In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Retrieval
2.2. Molecular Dynamics Simulations
2.3. Trajectory Analysis
2.4. In Silico Pharmacokinetics and Toxicological Assessment
3. Results and Discussion
3.1. Data Retrieval
3.2. Molecular Dynamics Simulations
3.3. In Silico Pharmacokinetics and Toxicological Assessment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linden Junior, E.; Becker, J.; Schestatsky, P.; Rotta, F.T.; Marrone, C.D.; Gomes, I. Prevalence of Amyotrophic Lateral Sclerosis in the City of Porto Alegre, in Southern Brazil. Arq. Neuropsiquiatr. 2014, 71, 959–962. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; De Azevedo Abrahim Vieira, B.; De Mesquita, J.F. Comprehensive in Silico Analysis and Molecular Dynamics of the Superoxide Dismutase 1 (SOD1) Variants Related to Amyotrophic Lateral Sclerosis. PLoS ONE 2021, 16, e0247841. [Google Scholar] [CrossRef]
- Mancuso, R.; Navarro, X. Amyotrophic Lateral Sclerosis: Current Perspectives from Basic Research to the Clinic. Prog. Neurobiol. 2015, 133, 1–26. [Google Scholar] [CrossRef]
- Arthur, K.C.; Calvo, A.; Price, T.R.; Geiger, J.T.; Chiò, A.; Traynor, B.J. Projected Increase in Amyotrophic Lateral Sclerosis from 2015 to 2040. Nat. Commun. 2016, 7, 12408. [Google Scholar] [CrossRef]
- Ingre, C.; Roos, P.M.; Piehl, F.; Kamel, F.; Fang, F. Risk Factors for Amyotrophic Lateral Sclerosis. Clin. Epidemiol. 2015, 7, 181–193. [Google Scholar] [CrossRef]
- Gladman, M.; Dharamshi, C.; Zinman, L. Economic Burden of Amyotrophic Lateral Sclerosis: A Canadian Study of out-of-Pocket Expenses. Amyotroph. Lateral Scler. Front. Degener. 2014, 15, 426–432. [Google Scholar] [CrossRef]
- Jaiswal, M.K. Riluzole and Edaravone: A Tale of Two Amyotrophic Lateral Sclerosis Drugs. Med. Res. Rev. 2018, 39, 733–748. [Google Scholar] [CrossRef]
- Tang, L.; Ma, Y.; Liu, X.L.; Chen, L.; Fan, D.S. Better Survival in Female SOD1-Mutant Patients with ALS: A Study of SOD1-Related Natural History. Transl. Neurodegener. 2019, 8, 2. [Google Scholar] [CrossRef]
- Ahmad, A.; Singhal, U.; Hossain, M.M.; Islam, N.; Rizvi, I. The Role of the Endogenous Antioxidant Enzymes and Malondialdehyde in Essential Hypertension. J. Clin. Diagnostic Res. 2013, 7, 987–990. [Google Scholar] [CrossRef]
- Bunton-Stasyshyn, R.K.A.; Saccon, R.A.; Fratta, P.; Fisher, E.M.C. SOD1 Function and Its Implications for Amyotrophic Lateral Sclerosis Pathology: New and Renascent Themes. Neurosci. 2015, 21, 519–529. [Google Scholar] [CrossRef]
- Gill, C.; Phelan, J.P.; Hatzipetros, T.; Kidd, J.D.; Tassinari, V.R.; Levine, B.; Wang, M.Z.; Moreno, A.; Thompson, K.; Maier, M.; et al. SOD1-Positive Aggregate Accumulation in the CNS Predicts Slower Disease Progression and Increased Longevity in a Mutant SOD1 Mouse Model of ALS. Sci. Rep. 2019, 9, 6724. [Google Scholar] [CrossRef] [PubMed]
