Abstract
Short-term expression of transgenes is one of the problems frequently associated with non-viral in vivo gene transfer. To obtain experimental evidence for the design of sustainable transgene expression systems, the contribution of epigenetic modifications to the decline in transgene expression needs to be investigated. Bisulfite sequencing and reactivation by hydrodynamic injection of isotonic solution were employed to investigate methylation statues of CpG in transiently expressing plasmid, pCMV-Luc, in mouse liver after hydrodynamic delivery. The cytosines of CpGs in the promoter region of pCMV-Luc were methylated in mouse liver, but the methylation was much later than the decline in the expression. The expression from pre-methylated pCMV-Luc was insensitive to reactivation. Neither an inhibitor of DNA methylation nor an inhibitor of histone deacetylation had significant effects on transgene expression after hydrodynamic injection of pCMV-Luc. Partial hepatectomy, which reduces the transgene expression from the non-integrated vector into the genome, significantly reduced the transgene expression of human interferon γ from a long-term expressing plasmid pCpG-Huγ, suggesting that the CpG-reduced plasmid was not significantly integrated into the genomic DNA. These results indicate that the CpG-reduced plasmids achieve prolonged transgene expression without integration into the host genome, although the methylation status of CpG sequences in plasmids will not be associated with the prolonged expression.
1. Introduction
Therapeutic effects of in vivo gene therapy depend on the spacio-temporal distribution of the transgene products. The most important parameter associated with the in vivo gene transfer of secretory proteins is the temporal profile of the transgene expression or, in other words, the level and duration of the expression [1,2], because the number of transfected cells would be of less importance for secretory proteins compared with intracellular proteins. The level and duration of the expression are the results of complicated intracellular events, including transcription and translation, and the mechanisms underlying them have not fully been explored yet. The development of hydrodynamic delivery of naked plasmid DNA has almost solved the problem of low level of transgene expression as demonstrated in a comparison with adenoviral vectors [3]. Unfortunately, hydrodynamic injections are frequently associated with transient transgene expression, especially when using plasmid vectors containing cytomegalovirus (CMV) promoter, one of the strongest promoters used to date.
Short-term expression can be due to the degradation or clearance of plasmid DNA. Because plasmid DNA consists of natural phosphodiester deoxypolynucleotides, it is easily degraded by DNases. Kawabata et al. reported that naked plasmid DNA degrades in mouse blood with a half-life of less than 10 min [4]. However, plasmid DNA could exist for a long period of time once it has entered into the nucleus. It has been reported that plasmid DNA is present in the nucleus of mouse liver for at least 28 days after in vivo gene transfer even when the transgene expression from the plasmid DNA has almost ceased [5,6]. In addition, silenced transgene expression can be reactivated by several stimuli, including viral infection, ischemia reperfusion, and hydrodynamic injections, even 6 months after the original in vivo gene transfer [2,7,8,9]. Integration of plasmid DNA into the genome would also result in a sustained transgene expression [10]. These experimental results indicate that the degradation of plasmid DNA is not a major mechanism at least for the rapid decline in transgene expression observed within several days of gene transfer.
The presence of silenced or inactive plasmid DNA suggests some mechanisms that inhibit transgene expression from plasmids in the nucleus. One of the most frequently discussed mechanisms is the methylation of deoxycytidyl-deoxyguanosine (CpG) dinucleotides in plasmid DNA [11]. Yew et al. developed the GZB plasmid vector in which a synthetic CMV promoter/enhancer with no CpG sequences was used to drive cDNA [12]. In addition, we reported that a plasmid expressing murine interferon (IFN)-γ under the control of CpG-free human elongation factor (EF)-1 promoter produced a sustained level of IFN-γ compared with that with a CMV promoter after hydrodynamic injection into mice [13,14,15,16,17,18]. Another suggested mechanism includes the interaction of histones with plasmid DNA. Genomic DNA binds to histones or other nuclear proteins in the nucleus to form the nucleosome [19]. Deacetylation of histone has also been suggested to be involved in the decline of transgene expression from plasmid vectors [20]. Finally, if plasmid vectors were inserted into the genome, the expression might be controlled differently from the expression from episomal plasmid vectors.
To answer these questions, we have examined whether epigenetic modifications take place in plasmid vectors that have been delivered to mouse liver. The methylation status of the plasmids has been evaluated, and the involvement of histone deacetylation and the integration into the genome have been examined in mice.
2. Experimental Section
2.1. Mice
Four-week-old male ICR mice, approximately 20 g in body weight, were purchased from Japan SLC, Inc. (Hamamatsu, Japan), and were maintained under conventional housing conditions. The protocol for the animal experiments was approved by the Animal Experimentation Committee of Graduate School of Pharmaceutical Sciences, Kyoto University (the protocol number, #2010-17; approval date, 10 March 2010).
2.2. Plasmid DNA
pcDNA3.1 and pcDNA3 were purchased from Invitrogen (Carlsbad, CA, USA) and pCpG-mcs was obtained from InvivoGen (San Diego, CA, USA). pCMV-Luc and pCpG-Luc were constructed as previously reported [13,21]. pCpG-Huγ was constructed by inserting the BglII/NheI human IFN-γ cDNA fragment amplified by PCR into the BglII/NheI site of pCpG-mcs [22]. pCMV-Huγ was constructed by inserting the human IFN-γ fragment from pCpG-Huγ digested by StuI/NheI into the EcoRV/XbaI site of pcDNA3. The plasmids used in this study are summarized in Table 1.

