Kinase Inhibitor Screening Displayed ALK as a Possible Therapeutic Biomarker for Gastric Cancer
Abstract
:1. Introduction
2. Materials and Method
2.1. Cell Culture
2.2. Alamar Blue Method
2.3. Cell Cycle Progression
2.4. Caspase 3/7 Activity
2.5. Wound Healing Assay
2.6. Gene Expression by Quantitative Real-Time PCR
2.7. PPI Network and GENE Ontology Enrichment
2.8. Survival Analysis
2.9. Statistical Analysis
3. Results
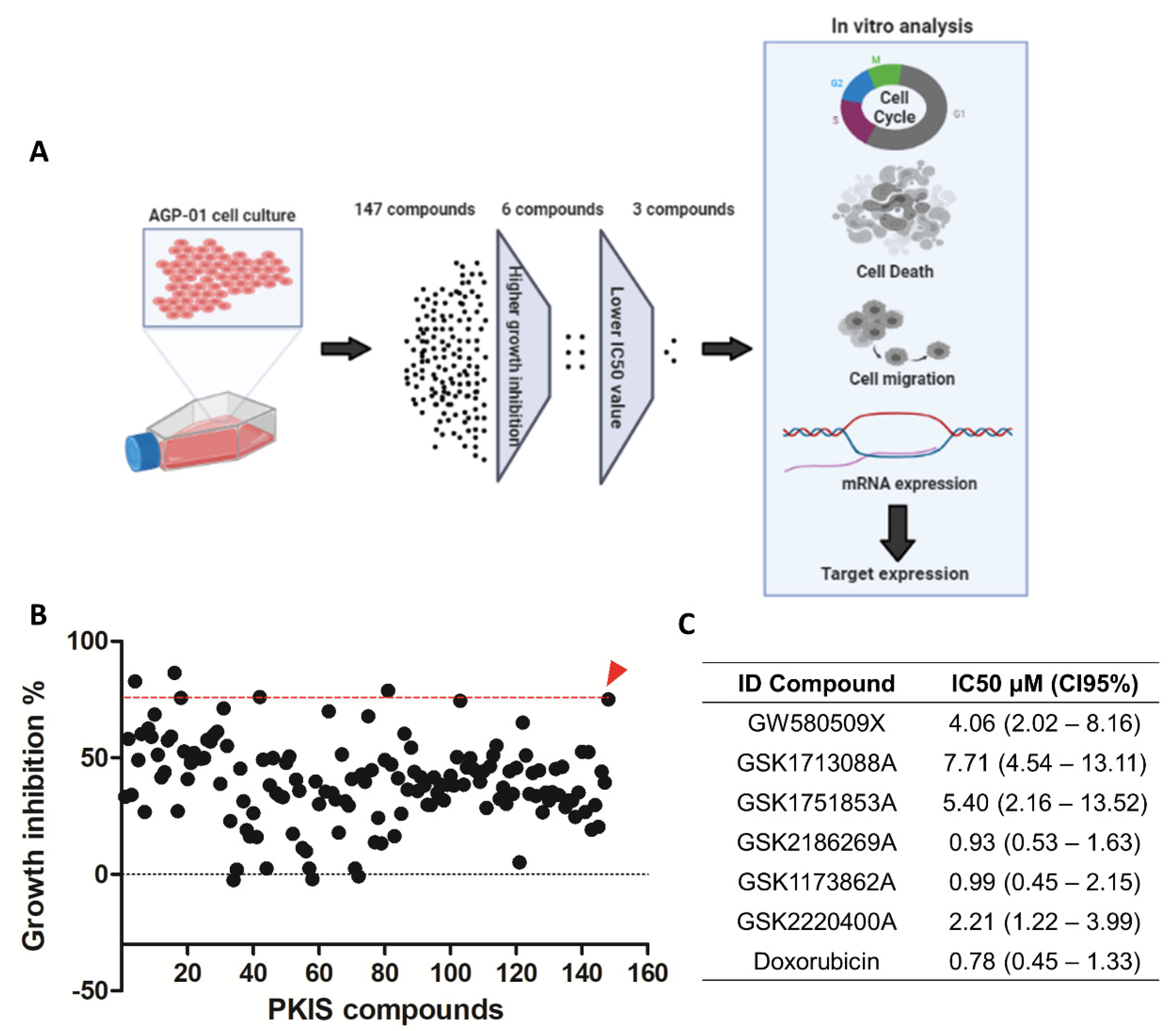
3.1. Screening of KIs Compounds against Metastatic Gastric Cancer Cell Line
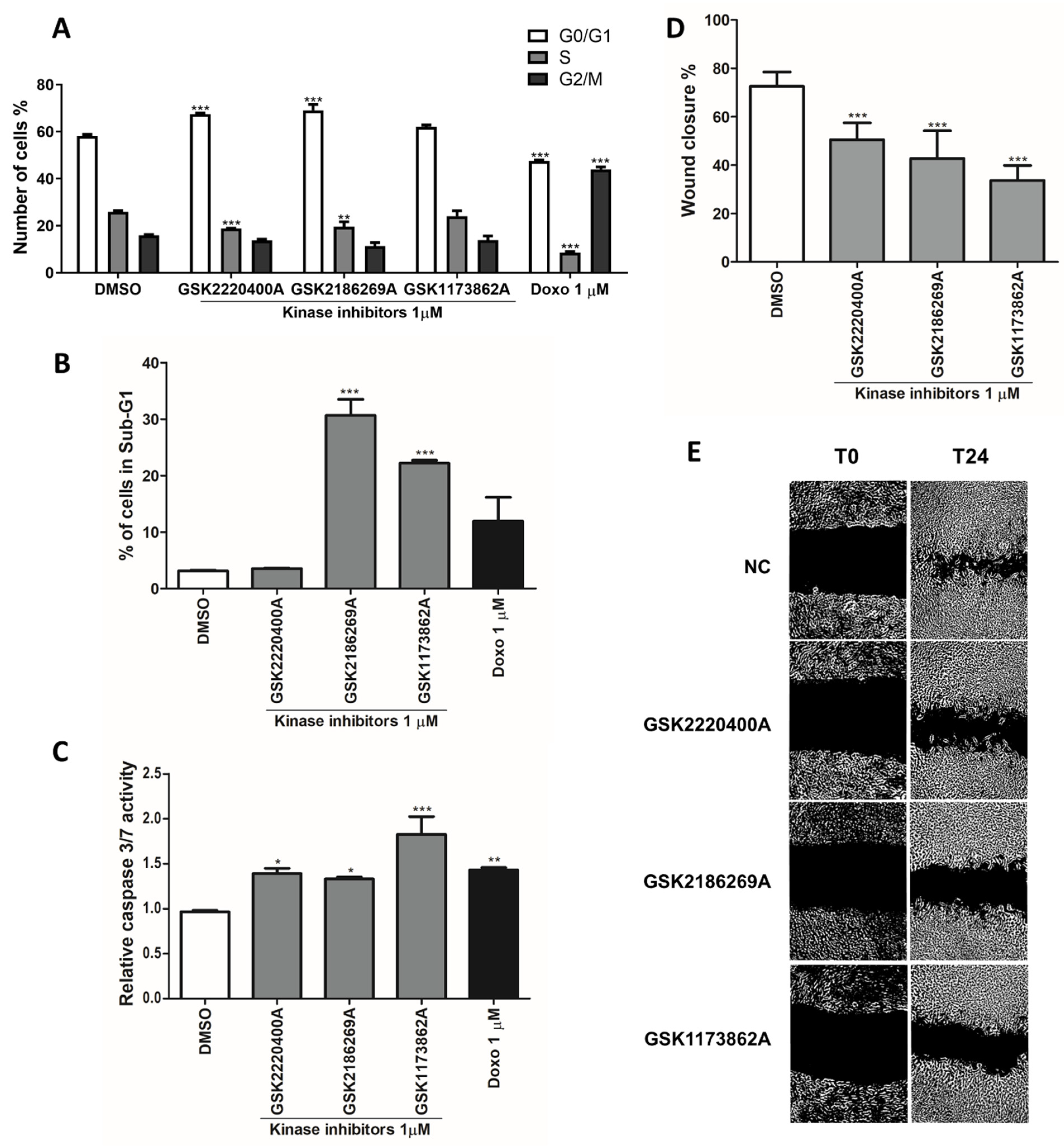
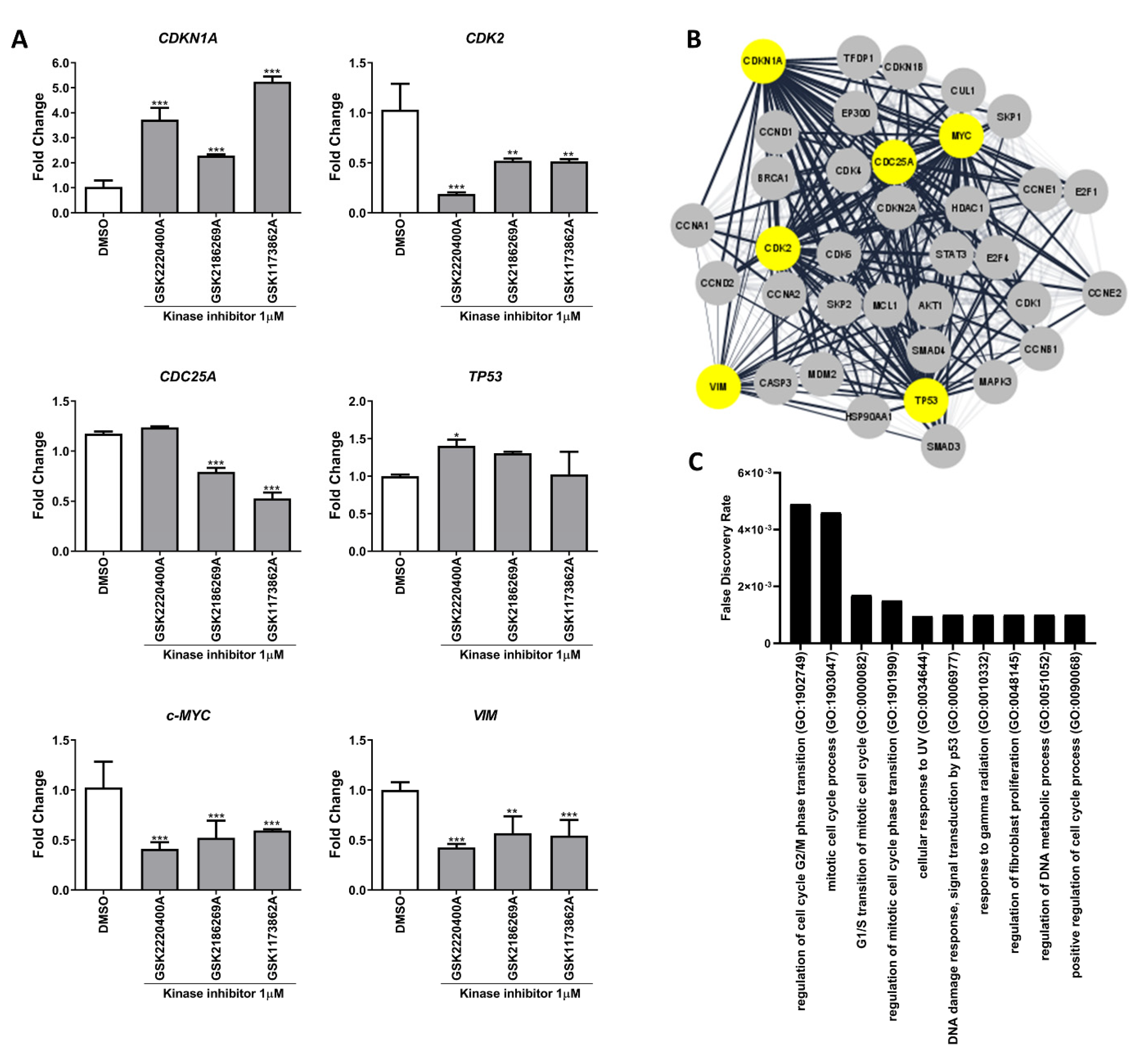
3.2. Kinase Inhibitors Act as Anti-Cancer Agents by Regulating Cell Cycle and Mesenchymal Genes
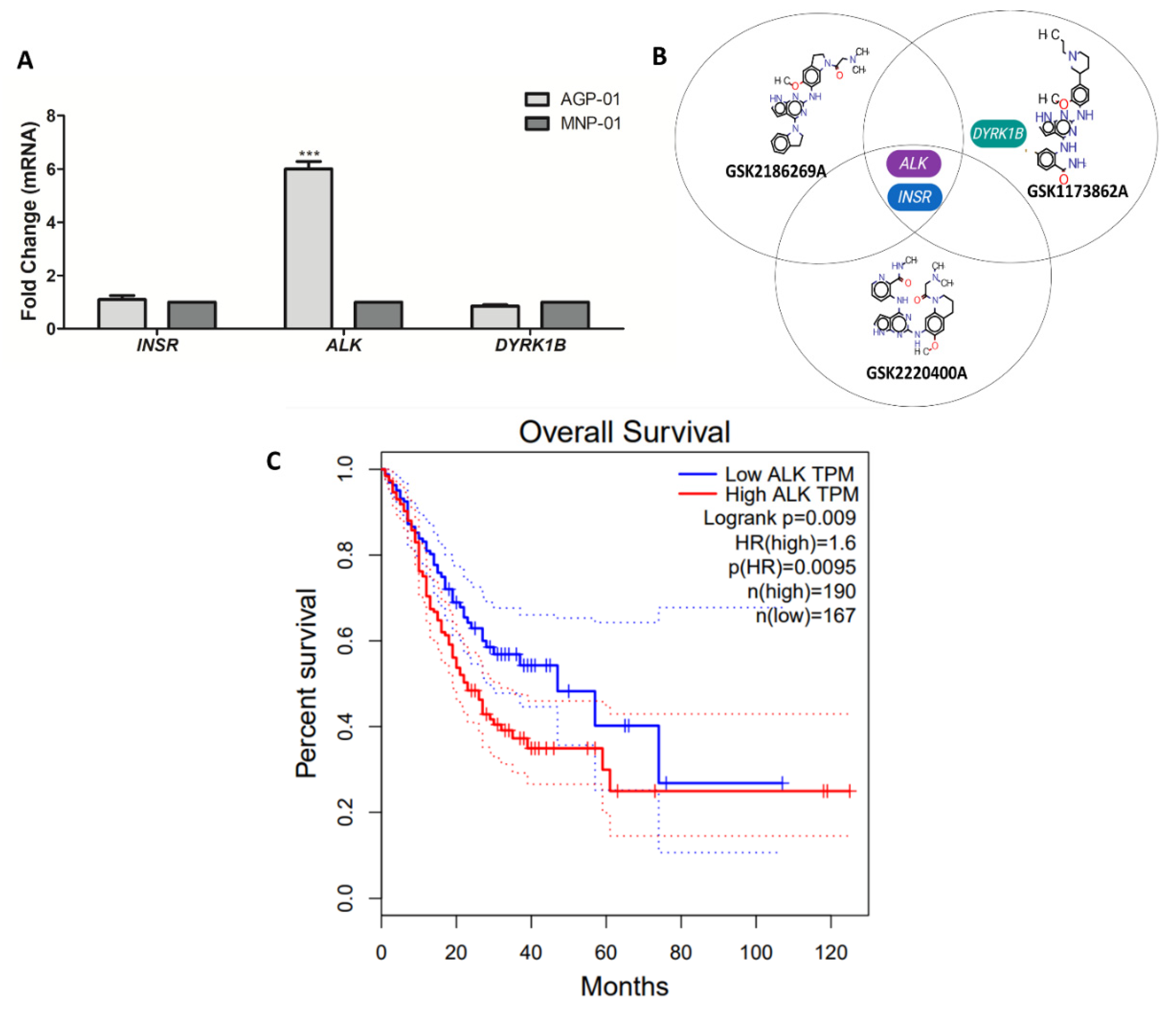
3.3. Kinase Target Expression in the Metastatic Gastric Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Lee, J.; Bass, A.J.; Ajani, J.A. Gastric Adenocarcinoma: An Update on Genomics, Immune System Modulations, and Targeted Therapy. Am. Soc. Clin. Oncol. Educ. book. Am. Soc. Clin. Oncol. Annu. Meet. 2016, 35, 104–111. [Google Scholar] [CrossRef]
- Eusebi, L.H.; Telese, A.; Marasco, G.; Bazzoli, F.; Zagari, R.M. Gastric Cancer Prevention Strategies: A Global Perspective. J. Gastroenterol. Hepatol. 2020, 35, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wu, X.; Li, S.; Li, C.; Guo, Z. Impact of Environmental Factors on Gastric Cancer: A Review of the Scientific Evidence, Human Prevention and Adaptation. J. Environ. Sci. 2020, 89, 65–79. [Google Scholar] [CrossRef]
- Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; Shen, H.; et al. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Jácome, A.A.; Coutinho, A.K.; Lima, E.M.; Andrade, A.C.; Dos Santos, J.S. Personalized Medicine in Gastric Cancer: Where Are We and Where Are We Going? World J. Gastroenterol. 2016, 22, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Labianca, R.; Beretta, G.D.; Gatta, G.; de Braud, F.; Van Cutsem, E. Gastric Cancer. Crit. Rev. Oncol. Hematol. 2009, 71, 127–164. [Google Scholar] [CrossRef]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in Combination with Chemotherapy versus Chemotherapy Alone for Treatment of HER2-Positive Advanced Gastric or Gastro-Oesophageal Junction Cancer (ToGA): A Phase 3, Open-Label, Randomised Controlled Trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Wagner, A.D.; Syn, N.L.; Moehler, M.; Grothe, W.; Yong, W.P.; Tai, B.C.; Ho, J.; Unverzagt, S. Chemotherapy for Advanced Gastric Cancer. Cochrane Database Syst. Rev. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-Targeted Cancer Therapies: Progress, Challenges and Future Directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Nielsen, T.E.; Clausen, M.H. FDA-Approved Small-Molecule Kinase Inhibitors. Trends Pharmacol. Sci. 2015, 36, 422–439. [Google Scholar] [CrossRef]
- Elkins, J.M.; Fedele, V.; Szklarz, M.; Abdul Azeez, K.R.; Salah, E.; Mikolajczyk, J.; Romanov, S.; Sepetov, N.; Huang, X.P.; Roth, B.L.; et al. Comprehensive Characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol. 2016, 34, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.F.; do Nascimento, J.L.M.; da Silva, C.E.A.; Lamarão, M.F.V.; Calcagno, D.Q.; Khayat, A.S.; Assumpção, P.P.; Cabral, I.R.; Smith, M.D.A.C.; Burbano, R.R. Establishment and Conventional Cytogenetic Characterization of Three Gastric Cancer Cell Lines. Cancer Genet. Cytogenet. 2009, 195, 85–91. [Google Scholar] [CrossRef]
