Peg-Grafted Liposomes for L-Asparaginase Encapsulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Liposomes Preparation
2.3. Dynamic Light Scattering (DLS), Zeta Potential (ζ) and Stability Analysis
2.4. Asparaginase Encapsulation
2.5. Determination of Encapsulation Efficiency
2.6. Transmission Electron Microscopy Analysis
2.7. Liposomes Stability over the Time
2.8. Liposome Permeability Assay
2.9. In Vitro Cytotoxicity against MOLT-4 Cells
2.10. Statistical Analysis
3. Results and Discussion
3.1. Liposomes Preparation and Characterization
3.2. Liposomes Stability
3.3. L-Asparaginase Encapsulation in Liposomes
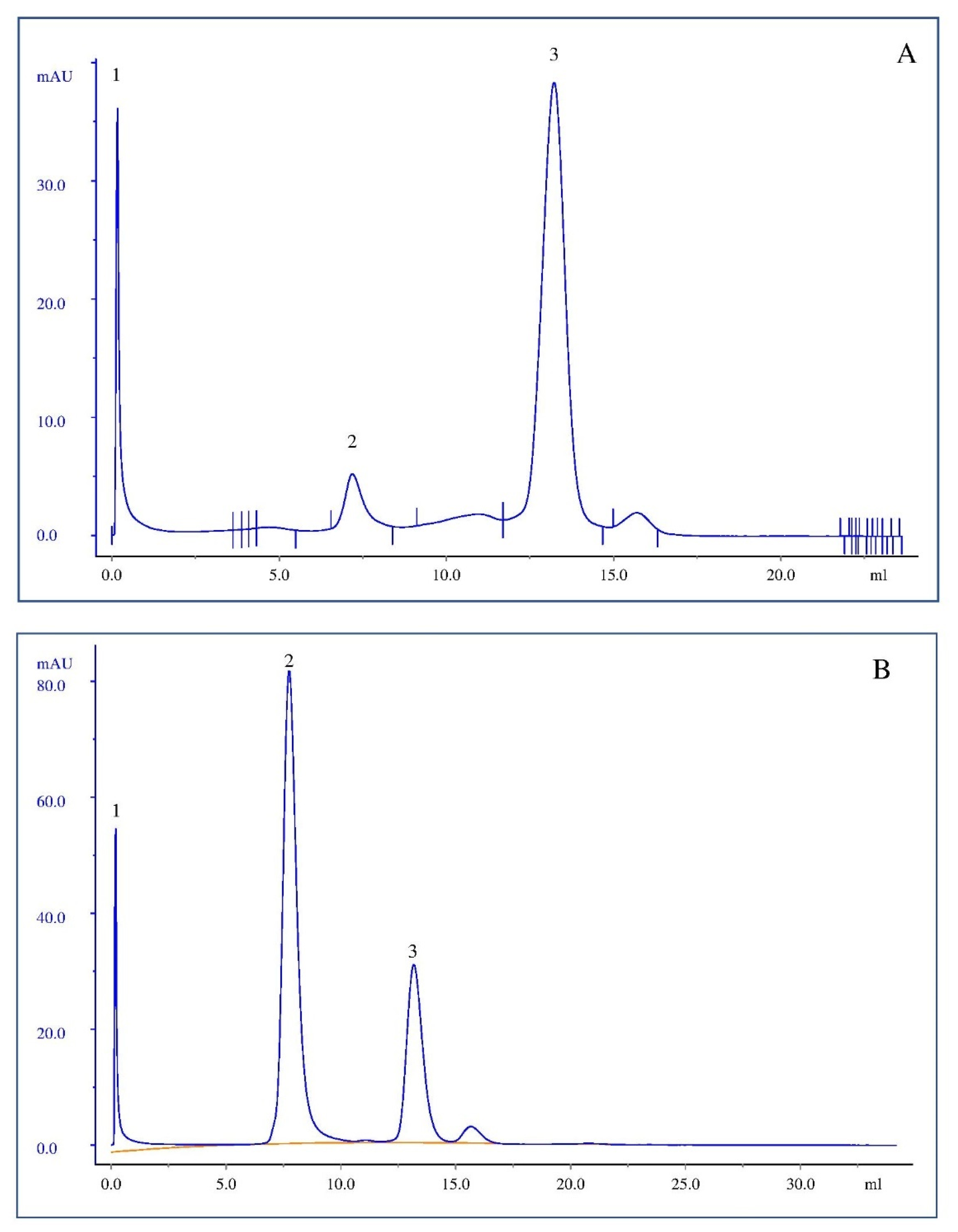
3.4. Transmission Electronic Microscopy (TEM) and Cryogenic Electron Microscopy (Cryo-EM)
3.5. Liposomes Permeability to Asparagine
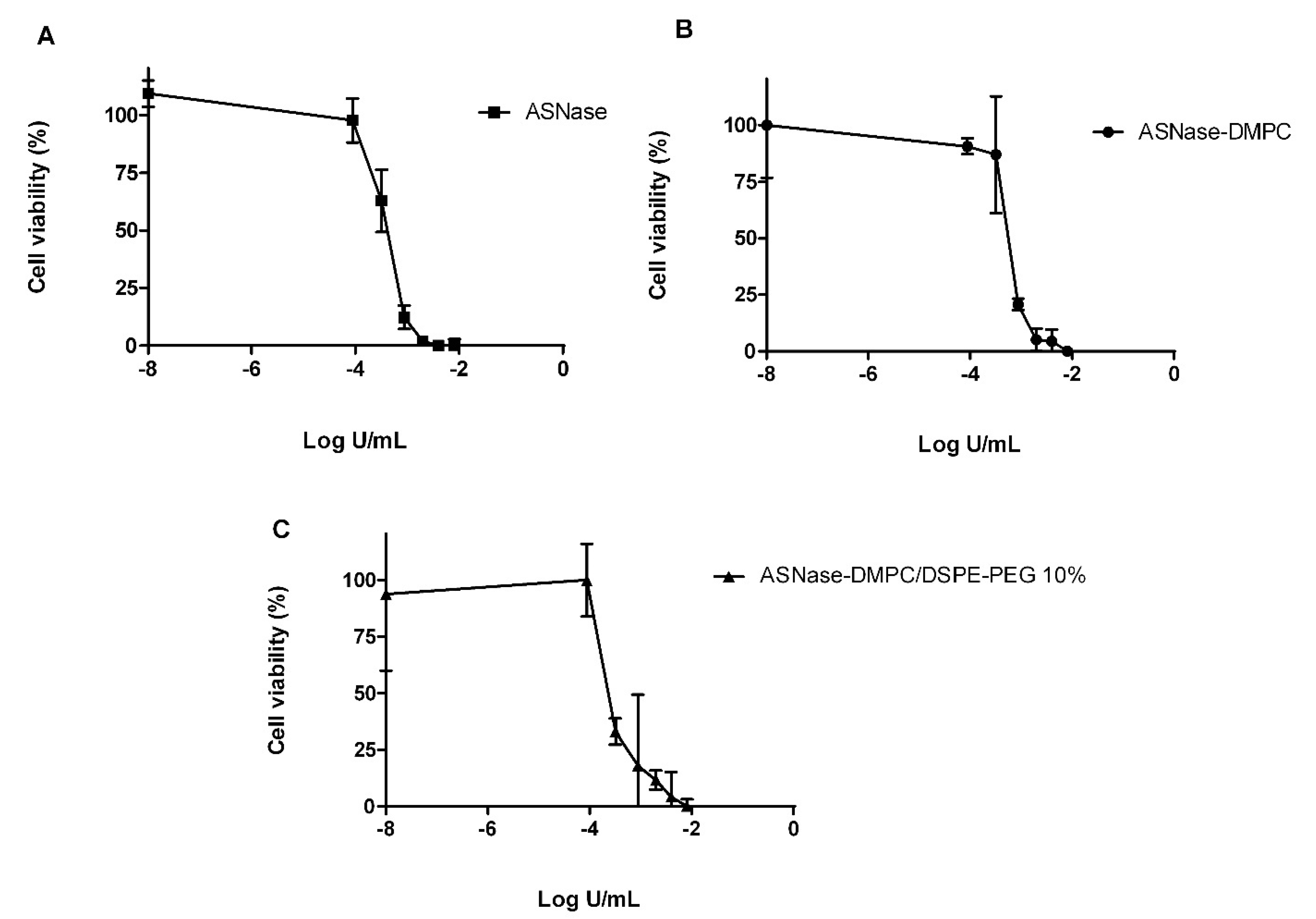
3.6. In Vitro Cytotoxicity against MOLT-4 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Terwilliger, T.; Abdul-Hay, M. Acute Lymphoblastic Leukemia: A Comprehensive Review and 2017 Update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.P.M.; Costa, I.M.; Molino, J.V.D.; Leite, M.S.M.; Pimenta, M.V.; Coutinho, J.A.P.; Pessoa, A.; Ventura, S.P.M.; Lopes, A.M.; Monteiro, G. Heterologous Expression and Purification of Active L-Asparaginase I of Saccharomyces Cerevisiae in Escherichia Coli Host. Biotechnol. Prog. 2017, 33, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Story, M.D.; Voehringer, D.W.; Stephens, L.C.; Meyn, R.E. L-Asparaginase Kills Lymphoma Cells by Apoptosis. Cancer Chemother. Pharmacol. 1993, 32, 129–133. [Google Scholar] [CrossRef]
- Egler, R.A.; Ahuja, S.P.; Matloub, Y. L-Asparaginase in the Treatment of Patients with Acute Lymphoblastic Leukemia. J. Pharmacol. Pharmacother. 2016, 7, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.H.G.; da Silva Fiúza, T.; de Morais, S.B.; de Souza, T.d.A.C.B.; Trevizani, R. Circumventing the Side Effects of L-Asparaginase. Biomed. Pharmacother. 2021, 139, 111616. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.P.; Reynolds, H.M.; Lumicisi, B.; Bryson, C.J. Immunogenicity of Protein Therapeutics: The Key Causes, Consequences and Challenges. Self Nonself 2010, 1, 314–322. [Google Scholar] [CrossRef]
