Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment
Abstract
:1. Introduction
2. CRISPR/Cas9-Mediated Gene Editing System
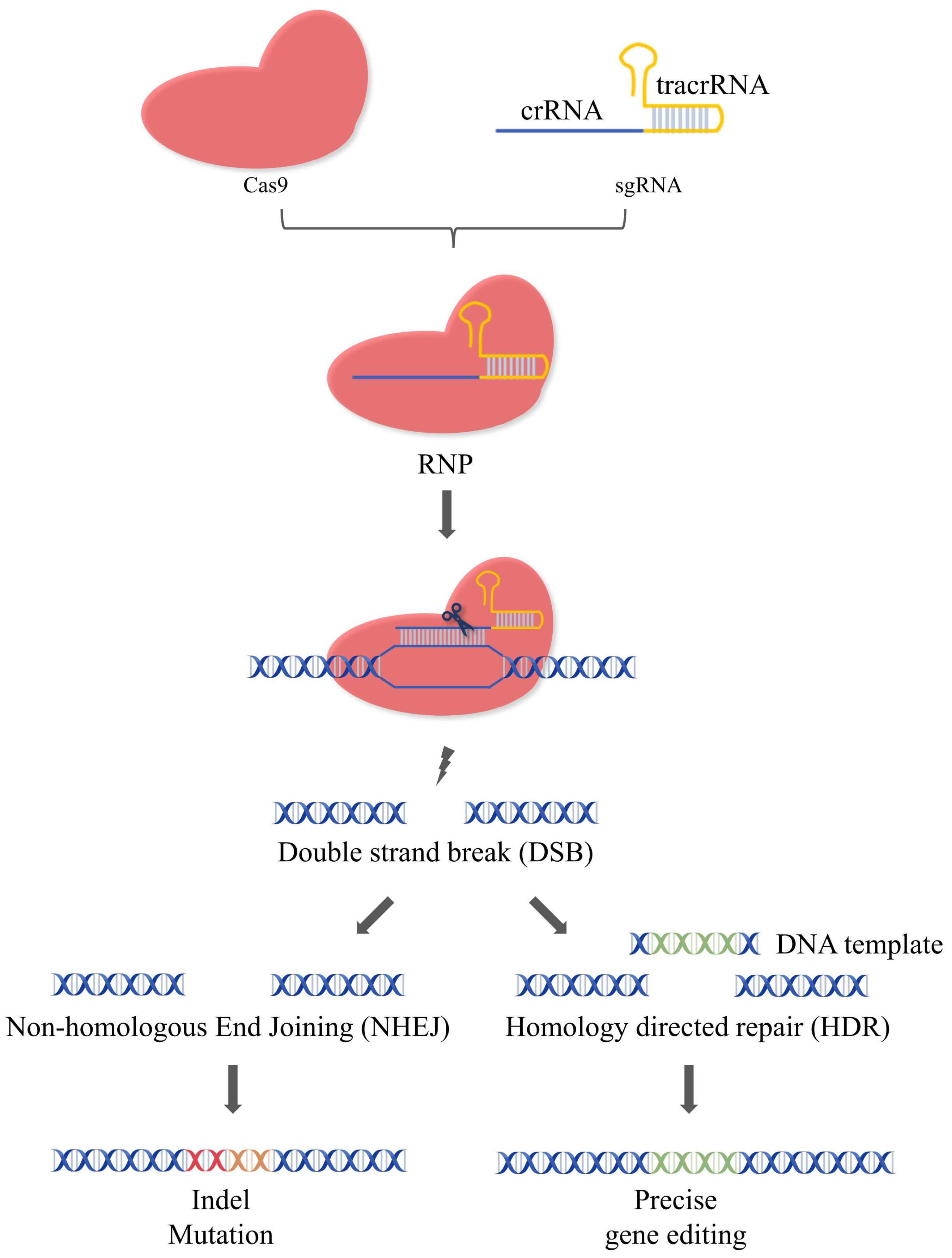
2.1. Mechanism of CRISPR/Cas9
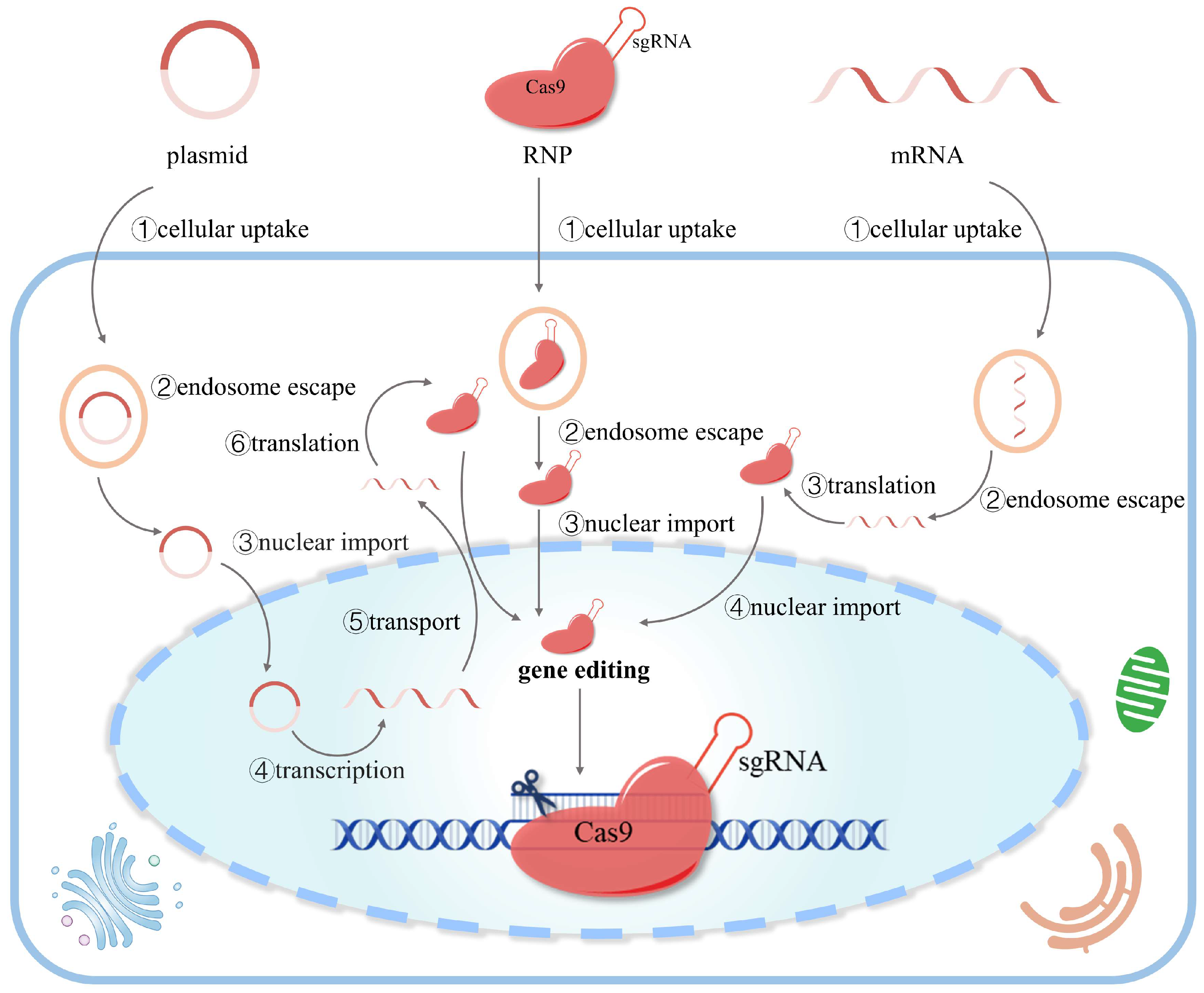
2.2. Three Forms of CRISPR/Cas9 Delivery
2.2.1. DNA (Plasmid)-Based Delivery
2.2.2. mRNA-Based Delivery
2.2.3. Protein-Based Delivery
2.3. Multiple Barriers of Delivery
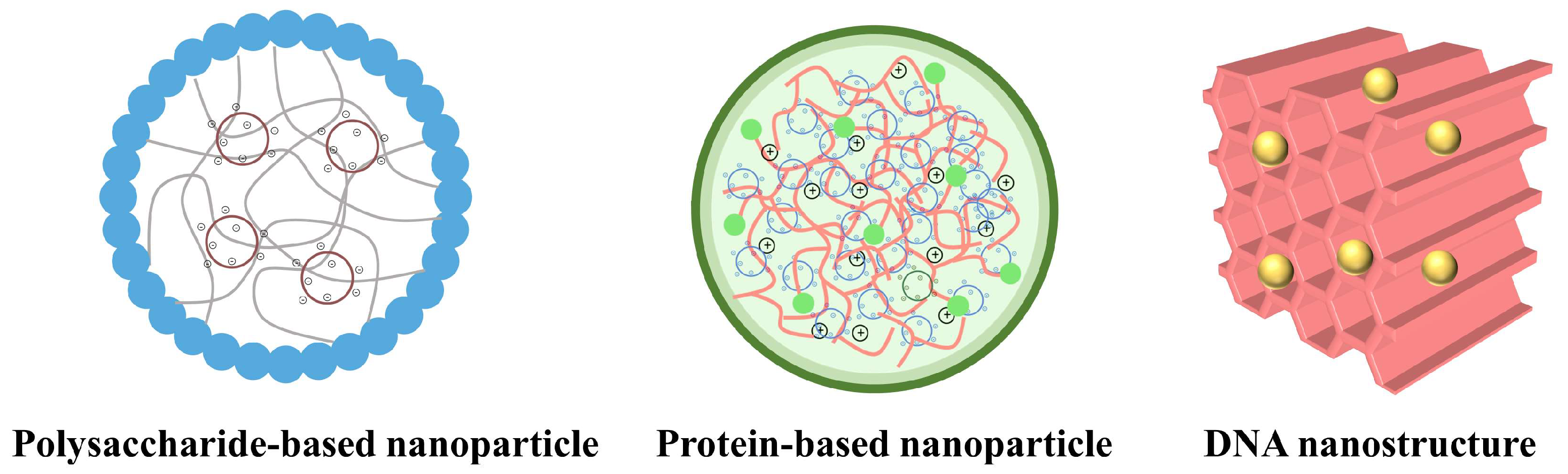
3. CRISPR/Cas9-Based Delivery Strategies
3.1. Polysaccharide-Based Delivery of CRISPR/Cas9
3.1.1. Chitosan
3.1.2. Alginate
3.1.3. Hyaluronic Acid
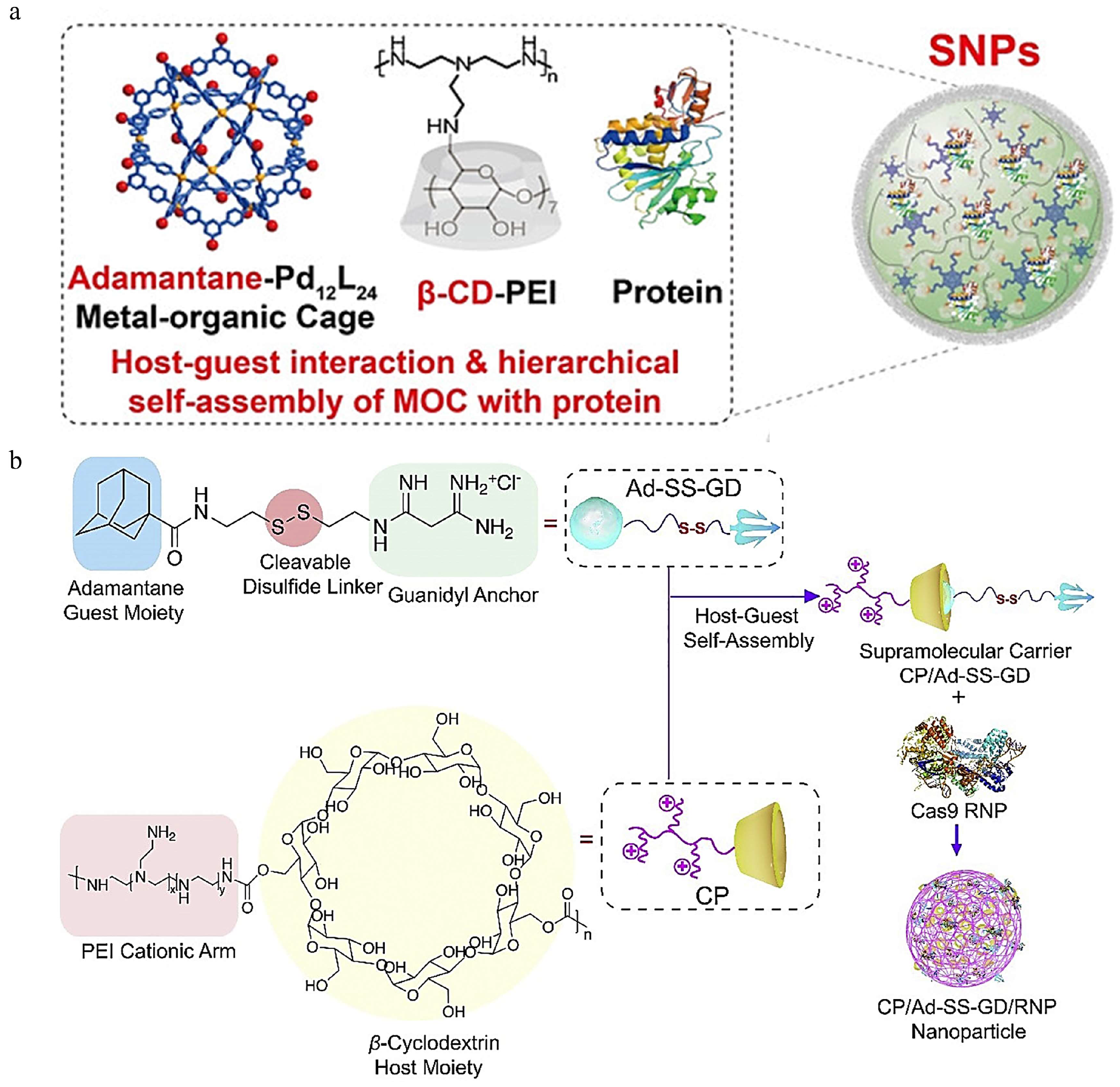
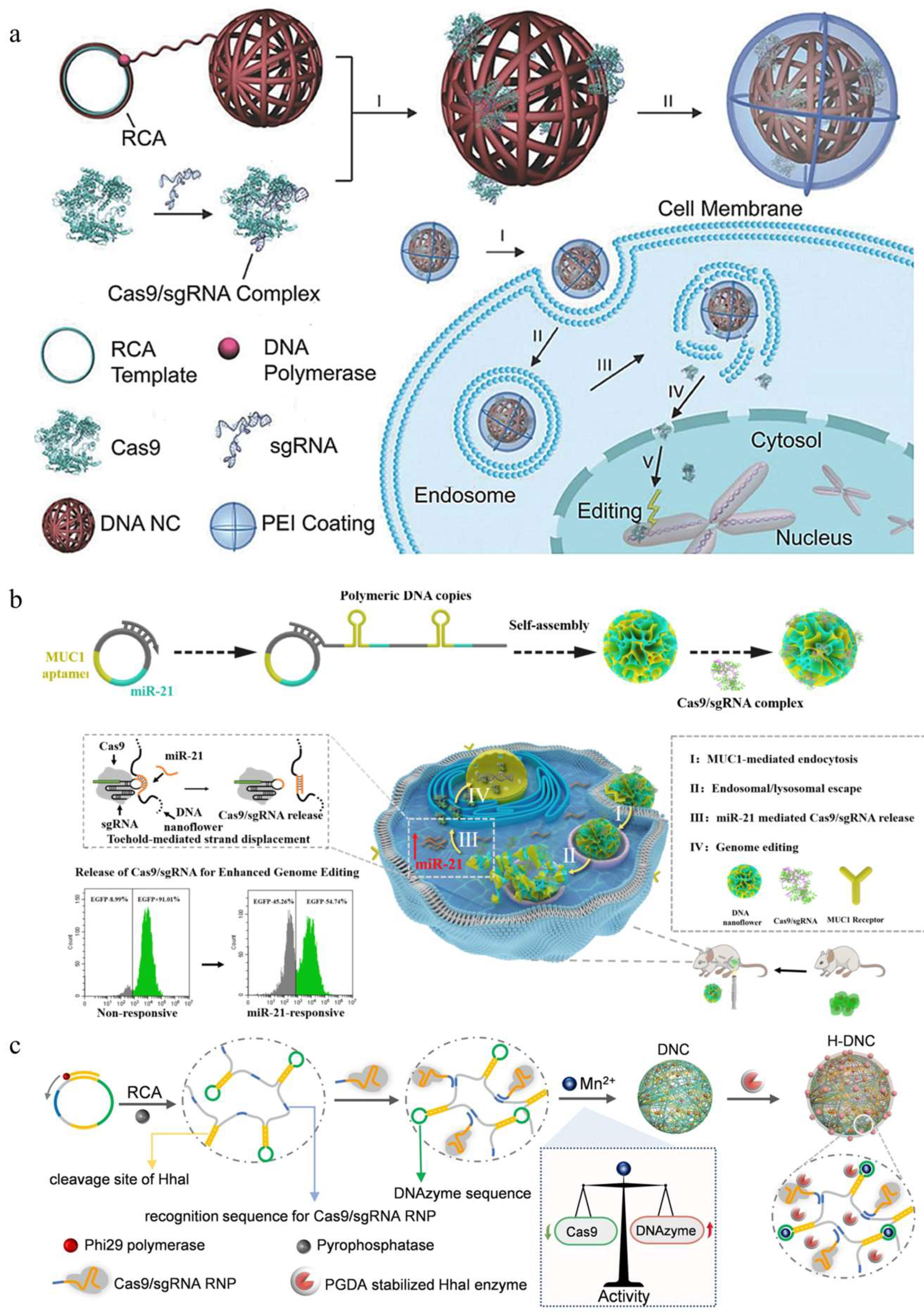
3.1.4. Cyclodextrin
3.2. Protein-Based Delivery of CRISPR/Cas9
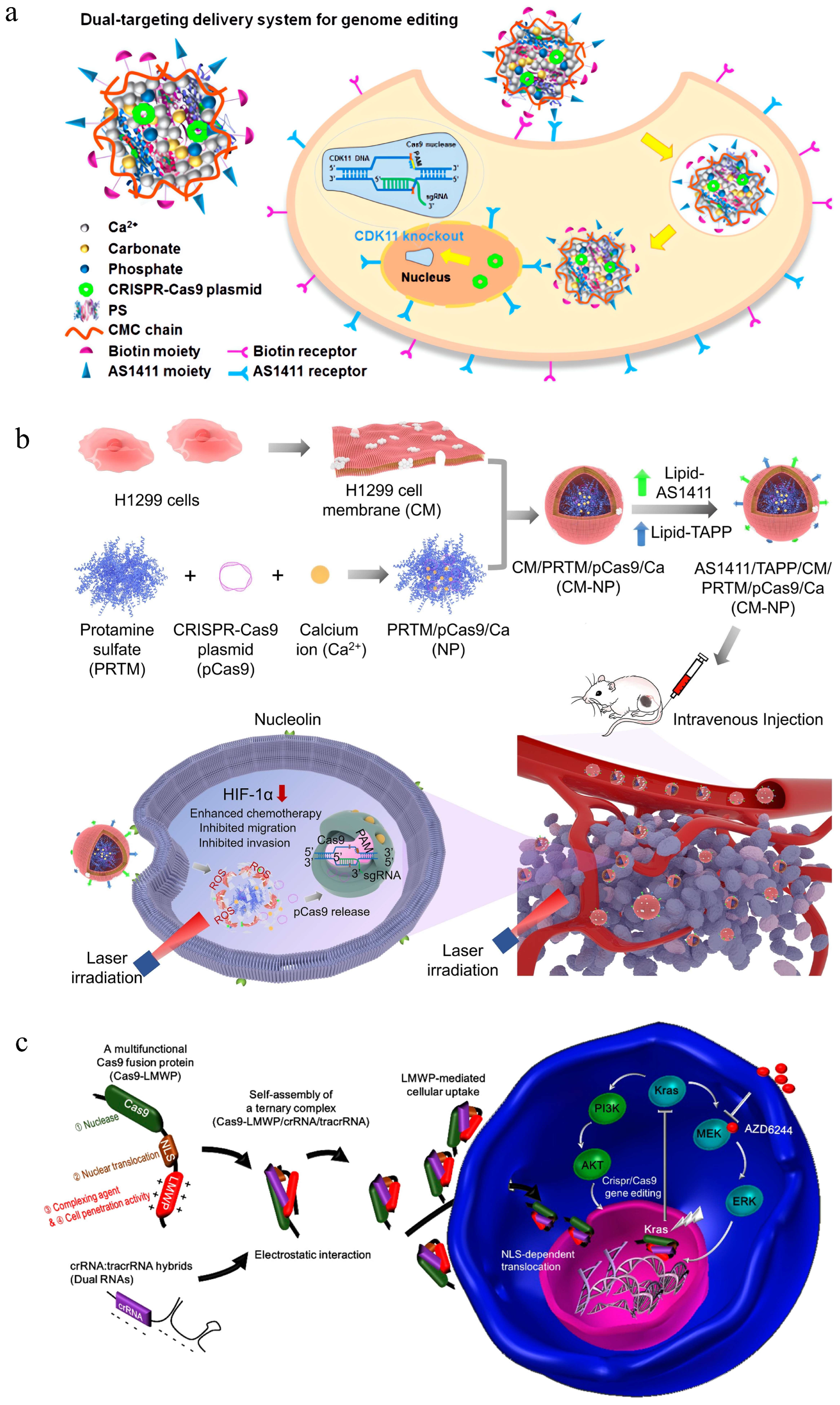
3.3. Polynucleotide-Based Delivery of CRISPR/Cas9
4. Conclusions and Outlooks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, Y.Q.; Feng, K.K.; Lu, J.Y.; Le, J.Q.; Li, W.L.; Zhang, B.C.; Li, C.L.; Song, X.H.; Tong, L.W.; Shao, J.W. CRISPR/Cas9-based application for cancer therapy: Challenges and solutions for non-viral delivery. J. Control. Release 2023, 361, 727–749. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Mao, A.; Xu, M.; Weng, Q.; Mao, J.; Ji, J. CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 2019, 447, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, C.; Wang, Y.; Koivisto, O.; Zhou, J.; Shu, Y.; Zhang, H. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv. Drug Deliv. Rev. 2021, 176, 113891. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Hazafa, A.; Mumtaz, M.; Farooq, M.F.; Bilal, S.; Chaudhry, S.N.; Firdous, M.; Naeem, H.; Ullah, M.O.; Yameen, M.; Mukhtiar, M.S.; et al. CRISPR/Cas9: A powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020, 263, 118525. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Kodama, M.; Kodama, T.; Newberg, J.Y.; Katayama, H.; Kobayashi, M.; Hanash, S.M.; Yoshihara, K.; Wei, Z.; Tien, J.C.; Rangel, R.; et al. In vivo loss-of-function screens identify kpnb1 as a new druggable oncogene in epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E7301–E7310. [Google Scholar] [CrossRef]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S.; Evan, G.I. Modelling myc inhibition as a cancer therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef]
- Li, L.; Song, L.; Liu, X.; Yang, X.; Li, X.; He, T.; Wang, N.; Yang, S.; Yu, C.; Yin, T.; et al. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 2017, 11, 95–111. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. Multiplex genome editing to generate universal car t cells resistant to pd1 inhibition. Clin. Cancer Res. 2017, 23, 2255–2266. [Google Scholar] [CrossRef]
