, which is a very concerning drawback. Most of the existing terminological services cause vendor lock-in. If a healthcare application currently using, for example, UMLS API wants to move to another service, say OntoServer, then the developers must reconfigure the application to work with the new service. This can be a time-consuming and expensive process. Using a terminology service compliant with FHIR, such as UMLS FHIR API, prevents vendor lock-ins, as any new FHIR-compliant terminology services can replace the existing one. This is a great benefit to the application and its stakeholders. Another hurdle to using a terminology is the value it provides as compared to the cost, as terminologies come with various technical challenges. One of them is terminology updates. An updated terminology allows users to access the newest semantic knowledge. As UMLS FHIR is a service, the users need not worry about UMLS terminology updates.
8. Conclusions
In this research, we have identified key challenges with UMLS and proposed an interoperable solution that uses the HL7 FHIR standard. Leveraging FHIR resources, extensions, and integrated support to REST, this research has designed and implemented a light-weight terminology service, UMLS FHIR API, which expresses UMLS knowledge as a FHIR ConceptMap. However, out of the box, the FHIR ConceptMap cannot express the UMLS knowledge. FHIR ConceptMap is profiled to align it with the UMLS knowledge structure and other requirements, identified by the critical gap analysis. The HAPI library, an implementation of the FHIR standard, enabled us to implement the profiled ConceptMap to develop the UMLS FHIR terminology service. The terminology server is available at http://umls.it.ilstu.edu/umlsfhir, whereas the UML FHIR API is available at http://umls.it.ilstu.edu/umlsfhir/fhir.
Author Contributions
Conceptualization, R.S.; software, R.S.; validation, R.S., M.S. and M.H.; investigation, R.S. and M.S.; resources, M.S. and M.H.; data curation, M.S. and M.H.; writing—original draft preparation, R.S.; writing—review and editing, M.S. and M.H.; visualization, R.S.; supervision, R.S.; project administration, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Figure A1.
UMLSRelationTypeCode ValueSet in JSON format.

Figure A2.
Profiled ConceptMap as FHIR structure.
References
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.W.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012, 40, 940. [Google Scholar] [CrossRef] [PubMed]
- Amos, L.; Anderson, D.; Brody, S.; Ripple, A.; Humphreys, B.L. UMLS users and uses: A current overview. J. Am. Med. Informatics Assoc. 2020, 27, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D26–D270. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, B.L.; Lindberg, D.A.B.; Schoolman, H.M.; Barnett, G.O. The Unified Medical Language System: An Informatics Research Collaboration. J. Am. Med. Inform. Assoc. 1997, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Medicine, N.L. Of UMLS® Reference Manual; National Library of Medicine: Bethesda, MD, USA, 2009. [Google Scholar]
- NLM UMLS REST API. Available online: https://documentation.uts.nlm.nih.gov/rest/home.html (accessed on 12 November 2020).
- Saripalle, R.K. Fast health interoperability resources (FHIR): Current status in the healthcare system. Int. J. E-Health Med. Commun. 2019, 10, 76–93. [Google Scholar] [CrossRef]
- Benson, T.; Grieve, G. Principles of Health Interoperability: SNOMED CT, HL7 and FHIR, 3rd ed.; Springer: Berlin, Germany, 2016. [Google Scholar]
- HL7 Fast Healthcare Interoperability Resources Documentation. Available online: https://www.hl7.org/fhir/documentation.html (accessed on 12 November 2020).
- HL HL7 FHIR Restful API. Available online: https://www.hl7.org/fhir/http.html (accessed on 5 February 2019).
- Boone, K.W. The CDATM Book, 2011th ed.; Springer: Berlin, Germany, 2011. [Google Scholar]
- Beale, T.; Heard, S. openEHR Architecture—Architecture Overview; openEHR Foundation, 2008; Available online: https://specifications.openehr.org/releases/1.0.2/architecture/overview.pdf (accessed on 12 November 2020).
- HAPI FHIR 2020. Available online: https://hapifhir.io/hapi-fhir/ (accessed on 12 November 2020).