- Cachumba, J.J.M.; Antunes, F.A.F.; Peres, G.F.D.; Brumano, L.P.; Santos, J.C.D.; Da Silva, S.S. Current Applications and Different Approaches for Microbial L-Asparaginase Production. Brazilian J. Microbiol. 2016, 47 (Suppl. S1), 77–85. [Google Scholar] [CrossRef]
- Apolinário, A.C.; Pachioni-Vasconcelos, J.A.; Pessoa, A., Jr.; Rangel-Yagui, C.O. Polimerossomos versus lipossomos: A evolução da “bala mágica”. Química Nova 2017, 40, 810–817. [Google Scholar] [CrossRef]
- Din, F.U.; Aman, W.; Ullah, I.; Qureshi, O.S.; Mustapha, O.; Shafique, S.; Zeb, A. Effective Use of Nanocarriers as Drug Delivery Systems for the Treatment of Selected Tumors. Int. J. Nanomed. 2017, 12, 7291–7309. [Google Scholar] [CrossRef]
- Chevalier, Y.; Zemb, T. The Structure of Micelles and Microemulsions. Rep. Prog. Phys. 1990, 53, 279–371. [Google Scholar] [CrossRef]
- Chamundeeswari, M.; Jeslin, J.; Verma, M.L. Nanocarriers for Drug Delivery Applications. Environ. Chem. Lett. 2019, 17, 849–865. [Google Scholar] [CrossRef]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in Cancer Therapy: Challenges, Opportunities, and Clinical Applications. J. Control. Release 2015, 200, 138–157. [Google Scholar] [CrossRef]
- Milla, P.; Dosio, F.; Cattel, L. PEGylation of Proteins and Liposomes: A Powerful and Flexible Strategy to Improve the Drug Delivery. Curr. Drug Metab. 2012, 13, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, D.; Calandra, P.; Barreca, D.; Magazù, S.; Kiselev, M.A. Soft Interaction in Liposome Nanocarriers for Therapeutic Drug Delivery. Nanomaterials 2016, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Franco, Y.L.; Vaidya, T.R.; Ait-Oudhia, S. Anticancer and Cardio-Protective Effects of Liposomal Doxorubicin in the Treatment of Breast Cancer. Breast Cancer (Dove Med. Press) 2018, 10, 131–141. [Google Scholar] [CrossRef]
