Computational Analysis of S1PR1 SNPs Reveals Drug Binding Modes Relevant to Multiple Sclerosis Treatment
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
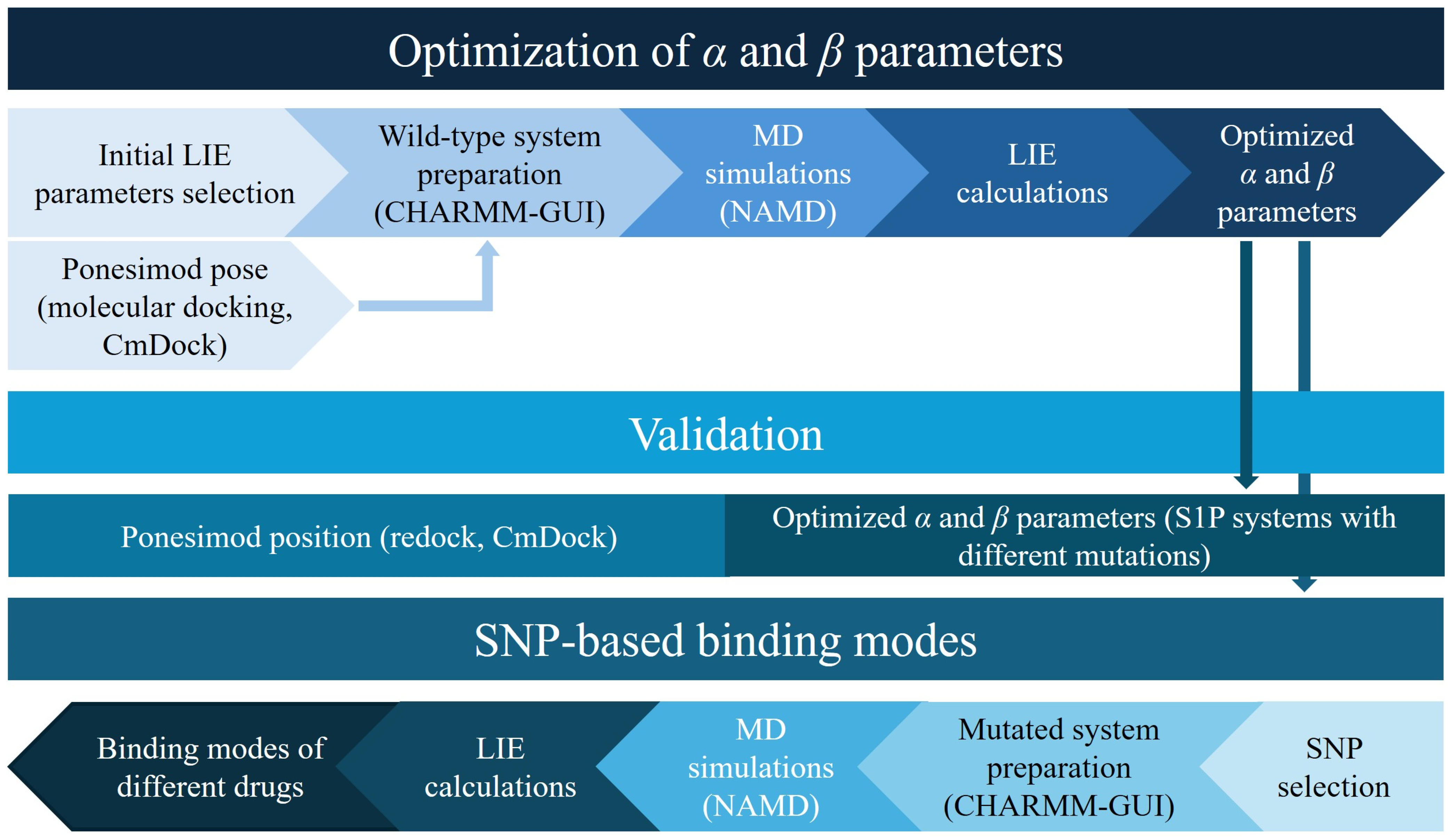
3.1. Empirical Parameter Optimization for LIE Calculations
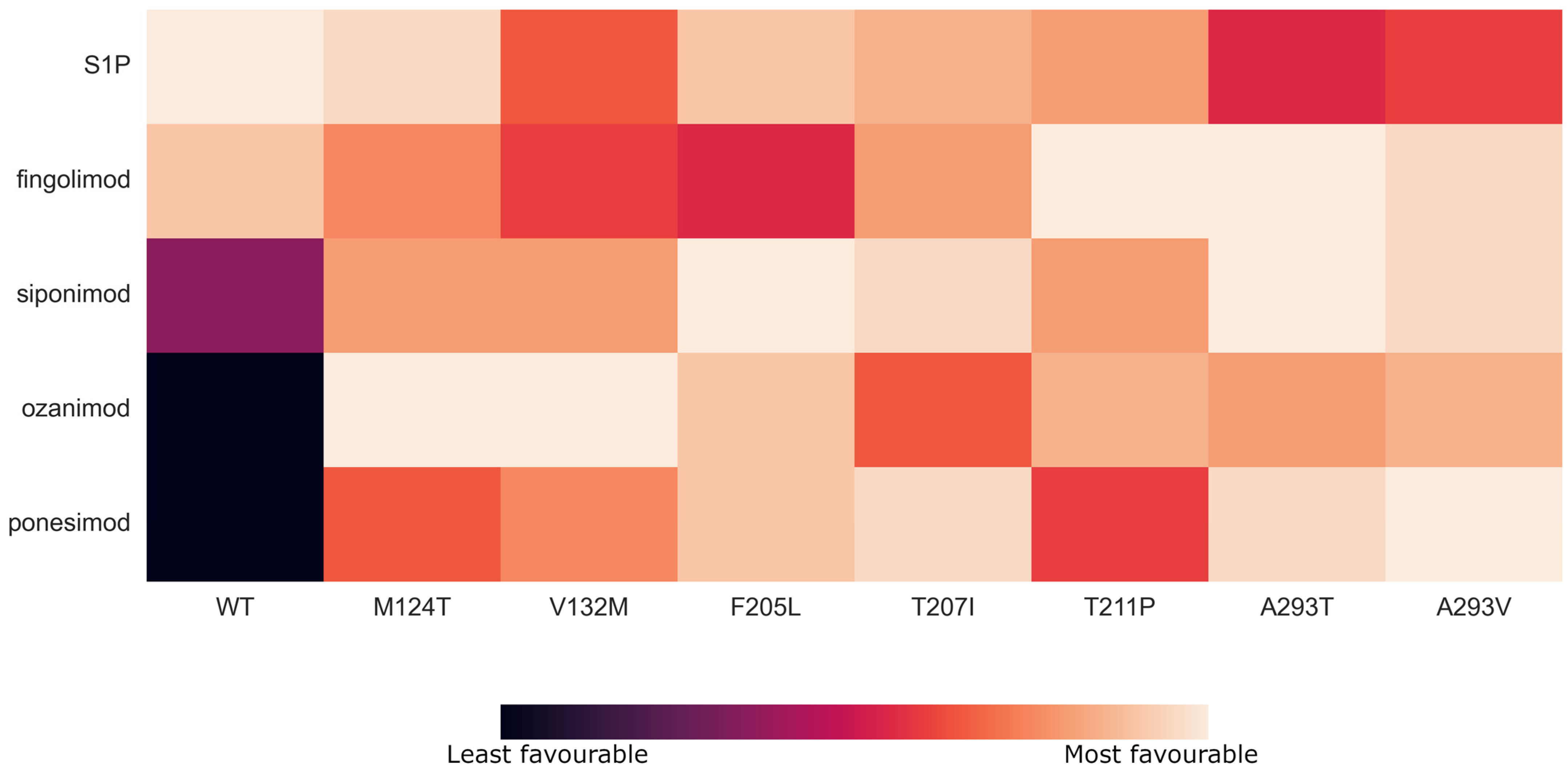
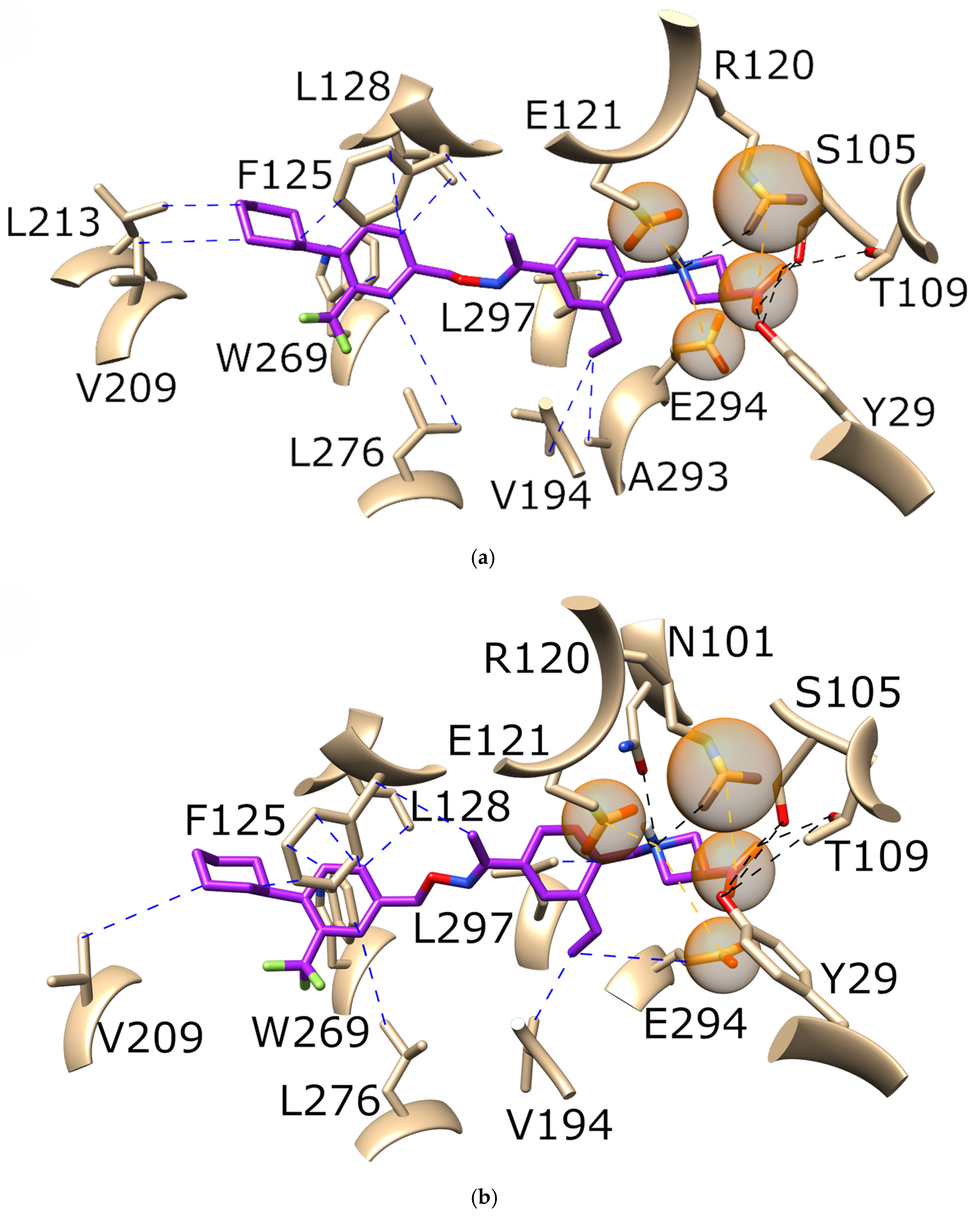
3.2. SNP-Based Binding Modes
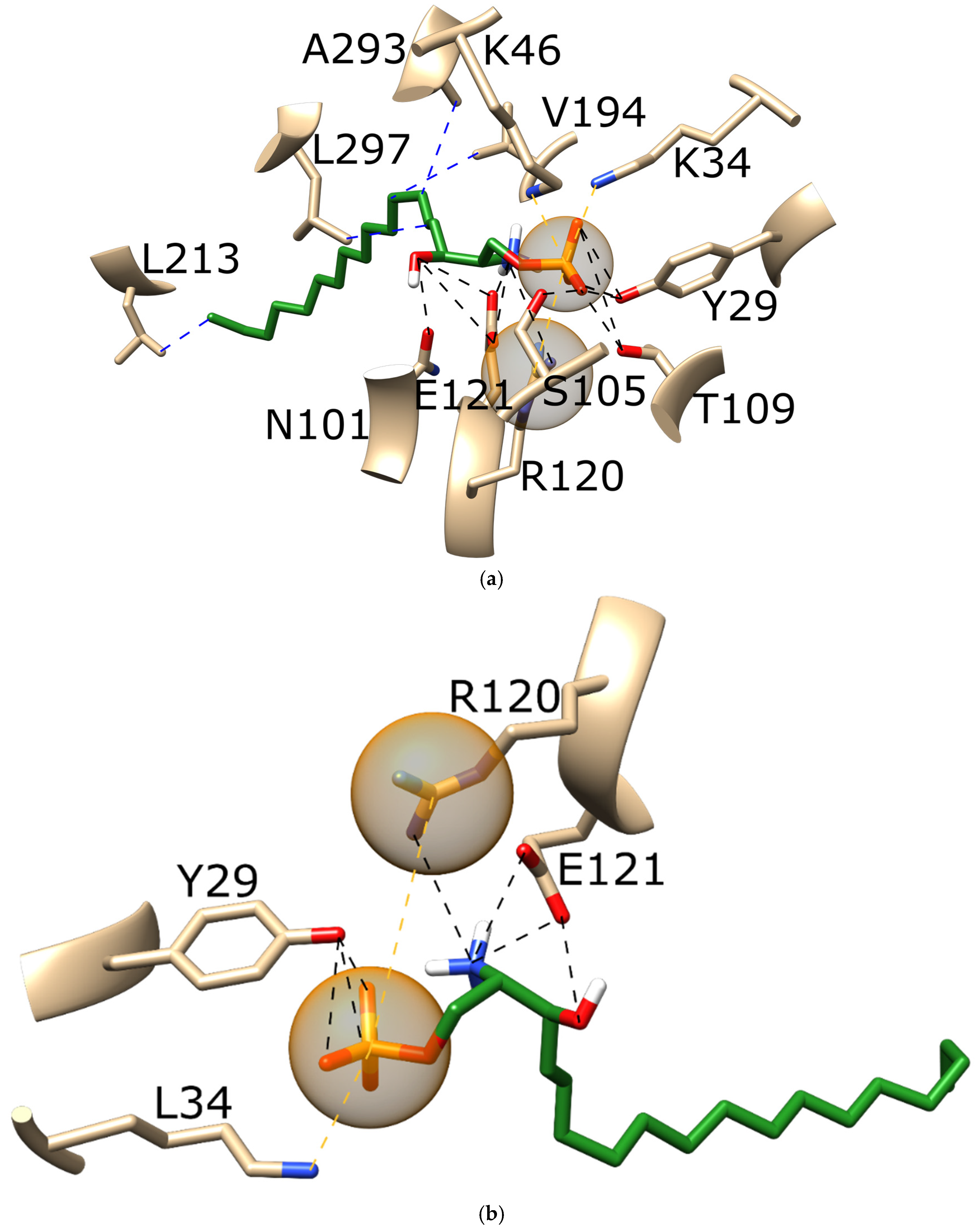
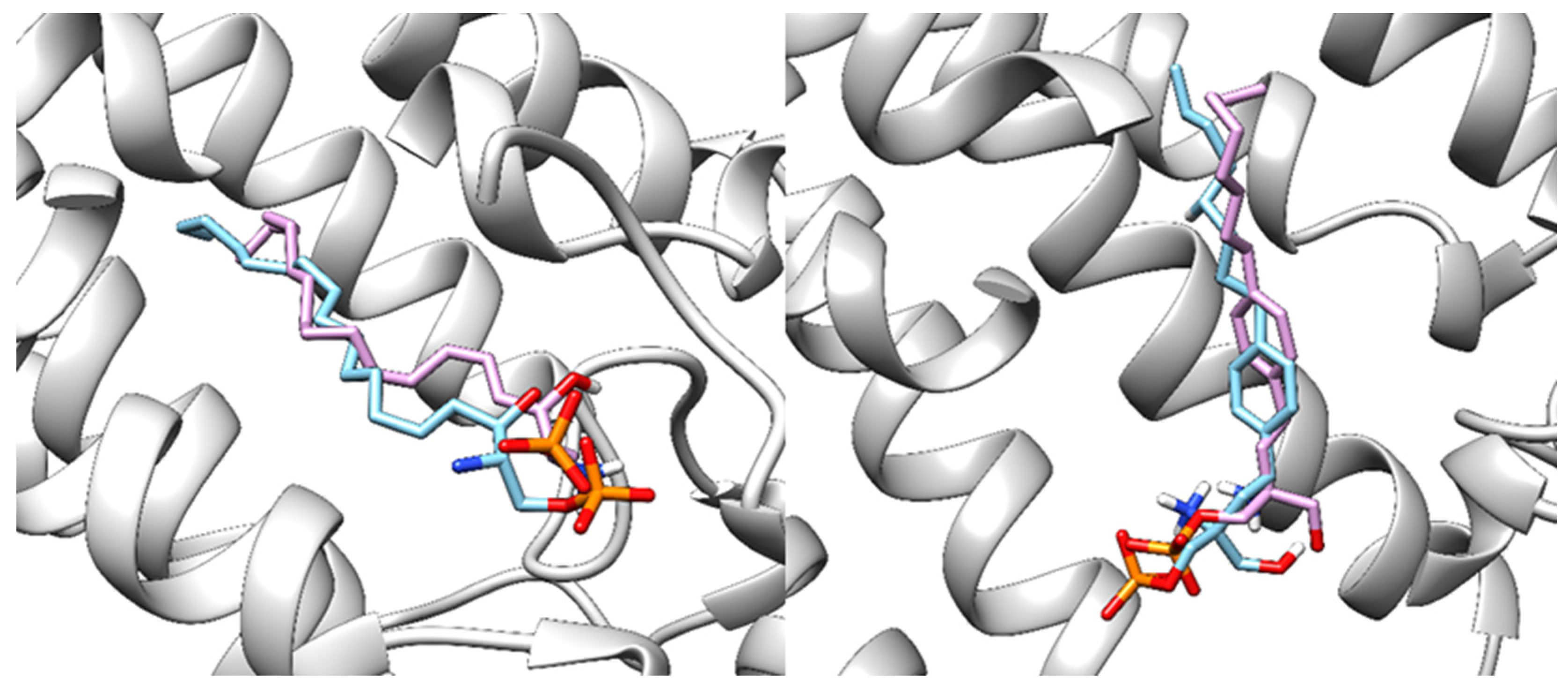
3.3. Ponesimod Pose Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Compston, A.; Coles, A. Multiple Sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, M.M. Multiple Sclerosis Review. Pharm. Ther. 2012, 37, 175–184. [Google Scholar]
- Hauser, S.L.; Cree, B.A.C. Treatment of Multiple Sclerosis: A Review. Am. J. Med. 2020, 133, 1380–1390.e2. [Google Scholar] [CrossRef] [PubMed]
- Piehl, F. Current and Emerging Disease-Modulatory Therapies and Treatment Targets for Multiple Sclerosis. J. Intern. Med. 2021, 289, 771–791. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.; Dev, K.K. The Structure and Function of the S1P1 Receptor. Trends Pharmacol. Sci. 2013, 34, 401–412. [Google Scholar] [CrossRef]
- Hla, T.; Brinkmann, V. Sphingosine 1-Phosphate (S1P). Neurology 2011, 76, S3–S8. [Google Scholar] [CrossRef]
- Wang, W.; Huang, M.-C.; Goetzl, E.J. Type 1 Sphingosine 1-Phosphate G Protein-Coupled Receptor (S1P1) Mediation of Enhanced IL-4 Generation by CD4 T Cells from S1P1 Transgenic Mice1. J. Immunol. 2007, 178, 4885–4890. [Google Scholar] [CrossRef]
- Shastry, B.S. SNPs: Impact on Gene Function and Phenotype. In Single Nucleotide Polymorphisms: Methods and Protocols; Komar, A.A., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 3–22. ISBN 978-1-60327-411-1. [Google Scholar]