- Leal, M.F.; Ribeiro, H.F.; Rey, J.A.; Pinto, G.R.; Smith, M.C.; Moreira-Nunes, C.A.; Assumpção, P.P.; Lamarão, L.M.; Calcagno, D.Q.; Montenegro, R.C.; et al. YWHAE Silencing Induces Cell Proliferation, Invasion and Migration through the up-Regulation of CDC25B and MYC in Gastric Cancer Cells: New Insights about YWHAE Role in the Tumor Development and Metastasis Process. Oncotarget 2016, 7, 85393–85410. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, F.P.; Silva, E.L.; Souza, P.F.N.; Lima, L.B.; Amaral, J.L.; Zuercher, W.; Albuquerque, L.M.; Rabenhorst, S.H.B.; Moreira-Nunes, C.A.; Moraes, M.E.; et al. Kinase Inhibitor Screening Reveals Aurora-a Kinase Is a Potential Therapeutic and Prognostic Biomarker of Gastric Cancer. J. Cell. Biochem. 2021, 122, 1376–1388. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Holla, V.R.; Elamin, Y.Y.; Bailey, A.M.; Johnson, A.M.; Litzenburger, B.C.; Khotskaya, Y.B.; Sanchez, N.S.; Zeng, J.; Shufean, M.A.; Shaw, K.R.; et al. ALK: A Tyrosine Kinase Target for Cancer Therapy. Cold Spring Harb. Mol. Case Stud. 2017, 3, a001115. [Google Scholar] [CrossRef]
- Hallberg, B.; Palmer, R.H. The Role of the ALK Receptor in Cancer Biology. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27 (Suppl. 3), iii4–iii15. [Google Scholar] [CrossRef] [PubMed]
- Schönherr, C.; Ruuth, K.; Yamazaki, Y.; Eriksson, T.; Christensen, J.; Palmer, R.H.; Hallberg, B. Activating ALK Mutations Found in Neuroblastoma Are Inhibited by Crizotinib and NVP-TAE684. Biochem. J. 2011, 440, 405–413. [Google Scholar] [CrossRef]