- Stewart, S.; Jablonowski, H.; Goebel, F.D.; Arasteh, K.; Spittle, M.; Rios, A.; Aboulafia, D.; Galleshaw, J.; Dezube, B.J. Randomized Comparative Trial of Pegylated Liposomal Doxorubicin versus Bleomycin and Vincristine in the Treatment of AIDS-Related Kaposi’s Sarcoma. International Pegylated Liposomal Doxorubicin Study Group. J. Clin. Oncol. 1998, 16, 683–691. [Google Scholar] [CrossRef]
- Santos, J.H.P.M.; Feitosa, V.A.; Meneguetti, G.P.; Carretero, G.; Coutinho, J.A.P.; Ventura, S.P.M.; Rangel-Yagui, C.O. Lysine-PEGylated Cytochrome C with Enhanced Shelf-Life Stability. Biosensors 2022, 12, 94. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, H.; Wu, J.; Wang, Z.; Sun, H.; Yuan, J.; Chen, S. A Novel Zwitterionic Copolymer with a Short Poly(Methyl Acrylic Acid) Block for Improving Both Conjugation and Separation Efficiency of a Protein without Losing Its Bioactivity. J. Mater. Chem. B 2013, 1, 2482–2488. [Google Scholar] [CrossRef]
- Tsao, C.; Zhang, P.; Yuan, Z.; Dong, D.; Wu, K.; Niu, L.; McMullen, P.; Luozhong, S.; Hung, H.-C.; Cheng, Y.-H.; et al. Zwitterionic Polymer Conjugated Glucagon-like Peptide-1 for Prolonged Glycemic Control. Bioconjug. Chem. 2020, 31, 1812–1819. [Google Scholar] [CrossRef]
- Grenier, P.; de Oliveira, P.; Viana, I.M.; Lima, E.M.; Bertrand, N. Anti-Polyethylene Glycol Antibodies Alter the Protein Corona Deposited on Nanoparticles and the Physiological Pathways Regulating Their Fate in Vivo. J. Control. Release 2018, 287, 121–131. [Google Scholar] [CrossRef]
- Nishimura, T.; Akiyoshi, K. Biotransporting Biocatalytic Reactors toward Therapeutic Nanofactories. Adv. Sci. 2018, 5, 1800801. [Google Scholar] [CrossRef] [PubMed]
- Municoy, S.; Bellino, M.G. A Liposome-Actuated Enzyme System and Its Capability as a Self-Biomineralized Silica Nanoreactor. RSC Adv. 2016, 7, 67–70. [Google Scholar] [CrossRef]