- Weiskopf, K.; Jahchan, N.S.; Schnorr, P.J.; Cristea, S.; Ring, A.M.; Maute, R.L.; Volkmer, A.K.; Volkmer, J.P.; Liu, J.; Lim, J.S.; et al. Cd47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J. Clin. Investig. 2016, 126, 2610–2620. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Li, F.; Wang, L.; Zhang, Z.K.; Wang, C.; He, B.; Li, J.; Chen, Z.; Shaikh, A.B.; Liu, J.; et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of vegfa in osteosarcoma. Biomaterials 2017, 147, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. CRISPR gene-editing tested in a person for the first time. Nature 2016, 539, 479. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Zhang, L.; Parvin, R.; Su, L.; Chi, J.; Shi, K.; Ye, F.; Huang, X. Progress and perspective of CRISPR-Cas9 technology in translational medicine. Adv. Sci. 2023, 10, e2300195. [Google Scholar] [CrossRef] [PubMed]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered t cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef] [PubMed]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef]
- Fu, B.; Liao, J.; Chen, S.; Li, W.; Wang, Q.; Hu, J.; Yang, F.; Hsiao, S.; Jiang, Y.; Wang, L.; et al. CRISPR-Cas9-mediated gene editing of the bcl11a enhancer for pediatric β(0)/β(0) transfusion-dependent β-thalassemia. Nat. Med. 2022, 28, 1573–1580. [Google Scholar] [CrossRef]
- Ishida, K.; Gee, P.; Hotta, A. Minimizing off-target mutagenesis risks caused by programmable nucleases. Int. J. Mol. Sci. 2015, 16, 24751–24771. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (sort) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Gutkin, A.; Kedmi, R.; Ramishetti, S.; Veiga, N.; Jacobi, A.M.; Schubert, M.S.; Friedmann-Morvinski, D.; Cohen, Z.R.; Behlke, M.A.; et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020, 6, eabc9450. [Google Scholar] [CrossRef]
- Wan, T.; Zhong, J.; Pan, Q.; Zhou, T.; Ping, Y.; Liu, X. Exosome-mediated delivery of Cas9 ribonucleoprotein complexes for tissue-specific gene therapy of liver diseases. Sci. Adv. 2022, 8, eabp9435. [Google Scholar] [CrossRef]
- Yang, C.; Fu, Y.; Huang, C.; Hu, D.; Zhou, K.; Hao, Y.; Chu, B.; Yang, Y.; Qian, Z. Chlorin e6 and CRISPR-Cas9 dual-loading system with deep penetration for a synergistic tumoral photodynamic-immunotherapy. Biomaterials 2020, 255, 120194. [Google Scholar] [CrossRef]
- Yang, S.; Wu, Y.; Zhong, W.; Chen, R.; Wang, M.; Chen, M. Gsh/ph dual activatable crosslinked and fluorinated pei for cancer gene therapy through endogenous iron de-hijacking and in situ ros amplification. Adv. Mater. 2023, 35, e2304098. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Xie, Y.; Wang, N.; Tang, R.; Zheng, W.; Jiang, X. Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core-shell nanocarrier. Adv. Sci. 2017, 4, 1700175. [Google Scholar] [CrossRef]
- Song, X.; Liu, C.; Wang, N.; Huang, H.; He, S.; Gong, C.; Wei, Y. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv. Drug Deliv. Rev. 2021, 168, 158–180. [Google Scholar] [CrossRef]
- Taha, E.A.; Lee, J.; Hotta, A. Delivery of CRISPR-Cas tools for in vivo genome editing therapy: Trends and challenges. J. Control. Release 2022, 342, 345–361. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Lee, Y.W.; Scaletti, F.; Rotello, V.M. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjugate Chem. 2017, 28, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 2017, 266, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.D.; Jasin, M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000, 19, 3398–3407. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Aravalli, R.N.; Steer, C.J. CRISPR/Cas9 therapeutics for liver diseases. J. Cell. Biochem. 2018, 119, 4265–4278. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Li, J. CRISPR-Cas9 therapeutics in cancer: Promising strategies and present challenges. Biochim. Biophys. Acta 2016, 1866, 197–207. [Google Scholar] [CrossRef]