- Murugan, A.K.; Xing, M.M. Anaplastic Thyroid Cancers Harbor Novel Oncogenic Mutations of the ALK Gene. Cancer Res. 2011, 71, 4403–4411. [Google Scholar] [CrossRef] [PubMed]
- Schoppmann, S.F.; Streubel, B.; Birner, P. Amplification but Not Translocation of Anaplastic Lymphoma Kinase Is a Frequent Event in Oesophageal Cancer. Eur. J. Cancer 2013, 49, 1876–1881. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-F.; Chen, T.-D.; Chang, I.-C.; Chen, Y.-T.; Liu, H.-P.; Wu, Y.-C.; Wang, C.-L.; Chen, Y.-R. Correlation of Anaplastic Lymphoma Kinase Overexpression and the EML4-ALK Fusion Gene in Non-Small Cell Lung Cancer by Immunohistochemical Study. Chang Gung Med. J. 2012, 35, 309–317. [Google Scholar] [CrossRef]
- Matsumoto, T.; Oda, Y.; Hasegawa, Y.; Hashimura, M.; Oguri, Y.; Inoue, H.; Yokoi, A.; Tochimoto, M.; Nakagawa, M.; Jiang, Z.; et al. Anaplastic Lymphoma Kinase Overexpression Is Associated with Aggressive Phenotypic Characteristics of Ovarian High-Grade Serous Carcinoma. Am. J. Pathol. 2021, 191, 1837–1850. [Google Scholar] [CrossRef]
- Fan, J.; Yang, M.; Huang, B.; Wang, Z.; Luo, D.; Zhang, J.; Zhang, P.; Shi, H.; Li, Y.; Nie, X. ALK Expressed in a Gastrointestinal Stromal Tumor Harboring PDGFRA p. D842V Mutation:A Case Report. Diagn. Pathol. 2020, 15, 8. [Google Scholar] [CrossRef]
- Suski, J.M.; Braun, M.; Strmiska, V.; Sicinski, P. Targeting Cell-Cycle Machinery in Cancer. Cancer Cell 2021, 39, 759–778. [Google Scholar] [CrossRef]