- Schork, N.J.; Fallin, D.; Lanchbury, J.S. Single Nucleotide Polymorphisms and the Future of Genetic Epidemiology. Clin. Genet. 2000, 58, 250–264. [Google Scholar] [CrossRef]
- Martinez-Forero, I.; Pelaez, A.; Villoslada, P. Pharmacogenomics of Multiple Sclerosis: In Search for a Personalized Therapy. Expert Opin. Pharmacother. 2008, 9, 3053–3067. [Google Scholar] [CrossRef]
- Laing, R.E.; Hess, P.; Shen, Y.; Wang, J.; Hu, S.X. The Role and Impact of SNPs in Pharmacogenomics and Personalized Medicine. Curr. Drug Metab. 2011, 12, 460–486. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Jafari, B.; Dastmalchi, S. Drug Repurposing for Identification of S1P1 Agonists with Potential Application in Multiple Sclerosis Using In Silico Drug Design Approaches. Adv. Pharm. Bull. 2023, 13, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Selkirk, J.V.; Bortolato, A.; Yan, Y.G.; Ching, N.; Hargreaves, R. Competitive Binding of Ozanimod and Other Sphingosine 1-Phosphate Receptor Modulators at Receptor Subtypes 1 and 5. Front. Pharmacol. 2022, 13, 892097. [Google Scholar] [CrossRef] [PubMed]
- Faissner, S.; Gold, R. Efficacy and Safety of Multiple Sclerosis Drugs Approved Since 2018 and Future Developments. CNS Drugs 2022, 36, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ikuta, T.; Kawakami, K.; Kise, R.; Qian, Y.; Xia, R.; Sun, M.-X.; Zhang, A.; Guo, C.; Cai, X.-H.; et al. Structural Basis of Sphingosine-1-Phosphate Receptor 1 Activation and Biased Agonism. Nat. Chem. Biol. 2022, 18, 281–288. [Google Scholar] [CrossRef]