Table 1.
Plasmid DNA used in this study.
| Plasmid | Promoter | Backbone | cDNA |
|---|---|---|---|
| pCMV-Luc | CMV | pcDNA3.1 | Luciferase |
| pCpG-Luc | EF1 | pCpG-mcs | Luciferase |
| pCMV-Huγ | CMV | pcDNA3 | Human IFN-γ |
| pCpG-Huγ | EF1 | pCpG-mcs | Human IFN-γ |
2.3. In Vivo Gene Transfer and Reactivation of Transgene Expression
For gene transfer to the liver, mice received a tail vein injection of plasmid DNA dissolved in a large volume of saline (8% volume of body weight) over 5 s [23]. In some cases, mice received another large volume injection of saline (8% volume of body weight) over 5 s to reactivate the silenced expression in mouse liver [2].
2.4. Luciferase Assay
At indicated periods after injection, mice were killed, and the livers were harvested and homogenized in 5 mL/g of a lysis buffer (0.1 M Tris, 0.05% TritonX-100, 2 mM EDTA, pH 7.8). The homogenates were centrifuged at 10,000× g for 10 min at 4 °C. Then, 5 μL of each supernatant was mixed with 50 μL luciferase assay buffer (Picagene, Tokyo Ink, Tokyo, Japan) and the chemiluminescence was measured with a luminometer (Lumat LB 9507, EG&G Berthold, Bad Wildbad, Germany).
2.5. Extraction of DNA
DNA was extracted from liver using a DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions.
2.6. Methylation of Cytosine Residues in Plasmid DNA
Plasmids (pCMV-Luc, pCpG-Luc) were treated with SssI CpG methylase (New England Biolabs, Ipswich, MA, USA), which methylates cytosine residues of CpG dinucleotides. Mock-methylated plasmids, which were treated in the same manner as methylated ones without addition of the methylase, were used as controls.
2.7. Bisulfite Sequencing
Methylation of plasmid DNA was determined by bisulfite sequencing [24]. Bisulfite modification was performed on samples containing 1 μg DNA (including genomic DNA and plasmid DNA) extracted from mouse liver using BisulFast® DNA modification kit (TOYOBO, Osaka, Japan) according to the manufacturer’s instructions. PCR primers were designed by changing all non-CpG cytosine residues in the sequence to thymine residues in order to amplify the bisulfite-treated DNA. Primers were selected from regions with no CpG nucleotides to amplify the methylated and unmethylated DNA equally. The following primers gave efficient and specific amplification of a part of the CMV promoter of pCMV-Luc containing 20CpGs: forward, 5′-GTTAATAGGGATTTTTTATTGA-3′; reverse, 5′-AAACTCTACTTATATAAACCTCCCACC-3′. Similarly, the following primers were used for the amplification of a part of the luciferase cDNA of pCMV-Luc containing 20 CpGs: forward, 5′-TGTTGGTGTTAATTTTATTTTTTTT-3′; reverse, 5′-CCAAAATATAACCATCCATCCTTAT-3′. Bisulfite-treated DNA was PCR-amplified using TaKaRa Ex Taq TM (Takara Bio Inc., Otsu, Japan). Cycling conditions were 5 min at 94 °C, then 40 cycles of 94 °C 30 s, 54 °C 30 s, 72 °C 30 s for the promoter part; and 5 min at 94 °C, then 40 cycles of 94 °C 30 s, 55 °C 30 s, 72 °C 30 s for the cDNA part. The PCR products were fractionated on 3% agarose gel and PCR products of the expected size were excised from the gel, purified and then cloned into pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Between 6–8 colonies that contained the insert were selected at random for each bisulfite reaction performed and sequenced to obtain a representative sampling DNA methylation pattern. The sequences of the amplified promoter and cDNA parts of pCMV-Luc were analyzed using a QUMA analysis tool (http://quma.cdb.riken.jp).
2.8. 5-aza-2’-Deoxycytidine and Trichostatin A Treatment
5-aza-2’-Deoxycytidine (5-AZA) (Sigma–Aldrich, St. Louis, MO, USA) was intraperitoneally injected into mice at a dose of 0.25 mg/kg body weight at 0, 2 and 4 days after hydrodynamic injection of pCMV-Huγ [25]. Trichostatin A (TSA) (Sigma–Aldrich) was dissolved in 50% DMSO to give a concentration of 1 mg/mL. Mice were given an intraperitoneal injection of TSA at a dose of 10 mg/kg body weight at 3 days after hydrodynamic injection of pCMV-Huγ. Control mice received an intraperitoneal injection of the same volume of the vehicle (50% DMSO).
2.9. Partial Hepatectomy
One week after hydrodynamic injection of pCpG-Huγ, mice were anesthetized with Nembutal (Dainihon Seiyaku, Osaka, Japan) and a midline incision was made on the abdomen. The left posterior lobes were excised to achieve a 37% hepatectomy, and the incision was closed with clips.
2.10. Measurement of Serum Concentration of Human IFN-γ
At indicated periods after gene transfer, 50 to 200 µL blood was collected from the tail vein. The blood samples were incubated at 4 °C for 2 h to allow clotting and then centrifuged at 8000× g to obtain serum. The concentration of human IFN-γ in the serum was analyzed by ELISA using commercial kits (Human IFN-γ Ready-SET-Go! ELISA kit, eBioscience, San Diego, CA, USA).