- Manjula, R.; Wright, G.S.A.; Strange, R.W.; Padmanabhan, B. Assessment of Ligand Binding at a Site Relevant to SOD1 Oxidation and Aggregation. FEBS Lett. 2018, 592, 1725–1737. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.M.; Gibbs, B.F.; Kabashi, E.; Minotti, S.; Durham, H.D.; Agar, J.N. Tryptophan 32 Potentiates Aggregation and Cytotoxicity of a Copper/Zinc Superoxide Dismutase Mutant Associated with Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2007, 282, 16329–16335. [Google Scholar] [CrossRef] [PubMed]
- Bosco, D.A.; Morfini, G.; Karabacak, N.M.; Song, Y.; Pasinelli, P.; Goolsby, H.; Fontaine, B.A.; Lemay, N.; Mckenna-yasek, D.; Frosch, M.P.; et al. Wild-Type and Mutant SOD1 Share an Aberrant Conformation and a Common Pathogenic Pathway in ALS. Nat. Neurosci. 2011, 13, 1396–1403. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 Mediated Pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef]
- Choudhury, A.R.; Cheng, T.; Phan, L.; Bryant, S.H.; Wang, Y. Supporting Precision Medicine by Data Mining across Multi-Disciplines: An Integrative Approach for Generating Comprehensive Linkages between Single Nucleotide Variants (SNVs) and Drug-Binding Sites. Bioinformatics 2017, 33, 1621–1629. [Google Scholar] [CrossRef]
- Zhang, W.; Roederer, M.W.; Chen, W.Q.; Fan, L.; Zhou, H.H. Pharmacogenetics of Drugs Withdrawn from the Market. Pharmacogenomics 2012, 13, 223–231. [Google Scholar] [CrossRef]
- Gellatly, N.; Sewell, F. Regulatory Acceptance of in Silico Approaches for the Safety Assessment of Cosmetic-Related Substances. Comput. Toxicol. 2019, 11, 82–89. [Google Scholar] [CrossRef]
- Pereira, G.R.C.; Gonçalves, L.M.; Abrahim-Vieira, D.A.B.; De Mesquita, J.F. In Silico Analyses of Acetylcholinesterase (AChE) and Its Genetic Variants in Interaction with the Anti—Alzheimer Drug Rivastigmine. J. Cell. Biochem. 2022, 123, 1259–1277. [Google Scholar] [CrossRef]
- Gonçalves, L.M.; Trevisol, E.T.V.; de Azevedo Abrahim Vieira, B.; De Mesquita, J.F. Trehalose Synthesis Inhibitor: A Molecular in Silico Drug Design. J. Cell. Biochem. 2020, 121, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Machado, T.R.; Ribeiro, M.M.J.; Sodero, A.C.R.; Domingos, T.F.S.; Rapozo, R.; de Paula, D.C.; Domingos, A.S.; Rodrigues, C.R.; Cabral, L.M.; de Souza, A.M.T.; et al. Evaluation of Chloroquine and Hydroxychloroquine as ACE-2 Inhibitors By In Silico Approaches: Cardiac Arrhythmia Cause? J. Mol. Struct. 2021, 1244, 130946. [Google Scholar] [CrossRef]
- Myatt, G.J.; Ahlberg, E.; Akahori, Y.; Allen, D.; Amberg, A.; Anger, L.T.; Aptula, A.; Auerbach, S.; Beilke, L.; Bellion, P.; et al. In Silico Toxicology Protocols. Regul. Toxicol. Pharmacol. 2018, 96, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rose, Y.; Duarte, J.M.; Lowe, R.; Segura, J.; Bi, C.; Bhikadiya, C.; Chen, L.; Rose, A.S.; Bittrich, S.; Burley, S.K.; et al. RCSB Protein Data Bank: Architectural Advances Towards Integrated Searching and Efficient Access to Macromolecular Structure Data from the PDB Archive. J. Mol. Biol. 2021, 433, 166704. [Google Scholar] [CrossRef]