- Kim, K.T.; Meeuwissen, S.A.; Nolte, R.J.M.; Hest, J.C.M. van Smart Nanocontainers and Nanoreactors. Nanoscale 2010, 2, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Bueno, C.Z.; Apolinário, A.C.; Duro-Castano, A.; Poma, A.; Pessoa, A.; Rangel-Yagui, C.O.; Battaglia, G. L-Asparaginase Encapsulation into Asymmetric Permeable Polymersomes. ACS Macro Lett. 2020, 9, 1471–1477. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.E.M.; Gaspar, M.M.; Lopes, F.; Jorge, J.S.; Perez-Soler, R. Liposomal L-Asparaginase: In Vitro Evaluation. Int. J. Pharm. 1993, 96, 67–77. [Google Scholar] [CrossRef]
- Jorge, J.C.S.; Perez-Soler, R.; Morais, J.G.; Cruz, M.E.M. Liposomal Palmitoyl-L-Asparaginase: Characterization and Biological Activity. Cancer Chemother. Pharmacol. 1994, 34, 230–234. [Google Scholar] [CrossRef]
- Gaspar, M.M.; Perez-Soler, R.; Cruz, M.E. Biological Characterization of L-Asparaginase Liposomal Formulations. Cancer Chemother. Pharmacol. 1996, 38, 373–377. [Google Scholar] [CrossRef]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. In Liposomes: Methods and Protocols; D’Souza, G.G.M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 17–22. ISBN 978-1-4939-6591-5. [Google Scholar]
- Robertson, J.D.; Rizzello, L.; Avila-Olias, M.; Gaitzsch, J.; Contini, C.; Magoń, M.S.; Renshaw, S.A.; Battaglia, G. Purification of Nanoparticles by Size and Shape. Sci. Rep. 2016, 6, 27494. [Google Scholar] [CrossRef]
- Bartenstein, J.E.; Robertson, J.; Battaglia, G.; Briscoe, W.H. Stability of Polymersomes Prepared by Size Exclusion Chromatography and Extrusion. Colloids Surf. A: Physicochem. Eng. Asp. 2016, 506, 739–746. [Google Scholar] [CrossRef]
- Kontogiannopoulos, K.N.; Tsermentseli, S.K.; Assimopoulou, A.N.; Papageorgiou, V.P. Sterically Stabilized Liposomes as a Potent Carrier for Shikonin. J. Liposome Res. 2014, 24, 230–240. [Google Scholar] [CrossRef]
- Imada, A.; Igarasi, S.; Nakahama, K.; Isono, M. Asparaginase and Glutaminase Activities of Micro-Organisms. J. Gen. Microbiol. 1973, 76, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Chibowski, E.; Szcześ, A. Zeta Potential and Surface Charge of DPPC and DOPC Liposomes in the Presence of PLC Enzyme. Adsorption 2016, 22, 755–765. [Google Scholar] [CrossRef]