- Min, Y.L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 corrects duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef]
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; Sharp, P.A.; Jacks, T.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553. [Google Scholar] [CrossRef]
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent alteration of pcsk9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014, 115, 488–492. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Luo, B.-Y.; Zou, J.-J.; Wu, P.-Y.; Jiang, J.-L.; Le, J.-Q.; Zhao, R.-R.; Chen, L.; Shao, J.-W. Co-delivery of sorafenib and CRISPR/Cas9 based on targeted core–shell hollow mesoporous organosilica nanoparticles for synergistic hcc therapy. ACS Appl. Mater. Interfaces 2020, 12, 57362–57372. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Zhang, Y.G.; Yang, Y.H.; Ma, C.C.; Wang, P.; Du, W.; Li, L.; Xiang, R.; Song, X.R.; Zhao, X.; et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum. Gene Ther. 2018, 29, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Yang, Z.; Yang, Y.; Peng, Y.; Li, J.; Du, Y.; Sun, Q.; Gao, D.; Yuan, Q.; Zhou, Y.; et al. CRISPR-based in situ engineering tumor cells to reprogram macrophages for effective cancer immunotherapy. Nano Today 2022, 42, 101359. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the human papillomavirus e6 or e7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014, 88, 11965–11972. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting hbv DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef]
- Tong, S.; Moyo, B.; Lee, C.M.; Leong, K.; Bao, G. Engineered materials for in vivo delivery of genome-editing machinery. Nat. Rev. Mater. 2019, 4, 726–737. [Google Scholar] [CrossRef]
- Wang, H.X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.W.; Bao, G.; Leong, K.W. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Behr, M.; Zhou, J.; Xu, B.; Zhang, H. In vivo delivery of CRISPR-Cas9 therapeutics: Progress and challenges. Acta Pharm. Sin. B 2021, 11, 2150–2171. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.J.; Majors, B.S.; Zelikin, A.; Putnam, D. Structure-function relationships of gene delivery vectors in a limited polycation library. J. Control. Release 2005, 103, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Shim, M.S.; Kwon, Y.J. Stimuli-responsive polymers and nanomaterials for gene delivery and imaging applications. Adv. Drug Deliv. Rev. 2012, 64, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Gao, W.; Zhang, Y.; Yang, M.; Yan, X.; Zhang, Y.; Li, G.; Liu, C.; Xu, C.; Zhang, M. Biomimetic mof nanoparticles delivery of c-dot nanozyme and CRISPR/Cas9 system for site-specific treatment of ulcerative colitis. ACS Appl. Mater. Interfaces 2022, 14, 6358–6369. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Y.; Xin, H.; Wan, T.; Ping, Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc. Natl. Acad. Sci. USA 2020, 117, 2395–2405. [Google Scholar] [CrossRef] [PubMed]