2.11. Statistical Analysis
Differences were statistically evaluated by Student’s t-test. The level of statistical significance was set at p < 0.05.
3. Results
3.1. Methylation Status of CMV Promoter and Luciferase cDNA of pCMV-Luc in Mouse Liver
It was observed that, on average, about 95% of cytosines were methylated in the SssI-treated plasmid DNA, whereas no cytosines were found to be methylated in the untreated one (data not shown), supporting the validity of the bisulfite sequencing method to estimate the methylation of plasmids. Figure 1a shows the methylation status of cytosine residues in the promoter region of pCMV-Luc. Few CpGs had been methylated 7 days after injection. On day 14, some of the CpGs in the region were detected to be methylated in samples isolated from two out of three mice. Several of the 6 colonies in the two mice showed methylation with a high frequency of 90%, 90% and 100%, respectively.
Then, the methylation status in the luciferase cDNA region was examined using the same DNA samples used in the above experiments (Figure 1b). The CpGs in this region were hardly methylated at least for the first 28 days after gene transfer.
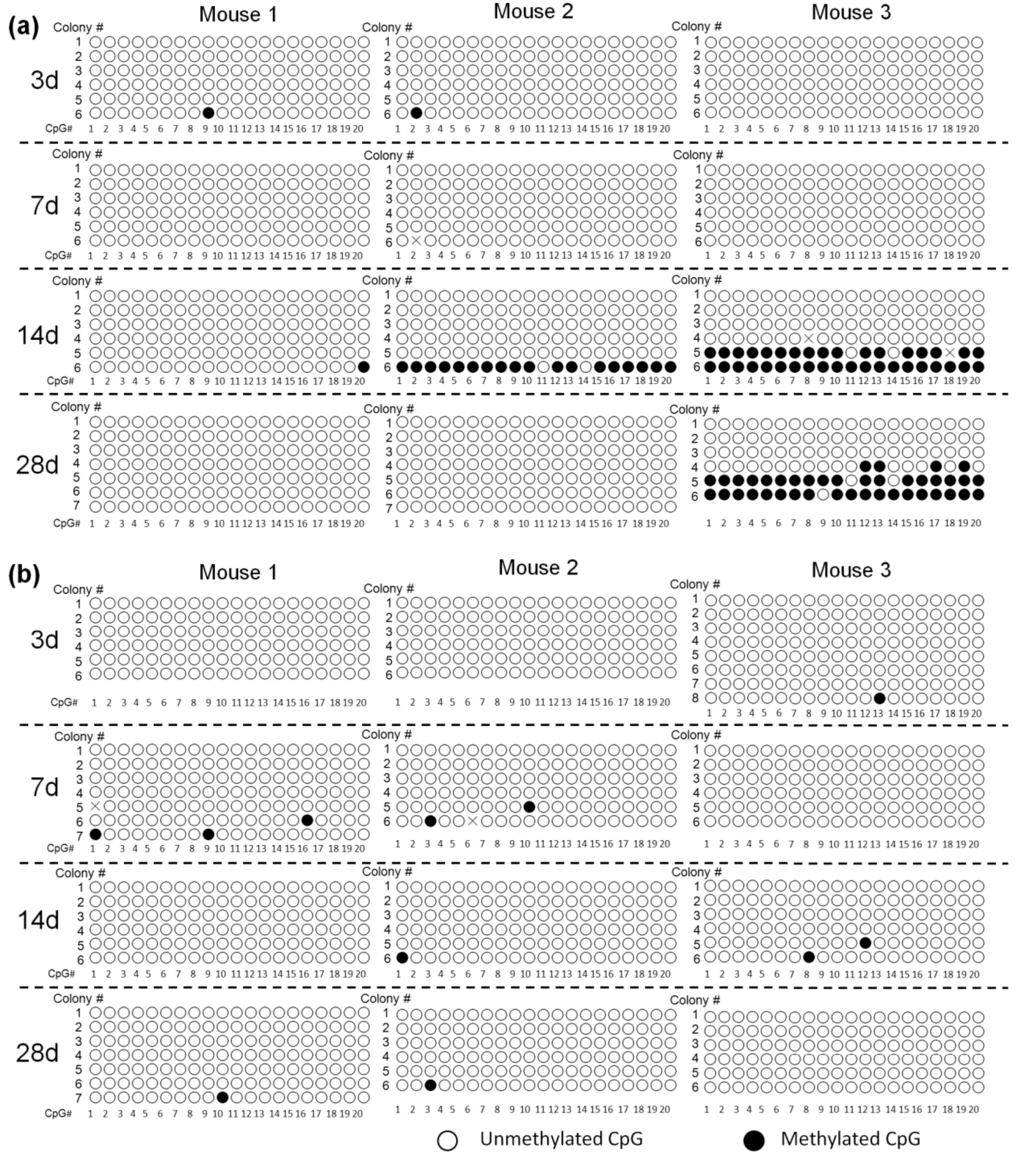
Figure 1.
Methylation status of cytomegalovirus (CMV) promoter (a) and luciferase cDNA (b) in pCMV-Luc. Mice were hydrodynamically injected with 100 μg pCMV-Luc. DNA was extracted from mouse liver on day 3, 7 14 and 28 after the hydrodynamic injection of pCMV-Luc and subjected to bisulfite sequencing. Each circle represents one CpG dinucleotide. Six to eight colonies were randomly selected from the liver samples of each mouse, and were numbered. CpG # represents the location of CpGs in the amplified promoter or cDNA parts, with #1 being the nearest to the 5′-end. The positions that could not be read were indicated with X.