- Lebègue, E.; Barrière, F.; Bard, A.J. Lipid Membrane Permeability of Synthetic Redox DMPC Liposomes Investigated by Single Electrochemical Collisions. Anal. Chem. 2020, 92, 2401–2408. [Google Scholar] [CrossRef] [PubMed]
- Tsermentseli, S.K.; Kontogiannopoulos, K.N.; Papageorgiou, V.P.; Assimopoulou, A.N. Comparative Study of PEGylated and Conventional Liposomes as Carriers for Shikonin. Fluids 2018, 3, 36. [Google Scholar] [CrossRef]
- Sandström, M.C.; Johansson, E.; Edwards, K. Structure of Mixed Micelles Formed in PEG-Lipid/Lipid Dispersions. Langmuir 2007, 23, 4192–4198. [Google Scholar] [CrossRef]
- Viitala, L.; Pajari, S.; Gentile, L.; Määttä, J.; Gubitosi, M.; Deska, J.; Sammalkorpi, M.; Olsson, U.; Murtomäki, L. Shape and Phase Transitions in a PEGylated Phospholipid System. Langmuir 2019, 35, 3999–4010. [Google Scholar] [CrossRef]
- Edwards, K.; Johnsson, M.; Karlsson, G.; Silvander, M. Effect of Polyethyleneglycol-Phospholipids on Aggregate Structure in Preparations of Small Unilamellar Liposomes. Biophys. J. 1997, 73, 258–266. [Google Scholar] [CrossRef]
- Marsh, D.; Bartucci, R.; Sportelli, L. Lipid Membranes with Grafted Polymers: Physicochemical Aspects. Biochim. Biophys. Acta (BBA)—Biomembr. 2003, 1615, 33–59. [Google Scholar] [CrossRef]
- Samuelsson, E.; Shen, H.; Blanco, E.; Ferrari, M.; Wolfram, J. Contribution of Kupffer Cells to Liposome Accumulation in the Liver. Colloids Surf. B Biointerfaces 2017, 158, 356–362. [Google Scholar] [CrossRef]
- Litzinger, D.C.; Buiting, A.M.; van Rooijen, N.; Huang, L. Effect of Liposome Size on the Circulation Time and Intraorgan Distribution of Amphipathic Poly(Ethylene Glycol)-Containing Liposomes. Biochim. Biophys. Acta 1994, 1190, 99–107. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Maity, P.; Saha, B.; Kumar, G.S.; Karmakar, S. Binding of Monovalent Alkali Metal Ions with Negatively Charged Phospholipid Membranes. Biochim. Biophys. Acta (BBA)—Biomembr. 2016, 1858, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Disalvo, E.A.; Bouchet, A.M. Electrophoretic Mobility and Zeta Potential of Liposomes Due to Arginine and Polyarginine Adsorption. Colloids Surf. A Physicochem. Eng. Asp. 2014, 440, 170–174. [Google Scholar] [CrossRef]