- Dumontel, B.; Conejo-Rodríguez, V.; Vallet-Regí, M.; Manzano, M. Natural biopolymers as smart coating materials of mesoporous silica nanoparticles for drug delivery. Pharmaceutics 2023, 15, 447. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Liu, B.Y.; Peng, Y.; Zhuo, R.X.; Cheng, S.X. Multifunctional vector for delivery of genome editing plasmid targeting β-catenin to remodulate cancer cell properties. ACS Appl. Mater. Interfaces 2019, 11, 226–237. [Google Scholar] [CrossRef]
- Li, Q.; Lv, X.; Tang, C.; Yin, C. Co-delivery of doxorubicin and CRISPR/Cas9 or RNAi-expressing plasmid by chitosan-based nanoparticle for cancer therapy. Carbohydr. Polym. 2022, 287, 119315. [Google Scholar] [CrossRef]
- Song, N.; Chu, Y.; Li, S.; Dong, Y.; Fan, X.; Tang, J.; Guo, Y.; Teng, G.; Yao, C.; Yang, D. Cascade dynamic assembly/disassembly of DNA nanoframework enabling the controlled delivery of CRISPR-Cas9 system. Sci. Adv. 2023, 9, eadi3602. [Google Scholar] [CrossRef]
- Botelho da Silva, S.; Krolicka, M.; van den Broek, L.A.M.; Frissen, A.E.; Boeriu, C.G. Water-soluble chitosan derivatives and ph-responsive hydrogels by selective c-6 oxidation mediated by tempo-laccase redox system. Carbohydr. Polym. 2018, 186, 299–309. [Google Scholar] [CrossRef]
- Krishnan, R.A.; Deshmukh, P.; Agarwal, S.; Purohit, P.; Dhoble, D.; Waske, P.; Khandekar, D.; Jain, R.; Dandekar, P. Proton play in the formation of low molecular weight chitosan (lwcs) by hydrolyzing chitosan with a carbon based solid acid. Carbohydr. Polym. 2016, 151, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Rana, D.; Salave, S.; Gupta, R.; Patel, P.; Karunakaran, B.; Sharma, A.; Giri, J.; Benival, D.; Kommineni, N. Chitosan: A potential biopolymer in drug delivery and biomedical applications. Pharmaceutics 2023, 15, 1313. [Google Scholar] [CrossRef] [PubMed]
- MacLaughlin, F.C.; Mumper, R.J.; Wang, J.; Tagliaferri, J.M.; Gill, I.; Hinchcliffe, M.; Rolland, A.P. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J. Control. Release 1998, 56, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Caprifico, A.E.; Foot, P.J.S.; Polycarpou, E.; Calabrese, G. Advances in chitosan-based CRISPR/Cas9 delivery systems. Pharmaceutics 2022, 14, 1840. [Google Scholar] [CrossRef] [PubMed]
- Saranya, N.; Moorthi, A.; Saravanan, S.; Devi, M.P.; Selvamurugan, N. Chitosan and its derivatives for gene delivery. Int. J. Biol. Macromol. 2011, 48, 234–238. [Google Scholar] [CrossRef]
- Zhang, H.; Bahamondez-Canas, T.F.; Zhang, Y.; Leal, J.; Smyth, H.D.C. Pegylated chitosan for nonviral aerosol and mucosal delivery of the CRISPR/Cas9 system in vitro. Mol. Pharm. 2018, 15, 4814–4826. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, N.; Zhou, J.K.; Xu, C.; Quek, C.H.; Zhu, Y.; Tang, D.; Hung, L.Y.; Najjar, S.A.; Shiu, C.Y.A.; Margolis, K.G.; et al. Phenylboronic acid-functionalized polyplexes tailored to oral CRISPR delivery. Nano Lett. 2023, 23, 757–764. [Google Scholar] [CrossRef]