- Parrill, A.L.; Wang, D.; Bautista, D.L.; Brocklyn, J.R.V.; Lorincz, Z.; Fischer, D.J.; Baker, D.L.; Liliom, K.; Spiegel, S.; Tigyi, G. Identification of Edg1 Receptor Residues That Recognize Sphingosine 1-Phosphate. J. Biol. Chem. 2000, 275, 39379–39384. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Osborne, D.A.; Walker, M.D.; Wang, D.; Bautista, D.A.; Liliom, K.; Brocklyn, J.R.V.; Parrill, A.L.; Tigyi, G. Identification of the Hydrophobic Ligand Binding Pocket of the S1P1 Receptor. J. Biol. Chem. 2007, 282, 2374–2385. [Google Scholar] [CrossRef]
- Aqvist, J.; Medina, C.; Samuelsson, J.E. A New Method for Predicting Binding Affinity in Computer-Aided Drug Design. Protein Eng. 1994, 7, 385–391. [Google Scholar] [CrossRef]
- Almlöf, M.; Carlsson, J.; Åqvist, J. Improving the Accuracy of the Linear Interaction Energy Method for Solvation Free Energies. J. Chem. Theory Comput. 2007, 3, 2162–2175. [Google Scholar] [CrossRef]
- Mitrovič, M.; Patsopoulos, N.A.; Beecham, A.H.; Dankowski, T.; Goris, A.; Dubois, B.; D’hooghe, M.B.; Lemmens, R.; Damme, P.V.; Søndergaard, H.B.; et al. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell 2018, 175, 1679–1687.e7. [Google Scholar] [CrossRef]
- Everest, E.; Ahangari, M.; Uygunoglu, U.; Tutuncu, M.; Bulbul, A.; Saip, S.; Duman, T.; Sezerman, U.; Reich, D.S.; Riley, B.P.; et al. Investigating the Role of Common and Rare Variants in Multiplex Multiple Sclerosis Families Reveals an Increased Burden of Common Risk Variation. Sci. Rep. 2022, 12, 16984. [Google Scholar] [CrossRef]
- Lešnik, S.; Bren, U.; Domratcheva, T.; Bondar, A.-N. Fentanyl and the Fluorinated Fentanyl Derivative NFEPP Elicit Distinct Hydrogen-Bond Dynamics of the Opioid Receptor. J. Chem. Inf. Model. 2023, 63, 4732–4748. [Google Scholar] [CrossRef] [PubMed]