- Stierand, K.; Rarey, M. Drawing the PDB: Protein-Ligand Complexes in Two Dimensions. ACS Med. Chem. Lett. 2010, 1, 540–545. [Google Scholar] [CrossRef]
- Makarewicz, T.; Kaźmierkiewicz, R. Improvements in GROMACS Plugin for PyMOL Including Implicit Solvent Simulations and Displaying Results of PCA Analysis. J. Mol. Model. 2016, 22, 109. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38, 27–28. [Google Scholar] [CrossRef]
- Bietz, S.; Urbaczek, S.; Schulz, B.; Rarey, M. Protoss: A Holistic Approach to Predict Tautomers and Protonation States in Protein-Ligand Complexes. J. Cheminform. 2014, 6, 12. [Google Scholar] [CrossRef]
- ZOETE, V.; CUENDET, M.A.; GROSDIDIER, A.; MICHIELIN, O. SwissParam: A Fast Force Field Generation Tool for Small Organic Molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rosell, G.; Giorgino, T.; De Fabritiis, G. PlayMolecule ProteinPrepare: A Web Application for Protein Preparation for Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 1511–1516. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- SAPAY, N.; TIELEMAN, D.P. Combination of the CHARMM27 Force Field with United-Atom Lipid Force Fields. J. Comput. Chem. 2011, 32, 1400–1410. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- De Freitas, R.F.; Schapira, M. A Systematic Analysis of Atomic Protein-Ligand Interactions in the PDB. Medchemcomm 2017, 8, 1970–1981. [Google Scholar] [CrossRef]
- Fassio, A.V.; Santos, L.H.; Silveira, S.A.; Ferreira, R.S.; De Melo-Minardi, R.C. NAPOLI: A Graph-Based Strategy to Detect and Visualize Conserved Protein-Ligand Interactions in Large-Scale. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1317–1328. [Google Scholar] [CrossRef]
- Alvarez, S. A Cartography of the van Der Waals Territories. Dalt. Trans. 2013, 42, 8617–8636. [Google Scholar] [CrossRef]
- Brylinski, M. Aromatic Interactions at the Ligand–Protein Interface: Implications for the Development of Docking Scoring Functions. Chem. Biol. Drug Des. 2018, 91, 380–390. [Google Scholar] [CrossRef]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation-π Interactions in Protein-Ligand Binding: Theory and Data-Mining Reveal Different Roles for Lysine and Arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, D.; Minervini, G.; Tosatto, S.C.E. The RING 2.0 Web Server for High Quality Residue Interaction Networks. Nucleic Acids Res. 2016, 44, W367–W374. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, E.; Mereghetti, P.; Fantucci, P.; Grandori, R.; De Gioia, L. Free-Energy Landscape, Principal Component Analysis, and Structural Clustering to Identify Representative Conformations from Molecular Dynamics Simulations: The Myoglobin Case. J. Mol. Graph. Model. 2009, 27, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. G-Mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Zeiger, E. The Test That Changed the World: The Ames Test and the Regulation of Chemicals. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2019, 841, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Adamson, R.H. The Acute Lethal Dose 50 (LD50) of Caffeine in Albino Rats. Regul. Toxicol. Pharmacol. 2016, 80, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Lepailleur, A.; Mignani, S.M.; Dallemagne, P.; Rochais, C. HERG Toxicity Assessment: Useful Guidelines for Drug Design. Eur. J. Med. Chem. 2020, 195, 112290. [Google Scholar] [CrossRef] [PubMed]
- El-Demerdash, A.; Metwaly, A.M.; Hassan, A.; El-Aziz, T.M.A.; Elkaeed, E.B.; Eissa, I.H.; Arafa, R.K.; Stockand, J.D. Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (COVID-19). Biomolecules 2021, 11, 460. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ge, S.; Tu, Y.; Hu, M. Challenges and Opportunities with Predicting In Vivo Phase II Metabolism via Glucuronidation From In Vitro Data. Curr. Pharmacol. Rep. 2016, 2, 326–338. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Nagai, T.; Srivastava, A.; Miyashita, O.; Tama, F. Role of Computational Methods in Going beyond X-Ray Crystallography to Explore Protein Structure and Dynamics. Int. J. Mol. Sci. 2018, 19, 3401. [Google Scholar] [CrossRef] [PubMed]
- Wereszczynski, J.; McCammon, J.A. Statistical Mechanics and Molecular Dynamics in Evaluating Thermodynamic Properties of Biomolecular Recognition. Q. Rev. Biophys. 2012, 45, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Moro, S. Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Sneha, P.; Priya Doss, C.G. Molecular Dynamics: New Frontier in Personalized Medicine, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2016; Volume 102, ISBN 9780128047958. [Google Scholar]
- Forrey, C.; Douglas, J.F.; Gilson, M.K. The Fundamental Role of Flexibility on the Strength of Molecular Binding. Soft Matter 2012, 8, 6385–6392. [Google Scholar] [CrossRef] [PubMed]
- Vanni, S.; Neri, M.; Tavernelli, I.; Rothlisberger, U. Predicting Novel Binding Modes of Agonists to β Adrenergic Receptors Using All-Atom Molecular Dynamics Simulations. PLoS Comput. Biol. 2011, 7, e1001053. [Google Scholar] [CrossRef]
- Martinez, L. Automatic Identification of Mobile and Rigid Substructures in Molecular Dynamics Simulations and Fractional Structural Fluctuation Analysis. PLoS ONE 2015, 10, e0119264. [Google Scholar] [CrossRef] [PubMed]
- Agrahari, A.K.; Sneha, P.; George Priya Doss, C.; Siva, R.; Zayed, H. A Profound Computational Study to Prioritize the Disease-Causing Mutations in PRPS1 Gene. Metab. Brain Dis. 2018, 33, 589–600. [Google Scholar] [CrossRef]