- Khademi, Z.; Ramezani, M.; Alibolandi, M.; Zirak, M.R.; Salmasi, Z.; Abnous, K.; Taghdisi, S.M. A novel dual-targeting delivery system for specific delivery of CRISPR/Cas9 using hyaluronic acid, chitosan and as1411. Carbohydr. Polym. 2022, 292, 119691. [Google Scholar] [CrossRef]
- Liu, B.Y.; He, X.Y.; Zhuo, R.X.; Cheng, S.X. Tumor targeted genome editing mediated by a multi-functional gene vector for regulating cell behaviors. J. Control. Release 2018, 291, 90–98. [Google Scholar] [CrossRef]
- Qiao, J.; Sun, W.; Lin, S.; Jin, R.; Ma, L.; Liu, Y. Cytosolic delivery of CRISPR/Cas9 ribonucleoproteins for genome editing using chitosan-coated red fluorescent protein. Chem. Commun. 2019, 55, 4707–4710. [Google Scholar] [CrossRef]
- Karim, A.; Rehman, A.; Feng, J.; Noreen, A.; Assadpour, E.; Kharazmi, M.S.; Lianfu, Z.; Jafari, S.M. Alginate-based nanocarriers for the delivery and controlled-release of bioactive compounds. Adv. Colloid Interface Sci. 2022, 307, 102744. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Mooney, D.J. Alginate: Properties and biomedical applications. Prog. Polym. Sci. 2012, 37, 106–126. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.H.; Xu, C.; Li, L.L.; Zuo, Y.; Han, D.; He, X.Y.; Cheng, S.X. A targeting delivery system for effective genome editing in leukemia cells to reverse malignancy. J. Control. Release 2022, 343, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.R.; Chen, P.; He, X.; Hei, M.W.; Zhang, J.; Yu, X.Q. Sodium alginate-doping cationic nanoparticle as dual gene delivery system for genetically bimodal therapy. Biomacromolecules 2022, 23, 5312–5321. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Yang, J.; Huang, J.; Auguste, D.T.; Moses, M.A. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc. Natl. Acad. Sci. USA 2019, 116, 18295–18303. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Dai, Y.; Gao, H. Development and application of hyaluronic acid in tumor targeting drug delivery. Acta Pharm. Sin. B 2019, 9, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Vogus, D.R.; Evans, M.A.; Pusuluri, A.; Barajas, A.; Zhang, M.; Krishnan, V.; Nowak, M.; Menegatti, S.; Helgeson, M.E.; Squires, T.M.; et al. A hyaluronic acid conjugate engineered to synergistically and sequentially deliver gemcitabine and doxorubicin to treat triple negative breast cancer. J. Control. Release 2017, 267, 191–202. [Google Scholar] [CrossRef]
- Chen, Z.-X.; Liu, M.-D.; Zhang, M.-K.; Wang, S.-B.; Xu, L.; Li, C.-X.; Gao, F.; Xie, B.-R.; Zhong, Z.-L.; Zhang, X.-Z. Interfering with lactate-fueled respiration for enhanced photodynamic tumor therapy by a porphyrinic mof nanoplatform. Adv. Funct. Mater. 2018, 28, 1803498. [Google Scholar] [CrossRef]
- Wang, L.; Liu, C.; Wang, X.; Ma, S.; Liu, F.; Zhang, Y.; Wang, Y.; Shen, M.; Wu, X.; Wu, Q.; et al. Tumor-specific activated nano-domino-CRISPR to amplify intrinsic oxidative and activate endogenous apoptosis for spatiotemporally specific therapy. Biomaterials 2023, 295, 122056. [Google Scholar] [CrossRef]