- Lešnik, S.; Bren, U. Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking. Foods 2022, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Mai, B.K.; Derreumaux, P.; Vu, V.V. Adequate Prediction for Inhibitor Affinity of Aβ40 Protofibril Using the Linear Interaction Energy Method. RSC Adv. 2019, 9, 12455–12461. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Hong, N.D.; Anh, L.H.Q.; Hiep, D.M.; Tung, N.T. Effective Estimation of the Inhibitor Affinity of HIV-1 Protease via a Modified LIE Approach. RSC Adv. 2020, 10, 7732–7739. [Google Scholar] [CrossRef] [PubMed]
- Jukič, M.; Škrlj, B.; Tomšič, G.; Pleško, S.; Podlipnik, Č.; Bren, U. Prioritisation of Compounds for 3CLpro Inhibitor Development on SARS-CoV-2 Variants. Molecules 2021, 26, 3003. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Huang, D.; Caflisch, A. Efficient Evaluation of Binding Free Energy Using Continuum Electrostatics Solvation. J. Med. Chem. 2004, 47, 5791–5797. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.J.; Neway, W.; Mills, S.G.; Hajdu, R.; Ann Keohane, C.; Rosenbach, M.; Milligan, J.; Shei, G.-J.; Chrebet, G.; Bergstrom, J.; et al. Potent S1P Receptor Agonists Replicate the Pharmacologic Actions of the Novel Immune Modulator FTY720. Bioorg. Med. Chem. Lett. 2004, 14, 3351–3355. [Google Scholar] [CrossRef]
- Saha, A.K.; Yu, X.; Lin, J.; Lobera, M.; Sharadendu, A.; Chereku, S.; Schutz, N.; Segal, D.; Marantz, Y.; McCauley, D.; et al. Benzofuran Derivatives as Potent, Orally Active S1P1 Receptor Agonists: A Preclinical Lead Molecule for MS. ACS Med. Chem. Lett. 2011, 2, 