- Kumar, C.V.; Swetha, R.G.; Anbarasu, A.; Ramaiah, S. Computational Analysis Reveals the Association of Threonine 118 Methionine Mutation in PMP22 Resulting in CMT-1A. Adv. Bioinform. 2014, 2014, 10. [Google Scholar] [CrossRef]
- Khan, F.I.; Wei, D.Q.; Gu, K.R.; Hassan, M.I.; Tabrez, S. Current Updates on Computer Aided Protein Modeling and Designing. Int. J. Biol. Macromol. 2016, 85, 48–62. [Google Scholar] [CrossRef]
- De Oliveira, C.C.S.; Pereira, G.R.C.; De Alcantara, J.Y.S.; Antunes, D.; Caffarena, E.R.; De Mesquita, J.F. In Silico Analysis of the V66M Variant of Human BDNF in Psychiatric Disorders: An Approach to Precision Medicine. PLoS ONE 2019, 14, e0215508. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Kokubo, H. Exploring the Stability of Ligand Binding Modes to Proteins by Molecular Dynamics Simulations: A Cross-Docking Study. J. Chem. Inf. Model. 2017, 57, 2514–2522. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ma, Z.; Sang, J.; Lu, F. Edaravone Inhibits the Conformational Transition of Amyloid-Β42: Insights from Molecular Dynamics Simulations. J. Biomol. Struct. Dyn. 2020, 38, 2377–2388. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen Bonds in Proteins: Role and Strength. In Encyclopedia of Life Sciences; American Cancer Society: Atlanta, GA, USA, 2010; Volume 1. [Google Scholar] [CrossRef]
- Dhanik, A.; McMurray, J.S.; Kavraki, L.E. Binding Modes of Peptidomimetics Designed to Inhibit STAT3. PLoS ONE 2012, 7, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Liu, W.F.; Megido, L.; Díez, P.; Fuentes, M.; Fager, C.; Olsson, E.; Gessner, I.; Mathur, S. Understanding and Utilizing the Biomolecule/Nanosystems Interface. In Nanotechnologies in Preventive and Regenerative Medicine; Elsevier: Amsterdam, The Netherlands, 2018; pp. 207–297. [Google Scholar] [CrossRef]
- Stank, A.; Kokh, D.B.; Fuller, J.C.; Wade, R.C. Protein Binding Pocket Dynamics. Acc. Chem. Res. 2016, 49, 809–815. [Google Scholar] [CrossRef]
- Wan, S.; Bhati, A.P.; Zasada, S.J.; Coveney, P.V. Rapid, Accurate, Precise and Reproducible Ligand-Protein Binding Free Energy Prediction. Interface Focus 2020, 10, 20200007. [Google Scholar] [CrossRef]
- Poongavanam, V.; Olsen, J.M.H.; Kongsted, J. Binding Free Energy Based Structural Dynamics Analysis of HIV-1 RT RNase H-Inhibitor Complexes. Integr. Biol. 2014, 6, 1010–1022. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Shashikala, H.B.M.; Chakravorty, A.; Alexov, E. Modeling Electrostatic Force in Protein-Protein Recognition. Front. Mol. Biosci. 2019, 6, 94. [Google Scholar] [CrossRef]