- Smith, M.C.; Crist, R.M.; Clogston, J.D.; McNeil, S.E. Zeta Potential: A Case Study of Cationic, Anionic, and Neutral Liposomes. Anal. Bioanal. Chem. 2017, 409, 5779–5787. [Google Scholar] [CrossRef]
- Anderson, M.; Omri, A. The Effect of Different Lipid Components on the In Vitro Stability and Release Kinetics of Liposome Formulations. Drug Deliv. 2004, 11, 33–39. [Google Scholar] [CrossRef]
- Guldiken, B.; Gibis, M.; Boyacioglu, D.; Capanoglu, E.; Weiss, J. Physical and Chemical Stability of Anthocyanin-Rich Black Carrot Extract-Loaded Liposomes during Storage. Food Res. Int. 2018, 108, 491–497. [Google Scholar] [CrossRef]
- Muppidi, K.; Pumerantz, A.S.; Wang, J.; Betageri, G. Development and Stability Studies of Novel Liposomal Vancomycin Formulations. ISRN Pharm. 2012, 2012, e636743. [Google Scholar] [CrossRef]
- Hincha, D.K. Effects of Alpha-Tocopherol (Vitamin E) on the Stability and Lipid Dynamics of Model Membranes Mimicking the Lipid Composition of Plant Chloroplast Membranes. FEBS Lett. 2008, 582, 3687–3692. [Google Scholar] [CrossRef]
- Urano, S.; Kitahara, M.; Kato, Y.; Hasegawa, Y.; Matsuo, M. Membrane Stabilizing Effect of Vitamin E: Existence of a Hydrogen Bond between α-Tocopherol and Phospholipids in Bilayer Liposomes. J. Nutr. Sci. Vitaminol. 1990, 36, 513–519. [Google Scholar] [CrossRef]
- Okamoto, Y.; Taguchi, K.; Yamasaki, K.; Sakuragi, M.; Kuroda, S.; Otagiri, M. Albumin-Encapsulated Liposomes: A Novel Drug Delivery Carrier With Hydrophobic Drugs Encapsulated in the Inner Aqueous Core. J. Pharm. Sci. 2018, 107, 436–445. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chierico, L.; Little, D.; Patikarnmonthon, N.; Yang, Z.; Azzouz, M.; Madsen, J.; Armes, S.P.; Battaglia, G. Encapsulation of Biomacromolecules within Polymersomes by Electroporation. Angew. Chem. Int. Ed. 2012, 51, 11122–11125. [Google Scholar] [CrossRef] [PubMed]