3.2. Correlation of Methylation and Reactivation of Silenced Transgene Expression
It has been reported that silenced transgene expression is reactivated by hydrodynamic injections of isotonic solutions [1,2]. However, it is not clear whether the reactivation is mediated by methylated or unmethylated plasmid DNA. To further clarify the correlation of methylation and silenced transgene expression, the reactivation experiment was carried out using both methylated and unmethylated plasmids. The pCMV-Luc or pCpG-Luc in either unmethylated or methylated form was injected into mice by the hydrodynamic injection method, and a large volume of saline was injected into the mice 3 days later to reactivate the silenced transgene expression (Figure 2). The level of expression from the methylated pCMV-Luc was much lower than that of the unmethylated counterpart, and the saline injection resulted in a moderate (about 3.7-fold) increase in the expression. On the other hand, the same treatment increased the expression up to about 40-fold when the unmethylated pCMV-Luc was administered. The increase in the expression produced by the second saline injection was 3.5- and 1.9-fold for the unmethylated and methylated pCpG-Luc, respectively. The difference in the luciferase activity between unmethylated and methylated pCpG-Luc would be due to the methylation of CpGs in the luciferase cDNA.
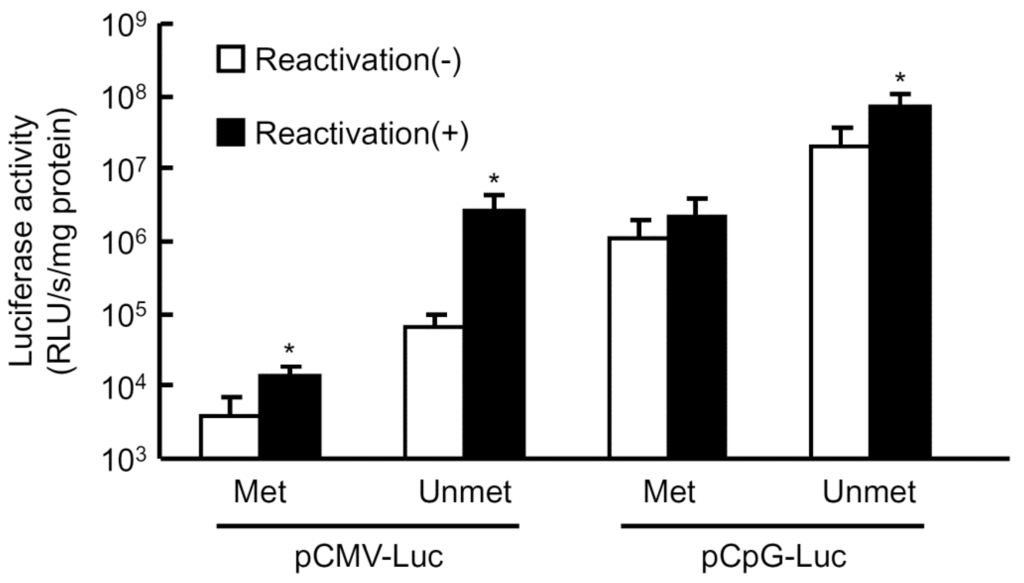
Figure 2.
Reactivation of transgene expression in mouse liver by large-volume injection of saline. Methylated or mock-methylated plasmid DNA was hydrodynamically injected into mice at a dose of 0.3 pmol/mouse. Three days later, groups of mice received a large-volume injection of saline (8% v/w of body weight) to reactivate the transgene expression in the liver, and the remaining groups were left untreated. The luciferase activity in the liver was evaluated 12 h after the saline injection. The results are expressed as the mean ± SD of four mice. * p < 0.05 compared with the untreated group.
3.3. Effect of 5-AZA on the Transgene Expression of pCMV-Huγ
The compound, 5-AZA, an inhibitor of DNA methylation, was employed to confirm whether plasmid DNA is methylated or not in mouse liver after hydrodynamic injection. pCMV-Huγ (a human IFN-γ expressing plasmid) was injected into mice by the hydrodynamic injection method with or without intraperitoneal injections of 5-AZA at a dose of 0.25 mg/kg every 2 days. Figure 3 shows the concentration of human IFN-γ in the mouse serum after gene transfer. There were no significant differences in the levels of human IFN-γ between the two groups, suggesting that pCMV-Huγ is hardly methylated during this period. Taken together with the results shown in Figure 1 and Figure 2, it is reasonable to conclude that plasmid DNA is hardly methylated in mouse liver after hydrodynamic injections at least for the first couple of weeks.