- Matysiak, J.; Karpińska, M.M.; Skrzypek, A.; Wietrzyk, J.; Kłopotowska, D.; Niewiadomy, A.; Paw, B.; Juszczak, M.; Rzeski, W. Design, Synthesis and Antiproliferative Activity against Human Cancer Cell Lines of Novel Benzo-, Benzofuro-, Azolo- and Thieno-1,3-Thiazinone Resorcinol Hybrids. Arab. J. Chem. 2019, 12, 2655–2667. [Google Scholar] [CrossRef]
- Sohlenius-Sternbeck, A.K.; Terelius, Y. Evaluation of ADMET Predictor in Early Discovery Drug Metabolism and Pharmacokinetics Project Work. Drug Metab. Dispos. 2022, 50, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Brüstle, M.; Beck, B.; Schindler, T.; King, W.; Mitchell, T.; Clark, T. Descriptors, Physical Properties, and Drug-Likeness. J. Med. Chem. 2002, 45, 3345–3355. [Google Scholar] [CrossRef] [PubMed]
- Neubert, D. Reproductive Toxicology: The Science Today. Teratog. Carcinog. Mutagen. 2002, 22, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the Chemical Beauty of Drugs Europe PMC Funders Group. Nat. Chem 2012, 4, 90–98. [Google Scholar] [CrossRef]
- Fonsi, M.; Orsale, M.V.; Monteagudo, E. High-Throughput Microsomal Stability Assay for Screening New Chemical Entities in Drug Discovery. J. Biomol. Screen. 2008, 13, 862–869. [Google Scholar] [CrossRef]
- Dorst, J.; Ludolph, A.C.; Huebers, A. Disease-Modifying and Symptomatic Treatment of Amyotrophic Lateral Sclerosis. Ther Adv. Neurol. Disord. 2018, 11, 1756285617734734. [Google Scholar] [CrossRef]
- Hakkola, J.; Hukkanen, J.; Turpeinen, M.; Pelkonen, O. Inhibition and Induction of CYP Enzymes in Humans: An Update. Arch. Toxicol. 2020, 94, 3671–3722. [Google Scholar] [CrossRef]
- Skvrce, N.M.; Macolic Sarinic, V.; Mucalo, I.; Krnic, D.; Bozina, N.; Tomic, S. Adverse Drug Reactions Caused by Drug-Drug Interactions Reported to Croatian Agency for Medicinal Products and Medical Devices: A Retrospective Observational Study. Croat. Med. J. 2011, 52, 604–614. [Google Scholar] [CrossRef]
- Polasek, T.M.; Lin, F.P.Y.; Miners, J.O.; Doogue, M.P. Perpetrators of Pharmacokinetic Drug-Drug Interactions Arising from Altered Cytochrome P450 Activity: A Criteria-Based Assessment. Br. J. Clin. Pharmacol. 2011, 71, 727–736. [Google Scholar] [CrossRef]
- Thakur, M.; Grossman, I.; McCrory, D.C.; Orlando, L.A.; Steffens, D.C.; Cline, K.E.; Gray, R.N.; Farmer, J.; DeJesus, G.; O’Brien, C.; et al. Review of Evidence for Genetic Testing for CYP450 Polymorphisms in Management of Patients with Nonpsychotic Depression with Selective Serotonin Reuptake Inhibitors. Genet. Med. 2007, 9, 826–835. [Google Scholar] [CrossRef]
- Ayano, G. Psychotropic Medications Metabolized by Cytochromes P450 (CYP) 2D6 Enzyme and Relevant Drug Interactions. Clin. Pharmacol. Biopharm. 2016, 5, 3–5. [Google Scholar] [CrossRef]
- Yoon, S.; Jeong, S.; Jung, E.; Kim, K.S.; Jeon, I.; Lee, Y.; Cho, J.Y.; Oh, W.Y.; Chung, J.Y. Effect of CYP3A4 Metabolism on Sex Differences in the Pharmacokinetics and Pharmacodynamics of Zolpidem. Sci. Rep. 2021, 11, 19150. [Google Scholar] [CrossRef] [PubMed]