- Huang, X.; Ou, C.; Shu, Y.; Wang, Y.; Gong, S.; Luo, R.; Chen, S.; Wu, Q.; Gong, C. A self-sustained nanoplatform reverses trail-resistance of pancreatic cancer through coactivating of exogenous and endogenous apoptotic pathway. Biomaterials 2021, 272, 120795. [Google Scholar] [CrossRef]
- Yang, J.; Song, L.; Shen, M.; Gou, X.; Bai, L.; Wang, L.; Zhang, W.; Wu, Q.; Gong, C. Hierarchically responsive tumor-microenvironment-activated nano-artificial virus for precise exogenous and endogenous apoptosis coactivation. Adv. Funct. Mater. 2021, 31, 2104423. [Google Scholar] [CrossRef]
- Yang, S.; Ou, C.; Wang, L.; Liu, X.; Yang, J.; Wang, X.; Wang, M.; Shen, M.; Wu, Q.; Gong, C. Virus-esque nucleus-targeting nanoparticles deliver trojan plasmid for release of anti-tumor shuttle protein. J. Control. Release 2020, 320, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Z.; Shen, M.; Wang, Y.; Wang, L.; Li, J.; Yang, W.; Li, J.; Li, H.; Wang, X.; et al. Programmable unlocking nano-matryoshka-CRISPR precisely reverses immunosuppression to unleash cascade amplified adaptive immune response. Adv. Sci. 2021, 8, 2100292. [Google Scholar] [CrossRef] [PubMed]
- Ortiz Mellet, C.; García Fernández, J.M.; Benito, J.M. Cyclodextrin-based gene delivery systems. Chem. Soc. Rev. 2011, 40, 1586–1608. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, P.X. Cyclodextrin-based supramolecular systems for drug delivery: Recent progress and future perspective. Adv. Drug Deliv. Rev. 2013, 65, 1215–1233. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wu, M.; Fang, C.; Cheng, C.; Zhao, M.; Fang, W.; Chu, P.K.; Ping, Y.; Tang, G. Engineering nanoparticle-coated bacteria as oral DNA vaccines for cancer immunotherapy. Nano Lett. 2015, 15, 2732–2739. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.; Hu, Q.; Tang, G.; Li, J. Fgfr-targeted gene delivery mediated by supramolecular assembly between β-cyclodextrin-crosslinked pei and redox-sensitive peg. Biomaterials 2013, 34, 6482–6494. [Google Scholar] [CrossRef]
- Zhang, Z.; Wan, T.; Chen, Y.; Chen, Y.; Sun, H.; Cao, T.; Songyang, Z.; Tang, G.; Wu, C.; Ping, Y.; et al. Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid Commun. 2019, 40, e1800068. [Google Scholar] [CrossRef]
- Taharabaru, T.; Yokoyama, R.; Higashi, T.; Mohammed, A.F.A.; Inoue, M.; Maeda, Y.; Niidome, T.; Onodera, R.; Motoyama, K. Genome editing in a wide area of the brain using dendrimer-based ternary polyplexes of Cas9 ribonucleoprotein. ACS Appl. Mater. Interfaces 2020, 12, 21386–21397. [Google Scholar] [CrossRef]
- Liu, J.; Luo, T.; Xue, Y.; Mao, L.; Stang, P.J.; Wang, M. Hierarchical self-assembly of discrete metal-organic cages into supramolecular nanoparticles for intracellular protein delivery. Angew. Chem. Int. Ed. Engl. 2021, 60, 5429–5435. [Google Scholar] [CrossRef]