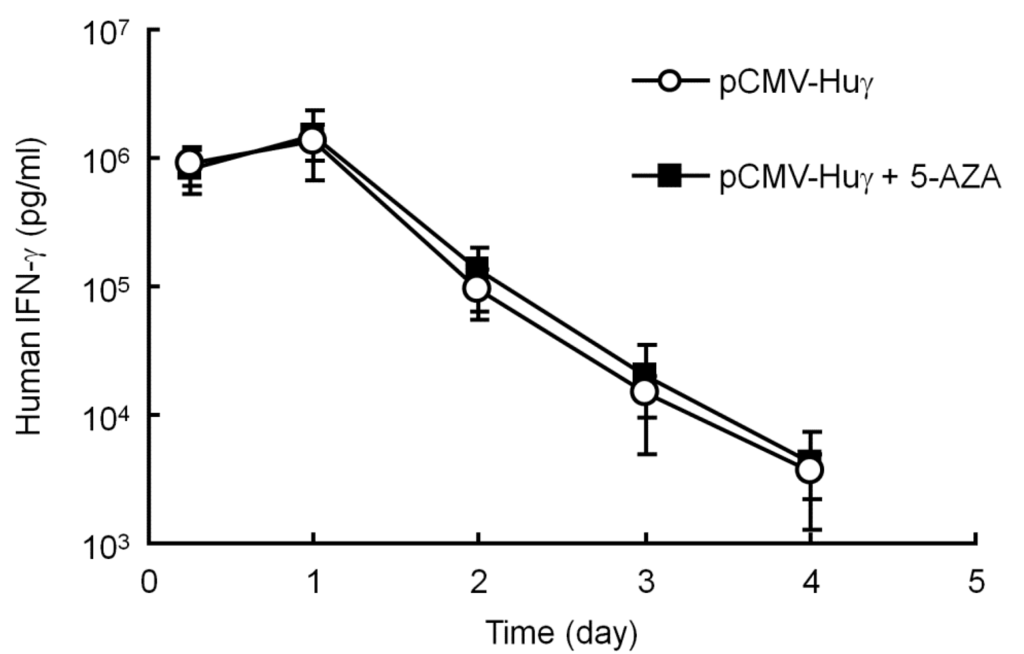
Figure 3.
Effect of 5-aza-2’-deoxycytidine (5-AZA) on the serum concentration of human interferon (IFN)-γ after injection of pCMV-Huγ. pCMV-Huγ was hydrodynamically injected into mice at a dose of 20 μg/mouse. A group of mice received intraperitoneal injections of 5-AZA at a dose of 0.25 mg/kg every 2 days. The concentrations of human IFN-γ in the serum were measured at the indicated time points. The results are expressed as the mean ± SD of four mice.
3.4. Effect of TSA on the Transgene Expression of pCMV-Huγ
To test the possibility of the involvement of histone deacetylation in silenced transgene expression, TSA, a histone deacetylase (HDAC) inhibitor, was used, as described in a previous report [26]. In the present study, TSA was injected into the peritoneal cavity of mice 3 days after hydrodynamic injection of pCMV-Huγ, when the transgene expression had dramatically decreased. The administration of TSA had no significant effects on the levels of human IFN-γ (Figure 4). These results exclude the possibility that histone deacetylation is involved in the silencing of the transgene expression of plasmids.
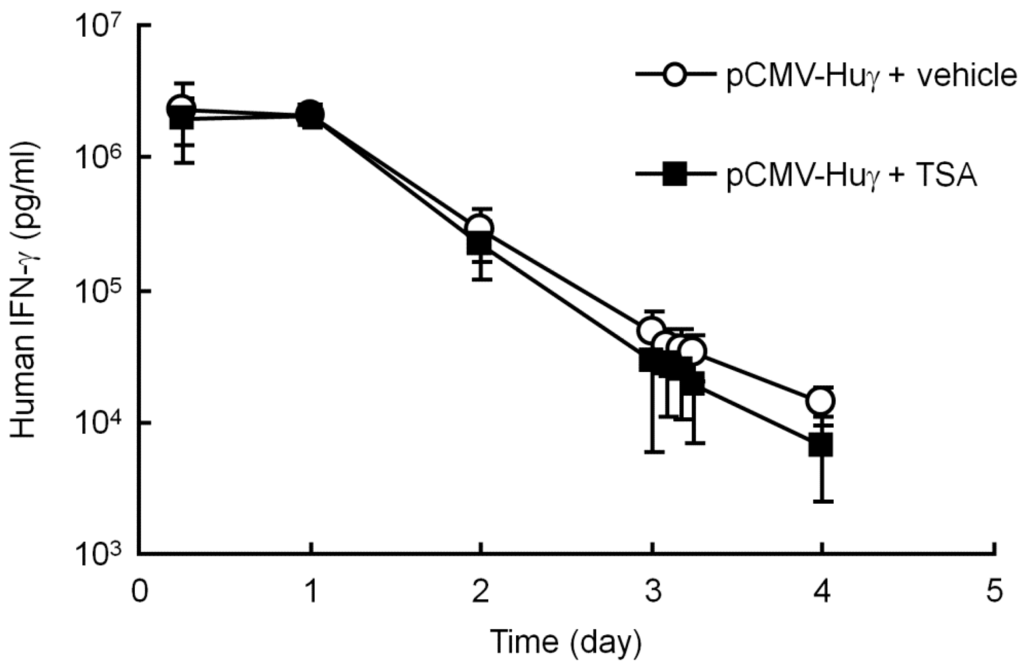
Figure 4.
Effect of trichostatin A (TSA) on the serum concentration of human IFN-γ after injection of pCMV-Huγ. pCMV-Huγ was hydrodynamically injected into mice at a dose of 20 μg/mouse. On day 3, mice received an intraperitoneal injection of vehicle or TSA at a dose of 10 mg/kg. The concentrations of human IFN-γ in the serum were measured at the indicated time points. The results are expressed as the mean ± SD of four mice.