97–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bell, M.; Foley, D.; Naylor, C.; Robinson, C.; Riley, J.; Epemolu, O.; Scullion, P.; Shishikura, Y.; Katz, E.; McLean, W.H.I.; et al. Discovery of Super Soft-Drug Modulators of Sphingosine-1-Phosphate Receptor 1. Bioorg. Med. Chem. Lett. 2018, 28, 3255–3259. [Google Scholar] [CrossRef] [PubMed]
- Evindar, G.; Bernier, S.G.; Kavarana, M.J.; Doyle, E.; Lorusso, J.; Kelley, M.S.; Halley, K.; Hutchings, A.; Wright, A.D.; Saha, A.K.; et al. Synthesis and Evaluation of Alkoxy-Phenylamides and Alkoxy-Phenylimidazoles as Potent Sphingosine-1-Phosphate Receptor Subtype-1 Agonists. Bioorg. Med. Chem. Lett. 2009, 19, 369–372. [Google Scholar] [CrossRef]
- He, X.; Wang, L.; Zheng, Z.; Xiao, J.; Zhong, W.; Li, S. Novel Immunomodulators Based on an Oxazolin-2-One-4-Carboxamide Scaffold. Bioorg. Med. Chem. Lett. 2012, 22, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.J.; Lynch, C.L.; Neway, W.; Mills, S.G.; Hajdu, R.; Keohane, C.A.; Rosenbach, M.J.; Milligan, J.A.; Shei, G.-J.; Parent, S.A.; et al. A Rational Utilization of High-Throughput Screening Affords Selective, Orally Bioavailable 1-Benzyl-3-Carboxyazetidine Sphingosine-1-Phosphate-1 Receptor Agonists. J. Med. Chem. 2004, 47, 6662–6665. [Google Scholar] [CrossRef]
- Luo, Z.; Liu, H.; Yu, Y.; Gropler, R.J.; Klein, R.S.; Tu, Z. Synthesis and Evaluation of Highly Selective Quinazoline-2,4-Dione Ligands for Sphingosine-1-Phosphate Receptor 2. RSC Med. Chem. 2022, 13, 202–207. [Google Scholar] [CrossRef]