- Wan, T.; Chen, Y.; Pan, Q.; Xu, X.; Kang, Y.; Gao, X.; Huang, F.; Wu, C.; Ping, Y. Genome editing of mutant kras through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J. Control. Release 2020, 322, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, B.; Abdeen, A.A.; Chen, G.; Xie, R.; Saha, K.; Gong, S. Versatile redox-responsive polyplexes for the delivery of plasmid DNA, messenger RNA, and CRISPR-Cas9 genome-editing machinery. ACS Appl. Mater. Interfaces 2018, 10, 31915–31927. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Ye, J.; Liu, E.; Liang, Q.; Liu, Q.; Yang, V.C. Low molecular weight protamine (lmwp): A nontoxic protamine substitute and an effective cell-penetrating peptide. J. Control. Release 2014, 193, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.Y.; He, X.Y.; Zhuo, R.X.; Cheng, S.X. Reversal of tumor malignization and modulation of cell behaviors through genome editing mediated by a multi-functional nanovector. Nanoscale 2018, 10, 21209–21218. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yi, K.; Hu, H.; Shao, D.; Li, M. Coassembly of nucleus-targeting gold nanoclusters with CRISPR/Cas9 for simultaneous bioimaging and therapeutic genome editing. J. Mater. Chem. B 2021, 9, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Tsuchiya, A.; Tamura, M.; Yamamoto, K.; Kawata, S.; Ishihara-Sugano, M.; Kato, M.; Kitamura, T.; Goyama, S. Runx1 inhibition using lipid nanoparticle-mediated silencing RNA delivery as an effective treatment for acute leukemias. Exp. Hematol. 2022, 112, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, Y.; Zhao, X.M.; Huang, G.; Gong, J.H.; Yang, S.D.; Li, H.; Wan, W.J.; Jia, C.H.; Chen, G.; et al. A multifunctional non-viral vector for the delivery of mth1-targeted CRISPR/Cas9 system for non-small cell lung cancer therapy. Acta Biomater. 2022, 153, 481–493. [Google Scholar] [CrossRef]
- Liu, B.Y.; He, X.Y.; Xu, C.; Xu, L.; Ai, S.L.; Cheng, S.X.; Zhuo, R.X. A dual-targeting delivery system for effective genome editing and in situ detecting related protein expression in edited cells. Biomacromolecules 2018, 19, 2957–2968. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Qiao, L.; Gao, M.; Yi, X.; Peng, H.; Zhang, R.; Yao, W.; Sun, G.; He, X. Biomimetic gene editing system for precise tumor cell reprogramming and augmented tumor therapy. J. Control. Release 2023, 356, 663–677. [Google Scholar] [CrossRef]
- Kim, S.M.; Shin, S.C.; Kim, E.E.; Kim, S.H.; Park, K.; Oh, S.J.; Jang, M. Simple in vivo gene editing via direct self-assembly of Cas9 ribonucleoprotein complexes for cancer treatment. ACS Nano 2018, 12, 7750–7760. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, S.; Wang, L.; Gu, H.; Fan, C. DNA nanostructure-based systems for intelligent delivery of therapeutic oligonucleotides. Adv. Healthc. Mater. 2018, 7, e1701153. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. Engl. 2015, 54, 12029–12033. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, X.; Li, Y.; Wang, D.; Liu, W.; Zhang, Z.; Liu, J.; Zhang, K. MicroRNA-responsive release of Cas9/sgRNA from DNA nanoflower for cytosolic protein delivery and enhanced genome editing. Biomaterials 2020, 256, 120221. [Google Scholar] [CrossRef]