- Bourin, M. Hypnotic Drugs: Pharmacological and Therapeutic Issues. SOJ Pharm. Pharm. Sci. 2018, 5, 1–4. [Google Scholar] [CrossRef]
- Iyanagi, T. Molecular Mechanism of Phase I and Phase II Drug-Metabolizing Enzymes: Implications for Detoxification. Int. Rev. Cytol. 2007, 260, 35–112. [Google Scholar] [CrossRef] [PubMed]
- VandenBrink, B.M.; Isoherranen, N. The Role of Metabolites in Predicting Drug-Drug Interactions: Focus on Irreversible Cytochrome P450 Inhibition. Curr. Opin. Drug Discov. Dev. 2010, 13, 66–77. [Google Scholar]
- Savjani, K.T.; Gajjar, A.K.; Savjani, J.K. Drug Solubility: Importance and Enhancement Techniques. ISRN Pharm. 2012, 2012, 195727. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Ritchie, T.J.; MacDonald, S.J.F.; Young, R.J.; Pickett, S.D. The Impact of Aromatic Ring Count on Compound Developability: Further Insights by Examining Carbo- and Hetero-Aromatic and -Aliphatic Ring Types. Drug Discov. Today 2011, 16, 164–171. [Google Scholar] [CrossRef]
- Garg, P.; Dhakne, R.; Belekar, V. Role of Breast Cancer Resistance Protein (BCRP) as Active Efflux Transporter on Blood-Brain Barrier (BBB) Permeability. Mol. Divers. 2015, 19, 163–172. [Google Scholar] [CrossRef]
- Liu, X.; Wright, M.; Hop, C.E.C.A. Rational Use of Plasma Protein and Tissue Binding Data in Drug Design. J. Med. Chem. 2014, 57, 8238–8248. [Google Scholar] [CrossRef]
- Gardiner, P.; Cox, R.J.; Grime, K. Plasma Protein Binding as an Optimizable Parameter for Acidic Drugs. Drug Metab. Dispos. 2019, 47, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Słoczyńska, K.; Gunia-Krzyzak, A.; Koczurkiewicz, P.; Wójcik-Pszczoła, K.; Zelaszczyk, D.; Popiół, J.; Pȩkala, E. Metabolic Stability and Its Role in the Discovery of New Chemical Entities. Acta Pharm. 2019, 69, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Negi, A.; Gupta, P.K.; Chauhan, M.; Kumar, R. Toxicophore Exploration as a Screening Technology for Drug Design and Discovery: Techniques, Scope and Limitations. Arch. Toxicol. 2016, 90, 1785–1802. [Google Scholar] [CrossRef] [PubMed]
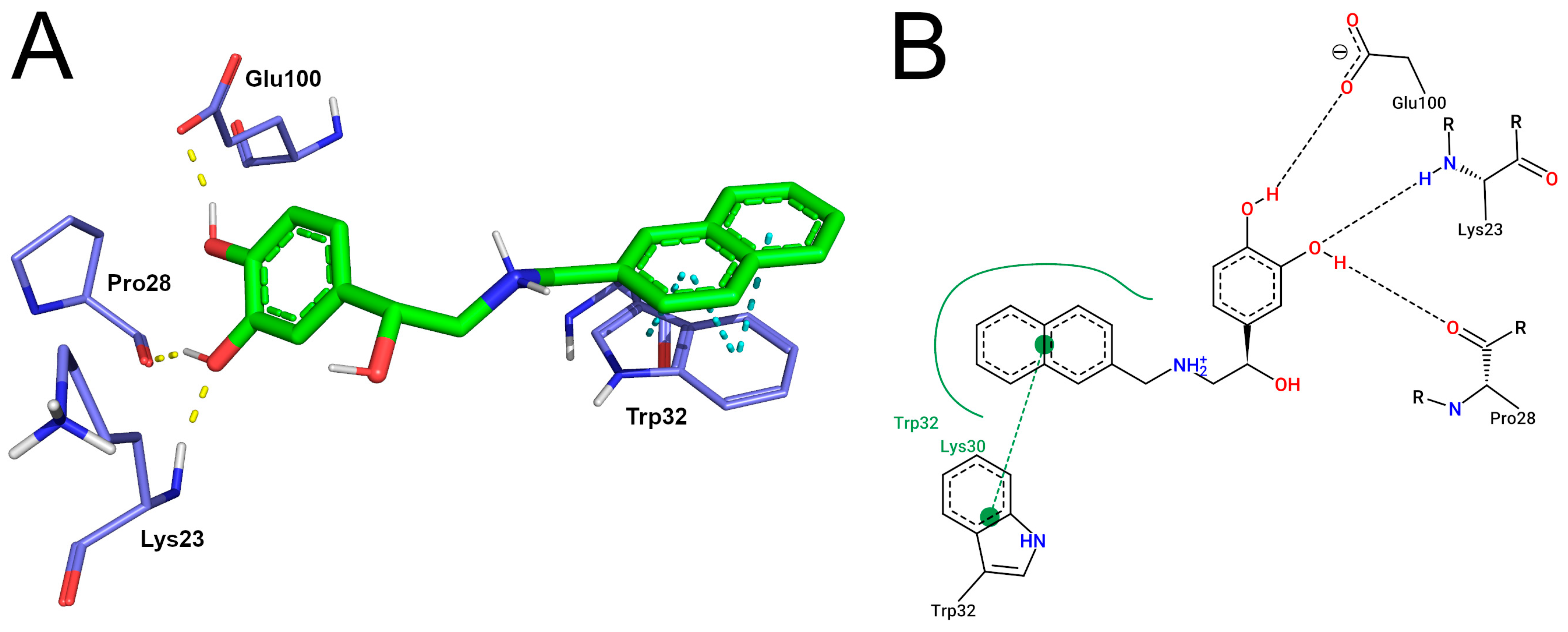

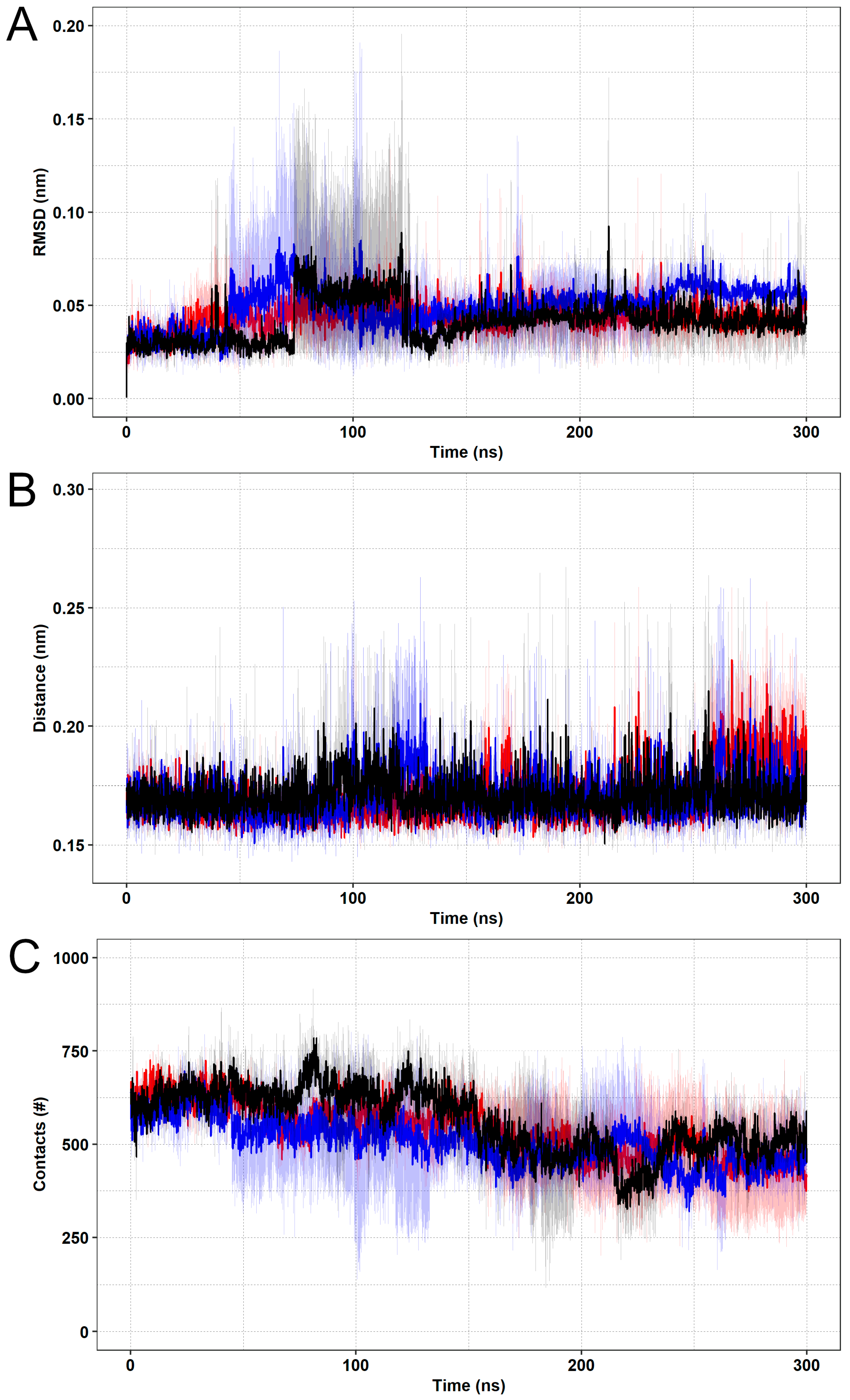
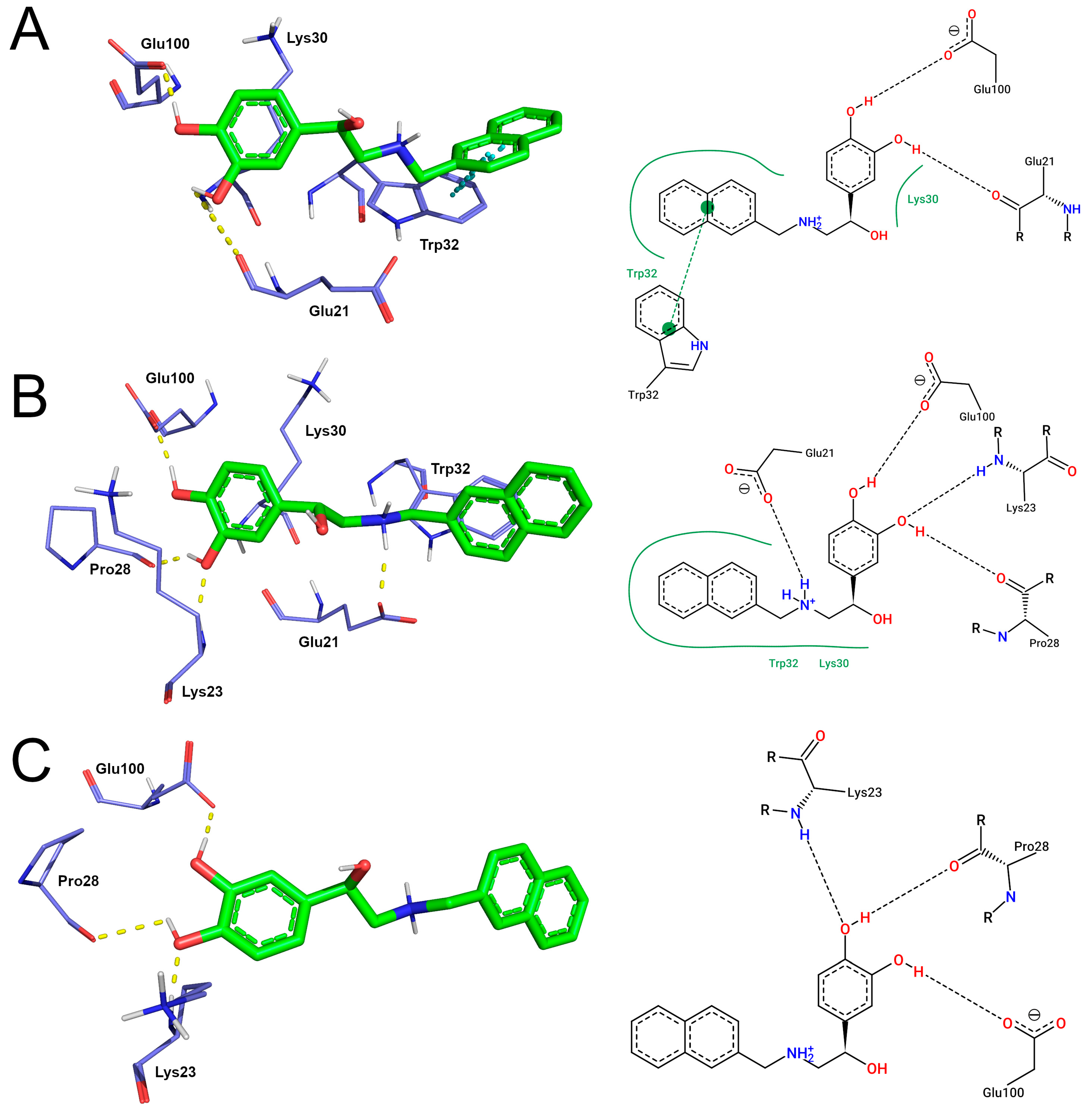
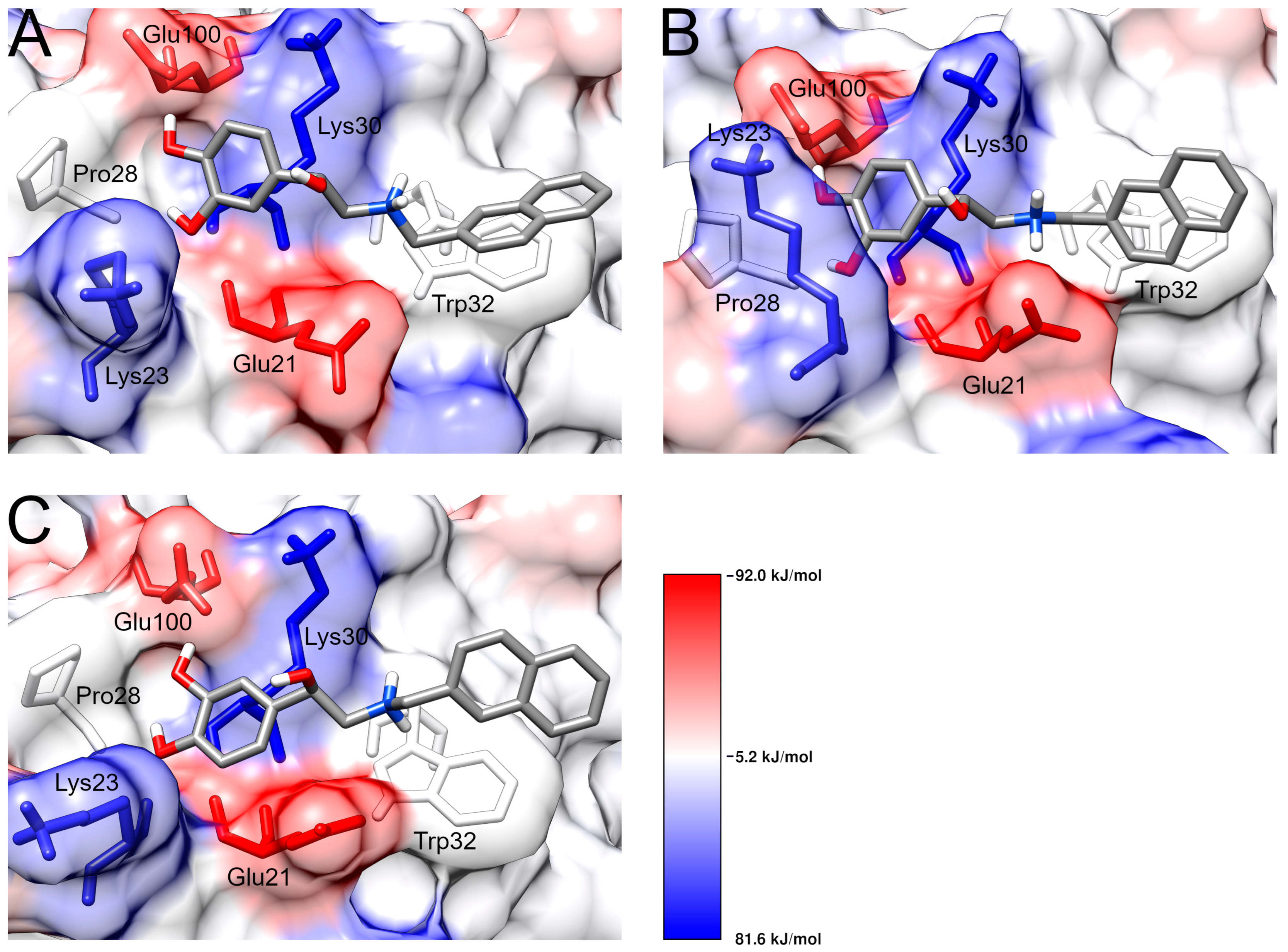

| Protein | π-Stacking 1 | Cation-π 1 | Hydrophobic Contacts 1 | Minimum Distance 2 |
|---|---|---|---|---|
| wild-type | 9.02% | 3.8% | 36.13% | 0.37 ± 0.10 nm |
| A4V | 16.88% | 38.46% | 89.55% | 0.26 ± 0.03 nm |
| D90A | 15.76% | 10.12% | 49.25% | 0.34 ± 0.08 nm |
| Protein | Hydrogen Bonds 1 | Hydrophobic Contacts 1 | Ionic Bonds 2 | Cation-π 2 |
|---|---|---|---|---|
| WT | 2.42 ± 1.07 | 10.07 ± 5.69 | 72.72% | 19.92% |
| A4V | 1.98 ± 1.25 | 11.81 ± 7.63 | 57.34% | 55.51% |
| D90A | 1.78 ± 0.91 | 8.91 ± 5.88 | 48.39% | 13.61% |