- Luo, Z.; Yue, X.; Yang, H.; Liu, H.; Klein, R.S.; Tu, Z. Design and Synthesis of Pyrazolopyridine Derivatives as Sphingosine 1-Phosphate Receptor 2 Ligands. Bioorg. Med. Chem. Lett. 2018, 28, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Rosenberg, A.; Liu, H.; Han, J. Compositions for Binding Sphingosine-1-Phosphate Receptor 1 (S1P1), Imaging of S1P1, and Methods of Use Thereof. US10676467B2, 9 June 2020. [Google Scholar]
- Rifai, E.A.; van Dijk, M.; Vermeulen, N.P.E.; Yanuar, A.; Geerke, D.P. A Comparative Linear Interaction Energy and MM/PBSA Study on SIRT1–Ligand Binding Free Energy Calculation. J. Chem. Inf. Model. 2019, 59, 4018–4033. [Google Scholar] [CrossRef] [PubMed]
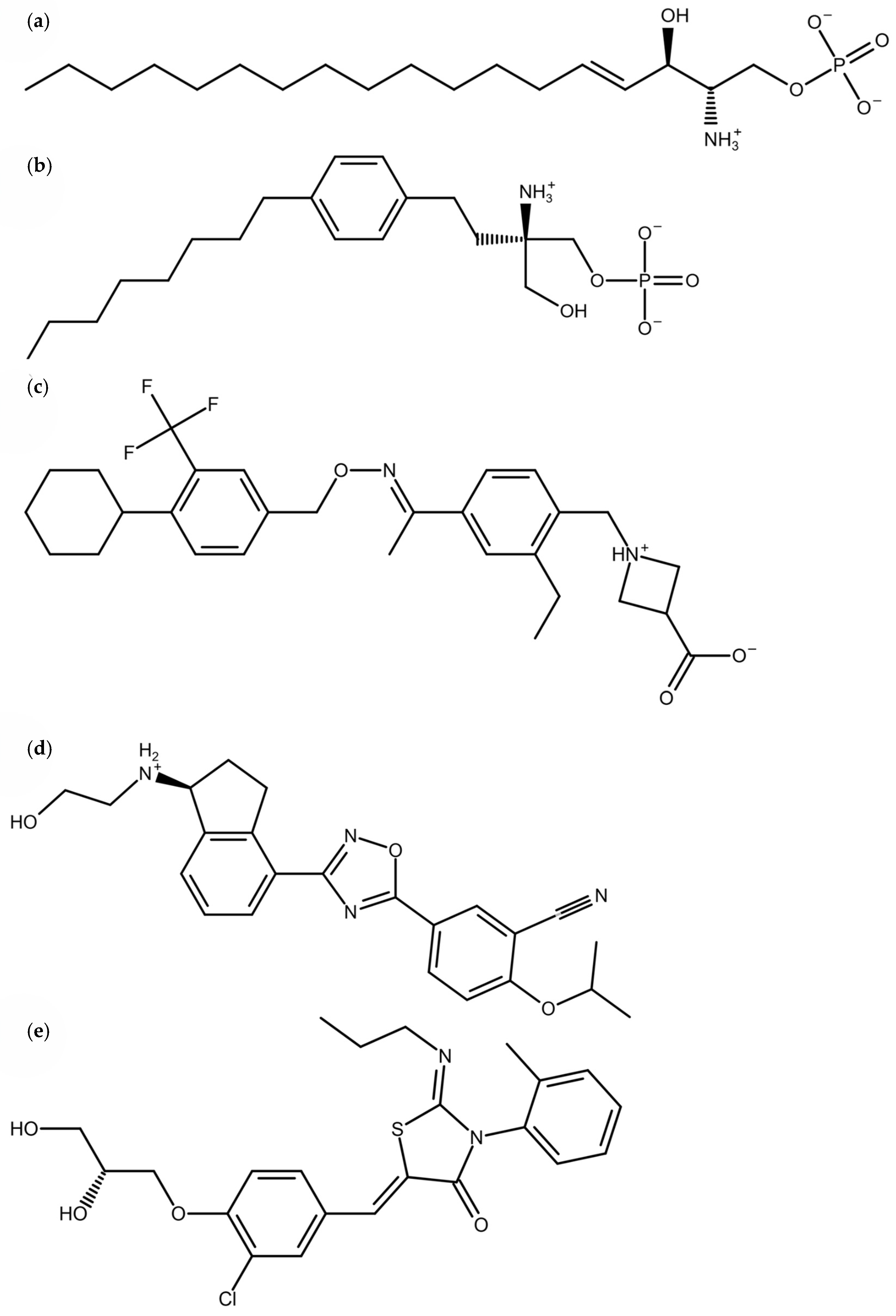

| SNPs | |||
|---|---|---|---|
| Reference SNP | Variant | Frequency | Potential Impact |
| rs1299231517 | M1243.32T | 0.000004 | possibly damaging |
| rs1323297044 | V1323.40M | 0.000004 | probably damaging |
| rs1223284736 | F2055.42L | 0.000004 | possibly damaging |
| rs1202284551 | T2075.44I | 0.00011 | benign |
| rs1209378712 | T2115.48P | 0.000008 | possibly damaging |
| rs201200746 | A2937.35T | 0.000004 | benign |
| rs1461490142 | A2937.35V | 0.000007 | benign |
| Ligand | Variant | Purpose |
|---|---|---|
| S1P | water | To obtain non-bound ligand energies (LIE calculations) |
| fingolimod | ||
| siponimod | ||
| ozanimod | ||
| ponesimod | ||
| S1P | WT | Optimization of α and β parameters (S1P, fingolimod, ponesimod), comparison of the mutation impact (all) |
| fingolimod | ||
| siponimod | ||
| ozanimod | ||
| ponesimod | ||
| S1P | N1012.60I | Validation of optimized α and β parameters |
| N1012.60K | ||
| E1213.29A | ||
| E1213.29Q | ||
| W2696.48A | ||
| W2696.48E | ||
| R2927.34A | ||
| R2927.34V | ||
| M1243.32T | Investigated SNPs | |
| S1P | V1323.40M | |
| fingolimod | F2055.42L | |