3.5. Effect of Partial Hepatectomy on the Transgene Expression of pCpG-Huγ
Partial hepatectomy induces the proliferation of hepatocytes, so that the size of the organ returns to normal within 21 days after the surgery [27]. This procedure has been used to discriminate between integrated and episomal DNA, because the plasmid integrated into the genome is replicated with the proliferation of hepatocytes. Therefore, this technique was used to see whether the pCpG vectors, the persistent expressing plasmid DNA, are integrated into the genome after hydrodynamic injection. About 37% of the liver was removed 7 days after the hydrodynamic injection of pCpG-Huγ. Figure 5 shows the concentration of human IFN-γ in the serum of mice with or without partial hepatectomy. The concentration in the hepatectomy group transiently increased soon after the partial hepatectomy, then decreased with time to a level much lower than that in the control group. The transgene expression was about 10% to 20% of that in the control mice at a steady-state 2 weeks after the partial hepatectomy. These results indicate that pCpG vectors remain in the episomal form, even though they are effective in producing transgenes for a long period of time.
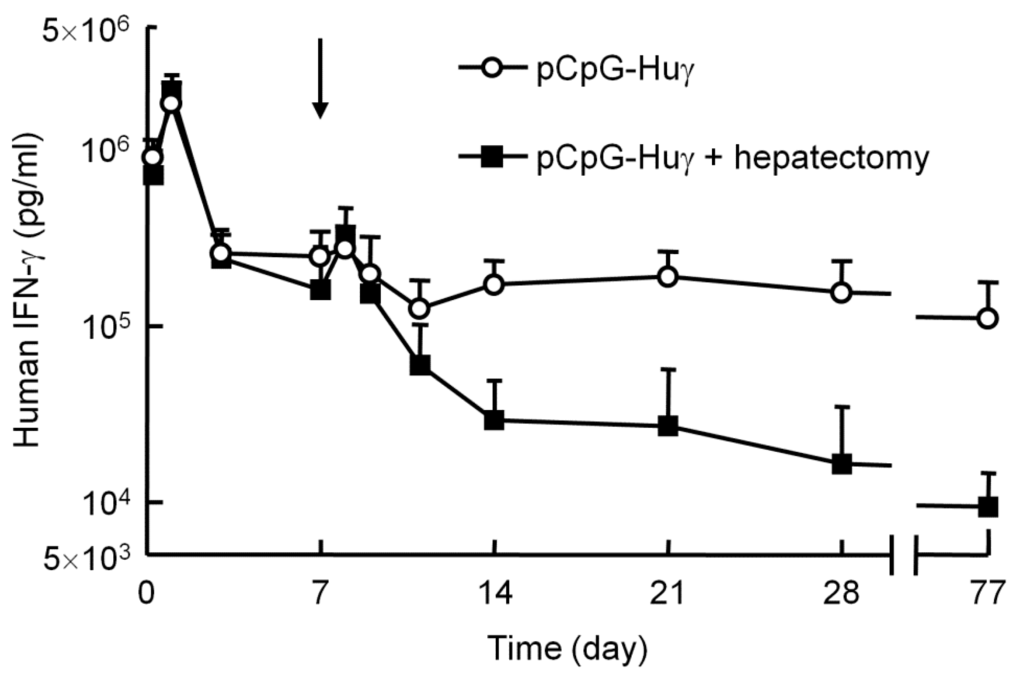
Figure 5.
Effect of partial hepatectomy on the serum concentration of human IFN-γ after injection of pCpG-Huγ. pCpG-Huγ was hydrodynamically injected into mice at a dose of 0.1 μg/mouse. On day 7, a group of mice received a partial hepatectomy (indicated with an arrow). The concentrations of human IFN-γ in the serum were measured at the indicated time points. The results are expressed as the mean + SD of four mice.
4. Discussion
The cytosines in CpG dinucleotides in mammalian DNA are mostly methylated by DNA methyltransferases. Because the methylation generally reduces the efficiency of gene expression, CpG depletion from plasmid vectors has been used to increase the duration of transgene expression [12]. We found in the present study that some CpGs in the promoter region of pCMV-Luc were methylated on and after day 14 post-injection. However, a great reduction in transgene expression of pCMV-Luc after hydrodynamic injection occurs by 3 days [13], when almost no CpG sequences were methylated (Figure 1), strongly suggesting that methylation is not the reason for the reduced transgene expression. Chen et al. also proved that CpG methylation status did not change after episomal DNA delivery into mouse liver [28]. On the other hand, we observed that the CpGs in the cDNA are less methylated than those in the promoter region, indicating that the methylation rate depends on the position of the CpGs.
Several research groups, including our own, have reported that the level of transgene expression from plasmid DNA can be significantly upregulated by some stimuli, including, but not limited to, large-volume injection of isotonic solutions, endotoxin, radiation and ischemia/reperfusion. The experiments using pre-methylated plasmid DNA showed that methylated plasmid DNA is much less sensitive to the reactivation stimulus of a large-volume injection (Figure 2). The levels of reactivated transgene expression from methylated pCMV-Luc and pCpG-Luc were lower than those from their unmethylated counterparts with no additional injections. Therefore, a significant level of reactivation of plasmid DNA by a large-volume injection of saline on day 3 (Figure 2) as well as at later time points [2] strongly suggests that pCMV-Luc and other plasmid DNA delivered by hydrodynamic injection are less methylated in mouse liver. An in vitro study reported that the intragenic CpG content influenced de novo transcriptional activity, but this regulation was not related to the level of CpG methylation [29]. Moreover, the administration of 5-AZA, an inhibitor of DNA methylation, hardly affected the level of transgene expression from pCMV-Huγ compared with that in the vehicle-treated group (Figure 3). Based on the results obtained so far, the major reason for the reduced transgene expression of CpG-replete plasmid DNA is not CpG methylation of the plasmid DNA. This is consistent with the conclusion made by Chen et al., that transcriptional activity of the transgene expression cassette was not influenced by the CpG methylation status of the plasmid bacterial backbone [28].