- Tran, L.T.; Divita, G.; Carter, M.E.; Judd, J.; Samore, M.H.; Gundlapalli, A.V. Exploiting the UMLS Metathesaurus for extracting and categorizing concepts representing signs and symptoms to anatomically related organ systems. J. Biomed. Inform. 2015, 58, 19–27. [Google Scholar] [CrossRef]
- Plaza, L.; Diaz, A. Retrieval of Similar Electronic Health Records Using UMLS Concept Graphs. In Lecture Notes in Computer Science; Hopfe, C., Rezgui, Y., Metais, E., Preece, A., Li, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6177, pp. 296–303. [Google Scholar] [CrossRef]
- Becker, M.; Bockmann, B. Extraction of UMLS Concepts Using Apache cTAKESTM for German Language. Stud. Health Technol. Inform. 2016, 223, 71–76. [Google Scholar]
- Shivade, C.; Malewadkar, P.; Fosler-Lussier, E.; Lai, A.M. Comparison of UMLS terminologies to identify risk of heart disease using clinical notes. J. Biomed. Inform. 2015, 58, S103–S110. [Google Scholar] [CrossRef]
- Divita, G.; Zeng, Q.T.; Gundlapalli, A.V.; Duvall, S.; Nebeker, J.; Samore, M.H. Sophia: A Expedient UMLS Concept Extraction Annotator. AMIA Annu. Symp. Proc. 2014, 2014, 467–476. [Google Scholar]
- Morrey, C.P.; Perl, Y.; Halper, M.; Chen, L.; Gu, H.H. A chemical specialty semantic network for the Unified Medical Language System. J. Cheminform. 2012, 4, 9. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, H.; Perl, Y.; Geller, J. Overcoming an obstacle in expanding a UMLS semantic type extent. J. Biomed. Inform. 2012, 45, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gu, H.; Perl, Y.; Halper, M.; Xu, J. Expanding the Extent of a UMLS Semantic Type via Group Neighborhood Auditing. J. Am. Med. Inform. Assoc. 2009, 16, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Uzuner, O. Semi-Supervised Learning to Identify UMLS Semantic Relations. AMIA Summits Transl. Sci. Proc. 2014, 2014, 67–75. [Google Scholar] [PubMed]
- Gu, H.; He, Z.; Wei, D.; Elhanan, G.; Chen, Y. Validating UMLS semantic type assignments using SNOMED CT semantic tags. Methods Inf. Med. 2018, 57, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Detmer, W.M.; Barnett, G.O.; Hersh, W.R. MedWeaver: Integrating decision support, literature searching, and Web exploration using the UMLS Metathesaurus. In Proceedings of the AMIA Annual Fall Symposium; American Medical Informatics Association: Bethesda, MD, USA, 1997; pp. 490–494. [Google Scholar]
- Atal, I.; Zeitoun, J.-D.; Neveol, A.; Ravaud, P.; Porcher, R.; Trinquart, L. Automatic classification of registered clinical trials towards the Global Burden of Diseases taxonomy of diseases and injuries. BMC Bioinform. 2016, 17, 392. [Google Scholar] [CrossRef] [PubMed]
- Saripalle, R.; Runyan, C.; Russell, M. Using HL7 FHIR to achieve interoperability in patient health record. J. Biomed. Inform. 2019, 94. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Kiefer, R.; Prud’hommeaux, E.; Solbrig, H.R. Building Interoperable FHIR-Based Vocabulary Mapping Services: A Case Study of OHDSI Vocabularies and Mappings. Stud. Health Technol. Inform. 2017, 245, 1327. [Google Scholar]
- Metke-Jimenez, A.; Steel, J.; Hansen, D.; Lawley, M. Ontoserver: A syndicated terminology server. J. Biomed. Semant. 2018, 9. [Google Scholar] [CrossRef]
- LONIC FHIR Terminology Server. Available online: https://loinc.org/fhir/ (accessed on 12 November 2020).
- Zong, N.; Wen, A.; Stone, D.J.; Sharma, D.K.; Wang, C.; Yu, Y.; Liu, H.; Shi, Q.; Jiang, G. Developing an FHIR-Based Computational Pipeline for Automatic Population of Case Report Forms for Colorectal Cancer Clinical Trials Using Electronic Health Records. JCO Clin. Cancer Informatics 2020, 4, 201–209. [Google Scholar] [CrossRef]
- Zong, N.; Sharma, D.K.; Yu, Y.; Egan, J.B.; Davila, J.I.; Wang, C.; Jiang, G. Developing a FHIR-based Framework for Phenome Wide Association Studies: A Case Study with A Pan-Cancer Cohort. AMIA Jt. Summits Transl. Sci. Proc. AMIA Jt. Summits Transl. Sci. 2020, 2020, 750–759. [Google Scholar]
- Bangalore, A.; Thorn, K.E.; Tilley, C.; Peters, L. The UMLS knowledge source server: An object model for delivering UMLS data. AMIA Annu. Symp. Proc. 2003, 2003, 51–55. [Google Scholar]
- UMLS Database Query Diagrams: How to Find All Information Associated with a Particular UMLS Concept. Available online: https://www.nlm.nih.gov/research/umls/implementation_resources/query_diagrams/er1.html (accessed on 12 November 2020).
- HL7 ConceptMap. Available online: http://hl7.org/implement/standards/fhir/conceptmap.html (accessed on 12 November 2020).
- HAPI Custom Structures. Available online: https://hapifhir.io/hapi-fhir/docs/model/custom_structures.html (accessed on 12 November 2020).
- HAPI REST Server Type. Available online: https://hapifhir.io/hapi-fhir/docs/server_plain/server_types.html (accessed on 12 November 2020).
- Resource Providers and Plan Providers—HAPI FHIR Documentation. Available online: https://hapifhir.io/hapi-fhir/docs/server_plain/resource_providers.html (accessed on 7 November 2020).
- Rocha, R.A.; Huff, S.M.; Haug, P.J.; Warner, H.R. Designing acontrolled medical vocabulary server: The VOSER project. Comput. Biomed. Res. 1994, 27, 472–507. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.L.; Solomon, W.D.; Nowlan, W.A.; Rush, T.W.; Zanstra, P.E.; Claassen, W.M. A Terminology Server for medical language and medical information systems. Methods Inf. Med. 1995, 34, 147–157. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).