| siponimod | T2075.44I | |
| ozanimod | T2115.48P | |
| ponesimod | A2937.35T | |
| A2937.35V |
| S1P | Fingolimod Phosphate | Ponesimod | |||
|---|---|---|---|---|---|
| IC50 [M] | ΔGexp [kcal/mol] | IC50 [M] | ΔGexp [kcal/mol] | IC50 [M] | ΔGexp [kcal/mol] |
| 1.60 x 10-10 [35] | −13.90 | 2.80 x 10-10 [36] | −13.56 | 1.30 x 10-8 [37] | −11.19 |
| 4.70 x 10-10 [38] | −13.24 | 2.10 x 10-9 [39] | −12.31 | ||
| 6.70 x 10-10 [40] | −13.02 | 2.20 x 10-9 [39] | −12.29 | ||
| 1.40 x 10-9 [41] | −12.56 | ||||
| 1.40 x 10-9 [42] | −12.56 | ||||
| 1.40 x 10-9 [43] | −12.56 | ||||
| average | −12.97 | average | −12.72 | ||
| st. dev. | 0.49 | st. dev. | 0.59 | ||
| Variant | Average Binding Free Energy [kcal/mol] | Standard Deviation |
|---|---|---|
| WT | −13.76 | 1.42 |
| N1012.60I | −12.11 | 1.66 |
| N1012.60K | −13.59 | 0.84 |
| E1213.29A | −11.58 | 3.32 |
| E1213.29Q | −12.24 | 3.11 |
| W2696.48A | −11.18 | 0.94 |
| W2696.48E | −14.15 | 2.14 |
| R2927.34A | −13.48 | 1.72 |
| R2927.34V | −13.23 | 1.77 |
| Variant | S1P | Fingolimod | Siponimod | Ozanimod | Ponesimod | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Average | St. dev. | Average | St. dev. | Average | St. dev. | Average | St. dev. | Average | St. dev. | |
| WT | −13.76 | 1.42 | −11.60 | 2.00 | −14.62 | 0.37 | −8.22 | 0.54 | −11.25 | 0.40 |
| M1243.32T | −13.56 | 1.03 | −11.29 | 0.68 | −15.99 | 0.62 | −12.72 | 0.26 | −13.18 | 0.50 |
| V1323.40M | −12.82 | 0.85 | −10.95 | 0.67 | −16.12 | 1.01 | −12.76 | 0.57 | −13.47 | 0.46 |
| F2055.42L | −13.51 | 1.54 | −10.83 | 1.19 | −16.72 | 0.49 | −12.56 | 0.74 | −13.90 | 0.95 |
| T2075.44I | −13.29 | 0.98 | −11.36 | 1.00 | −16.55 | 0.55 | −11.85 | 0.54 | −14.06 | 0.32 |