Some reports have raised the possibility that histone deacetylation is involved in the reduced transgene expression after gene transfer [20,29,30]. It has been reported that acetylated H3 and H4 histones in mouse liver increased after a single intraperitoneal injection of TSA at a dose of 10 mg/kg [26], and maximal gene activation occurred at approximately 2–4 h post-injection, with a return to base-line by 16 h [26,31]. However, the transgene expression from pCMV-Huγ was unaffected by administration of the same dose of TSA (Figure 4). This is consistent with the conclusion of another study, in which the relationship of histone modification and transgene expression silencing was investigated [5]. Therefore, we believe that the reduced transgene expression from hydrodynamically injected plasmid DNA was not directly caused by the deacetylation of histones.
Another possibility to explain the difference in the duration of transgene expression from CpG-replete and CpG-reduced plasmids involves the integration of plasmids into the genome. Integration of plasmid DNA into host genomic DNA has been challenged using transposon systems [10], because it is considered to increase the duration of transgene expression through integration into the host genome. It has long been known that the level of transgene expression is not significantly reduced by partial hepatectomy when the DNA delivered is integrated into genomic DNA, because hepatectomy-induced cell division increases the number of cells that have the integrated DNA [32,33]. However, the results of the present study showed that the long-expressing plasmid pCpG-Huγ is hardly integrated into genomic DNA (Figure 5).
The silencing of transgene expression from plasmid DNA should be due to complex intracellular events in which multiple factors are involved. Empirical studies have clearly demonstrated that the type of promoter, the bacterial backbone [34] and other epigenetic modifications except DNA methylation and histone deacetylation, such as histone methylation [35,36], at least partly explain the silencing of transgene expression from plasmid DNA.
5. Conclusions
We have demonstrated that the reduction in the transgene expression is not due to CpG methylation or histone deacetylation, and that genome integration is not a reason for prolonged transgene expression from CpG-reduced plasmids. These results indicate that the CpG-reduced plasmids are effective vectors that achieve prolonged transgene expression without integration into the host genome, although the CpG sequences in plasmids are less likely to be methylated in vivo.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan.
Author Contributions
Lei Zang, Makiya Nishikawa, Yuki Takahashi, and Yoshinobu Takakura designed the study, Lei Zang and Mitsuru Ando carried out the experiments. Lei Zang and Makiya Nishikawa analyzed the results. Lei Zang, Makiya Nishikawa, and Yuki Takahashi wrote the paper. All authors contributed to discussions and manuscript review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nishikawa, M.; Takakura, Y.; Hashida, M. Pharmacokinetic considerations regarding non-viral cancer gene therapy. Cancer Sci. 2008, 99, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, M.; Nakayama, A.; Takahashi, Y.; Fukuhara, Y.; Takakura, Y. Reactivation of silenced transgene expression in mouse liver by rapid, large-volume injection of isotonic solution. Hum. Gene Ther. 2008, 19, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.; Snoeys, J.; Feng, Y.; van Craeyveld, E.; Lievens, J.; Armentano, D.; Cheng, S.H.; De Geest, B. Direct comparison of hepatocyte-specific expression cassettes following adenoviral and nonviral hydrodynamic gene transfer. Gene Ther. 2008, 15, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, K.; Takakura, Y.; Hashida, M. The fate of plasmid DNA after intravenous injection in mice: Involvement of scavenger receptors in its hepatic uptake. Pharm. Res. 1995, 12, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, H.; Fujimuro, M.; Yokosawa, H.; Harashima, H.; Kamiya, H. Transient activation of transgene expression by hydrodynamics-based injection may cause rapid decrease in plasmid DNA expression. Gene Ther. 2007, 14, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Pringle, I.A.; Raman, S.; Sharp, W.W.; Cheng, S.H.; Hyde, S.C.; Gill, D.R. Detection of plasmid DNA vectors following gene transfer to the murine airways. Gene Ther. 2005, 12, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Loguercio, C.; Federico, A. Oxidative stress in viral and alcoholic hepatitis. Free Radic. Biol. Med. 2003, 34, 1–10. [Google Scholar] [CrossRef]
- Jaeschke, H. Molecular mechanisms of hepatic ischemia-reperfusion injury and preconditioning. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 284, G15–G26. [Google Scholar] [CrossRef] [PubMed]