- Strasser, A.; Vaux, D.L. Cell Death in the Origin and Treatment of Cancer. Mol. Cell 2020, 78, 1045–1054. [Google Scholar] [CrossRef]
- Razaghi, A.; Heimann, K.; Schaeffer, P.M.; Gibson, S.B. Negative Regulators of Cell Death Pathways in Cancer: Perspective on Biomarkers and Targeted Therapies. Apoptosis 2018, 23, 93–112. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting Apoptosis in Cancer Therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Debnath, P.; Huirem, R.S.; Dutta, P.; Palchaudhuri, S. Epithelial-Mesenchymal Transition and Its Transcription Factors. Biosci. Rep. 2022, 42, BSR20211754. [Google Scholar] [CrossRef]
- Chen, X.; Ji, J.; Chen, W.; Lian, W.; Chen, R.; Yang, J.; Zhang, Q.; Weng, Q.; Khan, Z.; Hu, J.; et al. (S)-Crizotinib Reduces Gastric Cancer Growth through Oxidative DNA Damage and Triggers pro-Survival Akt Signal. Cell Death Dis. 2018, 9, 660. [Google Scholar]
- Yang, Y.; Wu, N.; Shen, J.; Teixido, C.; Sun, X.; Lin, Z.; Qian, X.; Zou, Z.; Guan, W.; Yu, L.; et al. MET Overexpression and Amplification Define a Distinct Molecular Subgroup for Targeted Therapies in Gastric Cancer. Gastric Cancer 2016, 19, 778–788. [Google Scholar] [CrossRef]
- Montenegro, R.C.; Clark, P.G.K.; Howarth, A.; Wan, X.; Ceroni, A.; Siejka, P.; Nunez-Alonso, G.A.; Monteiro, O.; Rogers, C.; Gamble, V.; et al. BET Inhibition as a New Strategy for the Treatment of Gastric Cancer. Oncotarget 2016, 7, 43997. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.; Bertagnolli, M.M. Comparison of Gastric Cancer Survival between Caucasian and Asian Patients Treated in the United States: Results from the Surveillance Epidemiology and End Results (SEER) Database. Ann. Surg. Oncol. 