| T2115.48P | −13.24 | 0.43 | −11.88 | 0.85 | −15.98 | 0.79 | −12.37 | 0.76 | −13.12 | 0.46 |
| A2937.35T | −12.50 | 1.40 | −11.87 | 0.87 | −16.71 | 0.92 | −12.31 | 0.42 | −14.09 | 0.56 |
| A2937.35V | −12.67 | 0.78 | −11.72 | 0.99 | −16.52 | 0.44 | −12.38 | 0.69 | −14.21 | 0.52 |
| S1P | Fingolimod | Siponimod | Ozanimod | ||||
|---|---|---|---|---|---|---|---|
| Energy [kJ/mol] | RMSD [Å] | Energy [kJ/mol] | RMSD [Å] | Energy [kJ/mol] | RMSD [Å] | Energy [kJ/mol] | RMSD [Å] |
| −12.52 | 3.43 | −8.40 | 4.09 | −24.00 | 1.09 | −18.45 | 1.25 |
| −11.98 | 3.32 | −8.26 | 2.71 | −22.95 | 0.97 | −18.15 | 2.61 |
| −11.54 | 2.41 | −8.14 | 2.46 | −21.07 | 1.32 | −17.70 | 1.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kores, K.; Lešnik, S.; Bren, U. Computational Analysis of S1PR1 SNPs Reveals Drug Binding Modes Relevant to Multiple Sclerosis Treatment. Pharmaceutics 2024, 16, 1413. https://doi.org/10.3390/pharmaceutics16111413
Kores K, Lešnik S, Bren U. Computational Analysis of S1PR1 SNPs Reveals Drug Binding Modes Relevant to Multiple Sclerosis Treatment. Pharmaceutics. 2024; 16(11):1413. https://doi.org/10.3390/pharmaceutics16111413
Chicago/Turabian StyleKores, Katarina, Samo Lešnik, and Urban Bren. 2024. "Computational Analysis of S1PR1 SNPs Reveals Drug Binding Modes Relevant to Multiple Sclerosis Treatment" Pharmaceutics 16, no. 11: 1413. https://doi.org/10.3390/pharmaceutics16111413
APA StyleKores, K., Lešnik, S., & Bren, U. (2024). Computational Analysis of S1PR1 SNPs Reveals Drug Binding Modes Relevant to Multiple Sclerosis Treatment. Pharmaceutics, 16(11), 1413. https://doi.org/10.3390/pharmaceutics16111413