- Takiguchi, N.; Takahashi, Y.; Nishikawa, M.; Matsui, Y.; Fukuhara, Y.; Oushiki, D.; Kiyose, K.; Hanaoka, K.; Nagano, T.; Takakura, Y. Positive correlation between the generation of reactive oxygen species and activation/reactivation of transgene expression after hydrodynamic injections into mice. Pharm. Res. 2011, 28, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Argyros, O.; Wong, S.P.; Niceta, M.; Waddington, S.N.; Howe, S.J.; Coutelle, C.; Miller, A.D.; Harbottle, R.P. Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector. Gene Ther. 2008, 15, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Higuchi, Y.; Kawakami, S.; Yamashita, F.; Hashida, M. piggyBac transposon-mediated long-term gene expression in mice. Mol. Ther. 2010, 18, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Yew, N.S.; Zhao, H.; Przybylska, M.; Wu, I.H.; Tousignant, J.D.; Scheule, R.K.; Cheng, S.H. CpG-depleted plasmid DNA vectors with enhanced safety and long-term gene expression in vivo. Mol. Ther. 2002, 5, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, M.; Nishikawa, M.; Zang, L.; Ando, M.; Hattori, K.; Takahashi, Y.; Watanabe, Y.; Takakura, Y. Effect of the content of unmethylated CpG dinucleotides in plasmid DNA on the sustainability of transgene expression. J. Gene Med. 2009, 11, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Hattori, K.; Nishikawa, M.; Watcharanurak, K.; Ikoma, A.; Kabashima, K.; Toyota, H.; Takahashi, Y.; Takahashi, R.; Watanabe, Y.; Takakura, Y. Sustained exogenous expression of therapeutic levels of IFN-γ ameliorates atopic dermatitis in NC/Nga mice via Th1 polarization. J. Immunol. 2010, 184, 2729–2735. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Takahashi, Y.; Nishikawa, M.; Watanabe, Y.; Takakura, Y. Constant and steady transgene expression of interferon-γ by optimization of plasmid construct for safe and effective interferon-γ gene therapy. J. Gene Med. 2012, 14, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Ando, M.; Nishikawa, M.; Hiraga, N.; Imamura, M.; Chayama, K.; Takakura, Y. Long-term elimination of hepatitis C virus from human hepatocyte chimeric mice after interferon-γ gene transfer. Hum. Gene Ther. Clin. Dev. 2014, 25, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, T.; Takahashi, Y.; Watcharanurak, K.; Nishikawa, M.; Ohara, S.; Ando, M.; Watanabe, Y.; Takakura, Y. Enhancement of anticancer effect of interferon-γ gene transfer against interferon-γ-resistant tumor by depletion of tumor-associated macrophages. Mol. Pharm. 2014, 11, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Watcharanurak, K.; Zang, L.; Nishikawa, M.; Yoshinaga, K.; Yamamoto, Y.; Takahashi, Y.; Ando, M.; Saito, K.; Watanabe, Y.; Takakura, Y. Effects of upregulated indoleamine 2,3-dioxygenase 1 by interferon γ gene transfer on interferon γ-mediated antitumor activity. Gene Ther. 2014, 21, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [PubMed]
- Riu, E.; Chen, Z.Y.; Xu, H.; He, C.Y.; Kay, M.A. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol. Ther. 2007, 15, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Yasuda, K.; Yamada, T.; Okamoto, S.; Mahato, R.I.; Watanabe, Y.; Takakura, Y.; Hashida, M. Gene expression and antitumor effects following direct interferon (IFN)-γ gene transfer with naked plasmid DNA and DC-chol liposome complexes in mice. Gene Ther. 1999, 6, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Zang, L.; Nishikawa, M.; Machida, K.; Ando, M.; Takahashi, Y.; Watanabe, Y.; Takakura, Y. Inhibition of nuclear delivery of plasmid DNA and transcription by interferon γ: Hurdles to be overcome for sustained gene therapy. Gene Ther. 2011, 18, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Song, Y.; Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999, 6, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C.; Guldberg, P. DNA methylation analysis techniques. Biogerontology 2003, 4, 233–250. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Low, J.A.; Daignault, S.; Imperiale, M.J.; Wojno, K.J.; Day, M.L. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006, 66, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Avila, A.M.; Burnett, B.G.; Taye, A.A.; Gabanella, F.; Knight, M.A.; Hartenstein, P.; Cizman, Z.; Di Prospero, N.A.; Pellizzoni, L.; Fischbeck, K.H.; et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Investig. 2007, 117, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Nikfarjam, M.; Malcontenti-Wilson, C.; Fanartzis, M.; Daruwalla, J.; Christophi, C. A model of partial hepatectomy in mice. J. Investig. Surg. 2004, 17, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Riu, E.; He, C.Y.; Xu, H.; Kay, M.A. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol. Ther. 2008, 16, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.P.; Leikam, D.; Krinner, S.; Notka, F.; Ludwig, C.; Längst, G.; Wagner, R. The impact of intragenic CpG content on gene expression. Nucleic Acids Res. 2010, 38, 3891–3908. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Bailey, E.C.; McCune, S.L.; Dong, J.Y.; Townes, T.M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl. Acad. Sci. USA 1997, 94, 5798–5803. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, L.; Taylor, G.W.; Aboagye, E.O.; Alao, J.P.; Latigo, J.R.; Coombes, R.C.; Vigushin, D.M. Plasma pharmacokinetics and metabolism of the histone deacetylase inhibitor trichostatin a after intraperitoneal administration to mice. Drug Metab. Dispos. 2004, 32, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Olivares, E.C.; Hollis, R.P.; Chalberg, T.W.; Meuse, L.; Kay, M.A.; Calos, M.P. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 2002, 20, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Nakai, H.; Yant, S.R.; Storm, T.A.; Fuess, S.; Meuse, L.; Kay, M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001, 75, 6969–6976. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; He, C.Y.; Meuse, L.; Kay, M.A. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004, 11, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Bannister, A.J.; Myers, F.A.; Thorne, A.W.; Crane-Robinson, C.; Kouzarides, T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004, 6, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Okitsu, C.Y.; Hsieh, C.L. DNA methylation dictates histone H3K4 methylation. Mol. Cell Biol. 2007, 27, 2746–2757. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).