2015, 22, 2965–2971. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Pinheiro, P.S.; Callahan, K.E.; Altekruse, S.F. Examining the Gastric Cancer Survival Gap between Asians and Whites in the United States. Gastric Cancer 2017, 20, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Trumbull, D.; Lemini, R.; Attwood, K.; Kukar, M.; Gabriel, E. Gastric Cancer Disparities Among Asian American Subpopulations. Anticancer Res. 2020, 40, 6381–6385. [Google Scholar] [CrossRef]
- Alese, O.B.; Rayes, B.F.; Zhang, G.; Khuri, F.R.; Sica, G.; Adsay, N.V.; Alexis, D.; Rosa, F.G.; Varella Garcia, M.; Chen, Z.; et al. Anaplastic Lymphoma Kinase (ALK) Gene Alteration in Signet Ring Cell Carcinoma of the Gastrointestinal Tract. Ther. Adv. Med. Oncol. 2015, 7, 56–62. [Google Scholar] [CrossRef]
- Wen, Z.; Xiong, D.; Zhang, S.; Liu, J.; Li, B.; Li, R.; Zhang, H. Case Report: RAB10-ALK: A Novel ALK Fusion in a Patient With Gastric Cancer. Front. Oncol. 2021, 11, 645370. [Google Scholar] [CrossRef]
- Ying, J.; Lin, C.; Wu, J.; Guo, L.; Qiu, T.; Ling, Y.; Shan, L.; Zhou, H.; Zhao, D.; Wang, J.; et al. Anaplastic Lymphoma Kinase Rearrangement in Digestive Tract Cancer: Implication for Targeted Therapy in Chinese Population. PLoS ONE 2015, 10, e0144731. [Google Scholar]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving Genetic Heterogeneity in Cancer. Nat. Rev. Genet. 2019, 20, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Bauer, T.M.; de Marinis, F.; Felip, E.; Goto, Y.; Liu, G.; Mazieres, J.; Kim, D.-W.; Mok, T.; Polli, A.; et al. First-Line Lorlatinib or Crizotinib in Advanced ALK -Positive Lung Cancer. N. Engl. J. Med. 2020, 383, 2018–2029. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mesquita, F.P.; Souza, P.F.N.; da Silva, E.L.; Lima, L.B.; de Oliveira, L.L.B.; Moreira-Nunes, C.A.; Zuercher, W.J.; Burbano, R.M.R.; de Moraes, M.E.A.; Montenegro, R.C. Kinase Inhibitor Screening Displayed ALK as a Possible Therapeutic Biomarker for Gastric Cancer. Pharmaceutics 2022, 14, 1841. https://doi.org/10.3390/pharmaceutics14091841
Mesquita FP, Souza PFN, da Silva EL, Lima LB, de Oliveira LLB, Moreira-Nunes CA, Zuercher WJ, Burbano RMR, de Moraes MEA, Montenegro RC. Kinase Inhibitor Screening Displayed ALK as a Possible Therapeutic Biomarker for Gastric Cancer. Pharmaceutics. 2022; 14(9):1841. https://doi.org/10.3390/pharmaceutics14091841
Chicago/Turabian StyleMesquita, Felipe Pantoja, Pedro Filho Noronha Souza, Emerson Lucena da Silva, Luina Benevides Lima, Lais Lacerda Brasil de Oliveira, Caroline Aquino Moreira-Nunes, William J. Zuercher, Rommel Mario Rodríguez Burbano, Maria Elisabete Amaral de Moraes, and Raquel Carvalho Montenegro. 2022. "Kinase Inhibitor Screening Displayed ALK as a Possible Therapeutic Biomarker for Gastric Cancer" Pharmaceutics 14, no. 9: 1841. https://doi.org/10.3390/pharmaceutics14091841
APA StyleMesquita, F. P., Souza, P. F. N., da Silva, E. L., Lima, L. B., de Oliveira, L. L. B., Moreira-Nunes, C. A., Zuercher, W. J., Burbano, R. M. R., de Moraes, M. E. A., & Montenegro, R. C. (2022). Kinase Inhibitor Screening Displayed ALK as a Possible Therapeutic Biomarker for Gastric Cancer. Pharmaceutics, 14(9), 1841. https://doi.org/10.3390/pharmaceutics14091841