- Li, F.; Song, N.; Dong, Y.; Li, S.; Li, L.; Liu, Y.; Li, Z.; Yang, D. A proton-activatable DNA-based nanosystem enables co-delivery of CRISPR/Cas9 and dnazyme for combined gene therapy. Angew. Chem. Int. Ed. Engl. 2022, 61, e202116569. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Morgens, D.W.; Deans, R.M.; Li, A.; Bassik, M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016, 34, 634–636. [Google Scholar] [CrossRef]
| Types | Advantages and Disadvantages | CRISPR/Cas9 Systems | Targets | Cells | Refs. |
|---|---|---|---|---|---|
| Chitosan | Good adsorptive property Facile modification Poor solubility Low transfection efficiency | Plasmid | Survivin | 4T1 cells | [58] |
| Plasmid | PCSK9 and ANGPTL3 | HEK293T and HCT116 cells | [67] | ||
| Plasmid | FOXM1 | MCF-7, HeLa, HEK293 and SK-MES-1 cells | [68] | ||
| Plasmid | CDK11 | MCF-7 cells | [69] | ||
| RNP | PRDX4 | HEK293T, RAW264.7, HeLa, U2OS and A549 cells | [70] | ||
| Alginate | Stability Hydrogel formation in water Mucoadhesive and mucopenetrating FDA approved Low encapsulation efficiency | Plasmid | CXCR4 | THP-1 cells | [73] |
| Plasmid | Lcn2 | TNBC cells | [75] | ||
| High affinity to CD44 receptor Degraded by hyaluronidase Easy to functionalize Short lifetime | Plasmid | MTH1 | SKOV3 cells | [9] | |
| Plasmid | CD47 | B16F10 cells | [43] | ||
| Plasmid | Bcl-2 | B16F10 cells | [79] | ||
| Plasmid | PD-L1 and PTPN2 | B16F10 cells | [83] | ||
| Cyclodextrin | Facile modification Facile encapsulation of bioactive molecules Improve the stability of encapsulated cargoes Modification required for DNA condensation | Plasmid | HBB and RHBDF1 | HeLa cells | [88] |
| RNP | AAVS1 | SH-SY5Y cells | [89] | ||
| RNP | GFP | GFP-HEK293 cells | [90] | ||
| RNP | KRAS | SW-480 cells | [91] | ||
| RNP | BFP | BFP-expressing HEK293 cells | [92] | ||
| Protamine | DNA condensation ability FDA approved Inherent pharmacological activity Prone to aggregation in circulation | Plasmid | CTNNB1 | HeLa cells | [57] |
| Plasmid | FAK | HeLa cells | [94] | ||
| Plasmid | CDK11 | HEK293T and MCF-7 cells | [98] | ||
| Plasmid | PLK1 | A375, PC3, and MCF-7 cells | [99] | ||
| Plasmid | HIF-1α | H1299 cells | [100] | ||
| RNP | KRAS | A549 cells | [101] | ||
| DNA | Great stability Can be engineered with a defined structure High capacity for payloads Modification required to improve delivery efficiency | RNP | PLK1 | MCF-7 and BEAS-2B cells | [59] |
| RNP | EGFP | U2OS.EGFP cells | [103] | ||
| RNP | EGFP | HeLa.EGFP cells | [104] | ||
| RNP | PLK1 | MCF-7 cells | [105] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, M.; Wang, X. Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment. Pharmaceutics 2024, 16, 62. https://doi.org/10.3390/pharmaceutics16010062
Lin M, Wang X. Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment. Pharmaceutics. 2024; 16(1):62. https://doi.org/10.3390/pharmaceutics16010062
Chicago/Turabian StyleLin, Meng, and Xueyan Wang. 2024. "Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment" Pharmaceutics 16, no. 1: 62. https://doi.org/10.3390/pharmaceutics16010062
APA StyleLin, M., & Wang, X. (2024). Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment. Pharmaceutics, 16(1), 62. https://doi.org/10.3390/pharmaceutics16010062