| Protein | Lys23 | Glu21 | Pro28 | Glu100 |
|---|---|---|---|---|
| WT | 18.23% | 46.76% | 5.21% | 93.67% |
| A4V | 12.15% | 71.51% | 1.38% | 51.09% |
| D90A | 18.83% | 35.74% | 6.85% | 70.99% |
| Protein | ΔE Polar (kJ/mol) | ΔE Apolar (kJ/mol) | ΔE MM (kJ/mol) | Binding Energy (kJ/mol) |
|---|---|---|---|---|
| WT | 229 ± 35 | −9.41 ± 1.27 | −349 ± 65 | −129 ± 42 |
| A4V | 205 ± 85 | −8.80 ± 2.27 | −409 ± 118 | −209 ± 52 |
| D90A | 240 ± 31 | −9.22 ± 1.20 | −377 ± 61 | −146 ± 39 |
| Model | ADMET Risk | Absn Risk | CYP Risk | Tox Risk | MutRisk | Mutx Risk | Vd & Fu | Lipinski’s Rule | Repro Tox | CYP Inhibition |
|---|---|---|---|---|---|---|---|---|---|---|
| Penalty | 2.5 | 0.5 | 1 | 1 | 1.2 | 0.64 | 0 | 0 | _ | _ |
| Violation | HBD; MUT; 2D6 | HBD | 2D6 | MUT | S_97; NIHS | S_97; NIHS | No | No | No | CYP1A2, CYP2D6, CYP3A4 |
| Cut-off | ≤7 | ≤4 | ≤2 | ≤2 | ≤1 | ≤1 | _ | ≤2 | _ | _ |
| Module | Model | Parameter Value | Classification | Reference Value |
|---|---|---|---|---|
| DrugSafetyprofiling | Mutagenicity | 0.35 | Undefined | ≤0.33 |
| hERG inhibition | 0.31 | Non-inhibitor | ≤0.33 | |
| CYP inhibition | 0.33–0.64 | Undefined (3A4, 2D6, 2C19, 1A2); Non-inhibitor (2C9) | ≤0.33 | |
| Acute Toxicity | _ | No hazardous fragment | No hazardous fragment | |
| PhysChem profiling | Lipinski’s Rule | 0 violations | Optimal | _ |
| Lead-like Rule | 0 violations | Optimal | _ | |
| Solubility | 33.9 mg/mL | Soluble | ≥10 mg/mL | |
| Rotatable bonds | 5 | Optimal | ≤10 | |
| Nº of rings | 3 | Optimal | ≤4 | |
| ADME profiling | Bioavailability | 96% | High | ≥70% |
| Caco-2 | 16 × 10−6 cm/s | High | ≥7 × 10−6 cm/s | |
| HIA | 100% | High | ≥70% | |
| PPB | 81% | High | ≥40% & ≤80% | |
| Metabolic Stability | 0.49 | Undefined | ≤0.33 | |
| CNS-Access | −2.93 | Sufficient for CNS activity | >3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, G.R.C.; Abrahim-Vieira, B.d.A.; de Mesquita, J.F. In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein. Pharmaceutics 2023, 15, 1095. https://doi.org/10.3390/pharmaceutics15041095
Pereira GRC, Abrahim-Vieira BdA, de Mesquita JF. In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein. Pharmaceutics. 2023; 15(4):1095. https://doi.org/10.3390/pharmaceutics15041095
Chicago/Turabian StylePereira, Gabriel Rodrigues Coutinho, Bárbara de Azevedo Abrahim-Vieira, and Joelma Freire de Mesquita. 2023. "In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein" Pharmaceutics 15, no. 4: 1095. https://doi.org/10.3390/pharmaceutics15041095
APA StylePereira, G. R. C., Abrahim-Vieira, B. d. A., & de Mesquita, J. F. (2023). In Silico Analyses of a Promising Drug Candidate for the Treatment of Amyotrophic Lateral Sclerosis Targeting Superoxide Dismutase I Protein. Pharmaceutics, 15(4), 1095. https://doi.org/10.3390/pharmaceutics15041095