- Franken, L.E.; Boekema, E.J.; Stuart, M.C.A. Transmission Electron Microscopy as a Tool for the Characterization of Soft Materials: Application and Interpretation. Adv. Sci. 2017, 4, 1600476. [Google Scholar] [CrossRef] [PubMed]
- Bibi, S.; Kaur, R.; Henriksen-Lacey, M.; McNeil, S.E.; Wilkhu, J.; Lattmann, E.; Christensen, D.; Mohammed, A.R.; Perrie, Y. Microscopy Imaging of Liposomes: From Coverslips to Environmental SEM. Int. J. Pharm. 2011, 417, 138–150. [Google Scholar] [CrossRef]
- Tahara, Y.; Fujiyoshi, Y. A New Method to Measure Bilayer Thickness: Cryo-Electron Microscopy of Frozen Hydrated Liposomes and Image Simulation. Micron 1994, 25, 141–149. [Google Scholar] [CrossRef]
- Karve, S.; Bajagur Kempegowda, G.; Sofou, S. Heterogeneous Domains and Membrane Permeability in Phosphatidylcholine−Phosphatidic Acid Rigid Vesicles as a Function of PH and Lipid Chain Mismatch. Langmuir 2008, 24, 5679–5688. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, A.R.; Scott, M.J.; Kennedy, N.I.; Jones, M.N. Effect of Grafted Polyethylene Glycol (PEG) on the Size, Encapsulation Efficiency and Permeability of Vesicles. Biochim. Biophys. Acta 2000, 1463, 167–178. [Google Scholar] [CrossRef]
- Nikolova, A.N.; Jones, M.N. Effect of Grafted PEG-2000 on the Size and Permeability of Vesicles. Biochim. Biophys. Acta 1996, 1304, 120–128. [Google Scholar] [CrossRef]
- Chakrabarti, A.C.; Deamer, D.W. Permeability of Lipid Bilayers to Amino Acids and Phosphate. Biochim. Biophys. Acta (BBA)—Biomembr. 1992, 1111, 171–177. [Google Scholar] [CrossRef]
- Stepniewski, M.; Pasenkiewicz-Gierula, M.; Róg, T.; Danne, R.; Orlowski, A.; Karttunen, M.; Urtti, A.; Yliperttula, M.; Vuorimaa, E.; Bunker, A. Study of PEGylated Lipid Layers as a Model for PEGylated Liposome Surfaces: Molecular Dynamics Simulation and Langmuir Monolayer Studies. Langmuir 2011, 27, 7788–7798. [Google Scholar] [CrossRef]
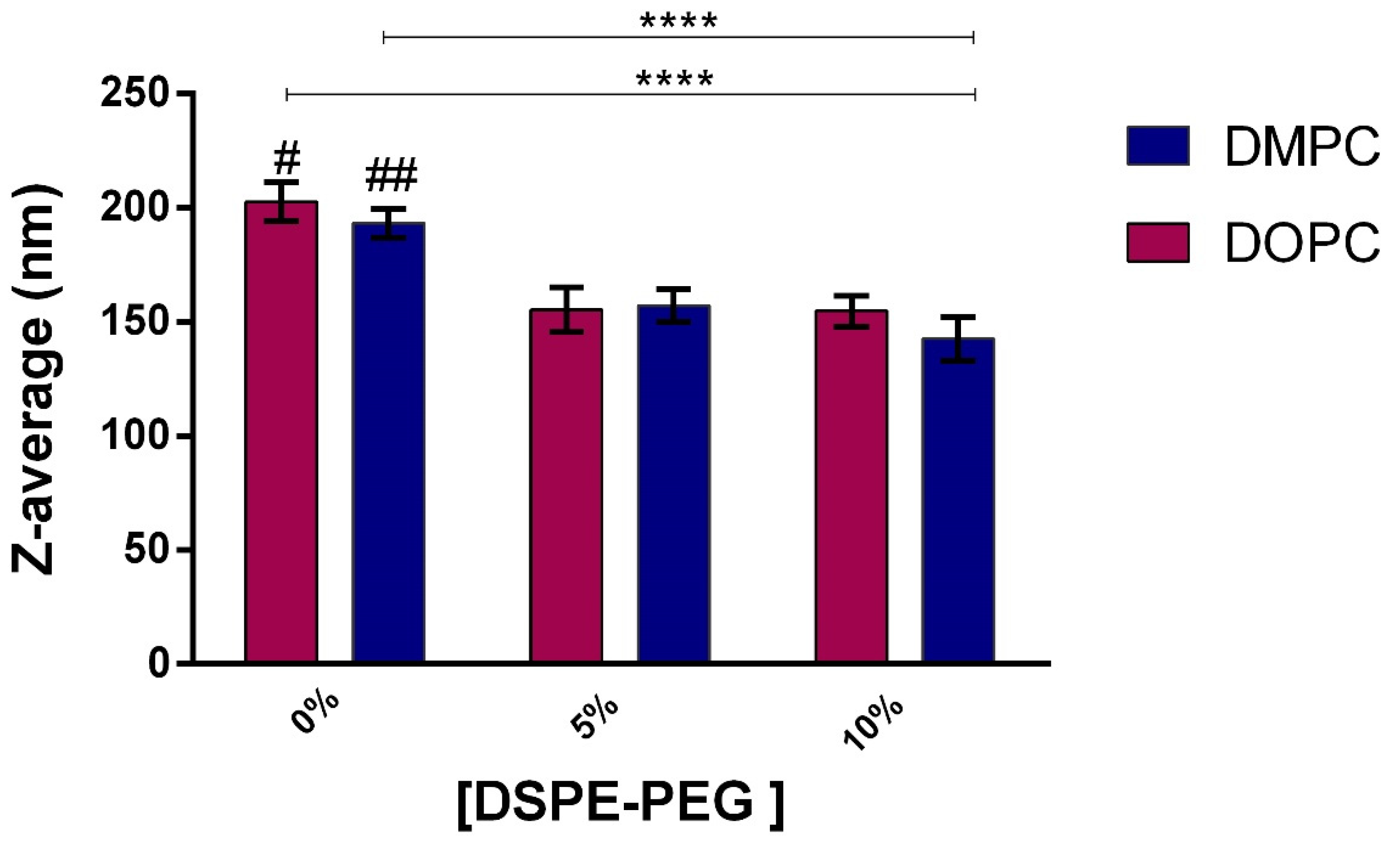
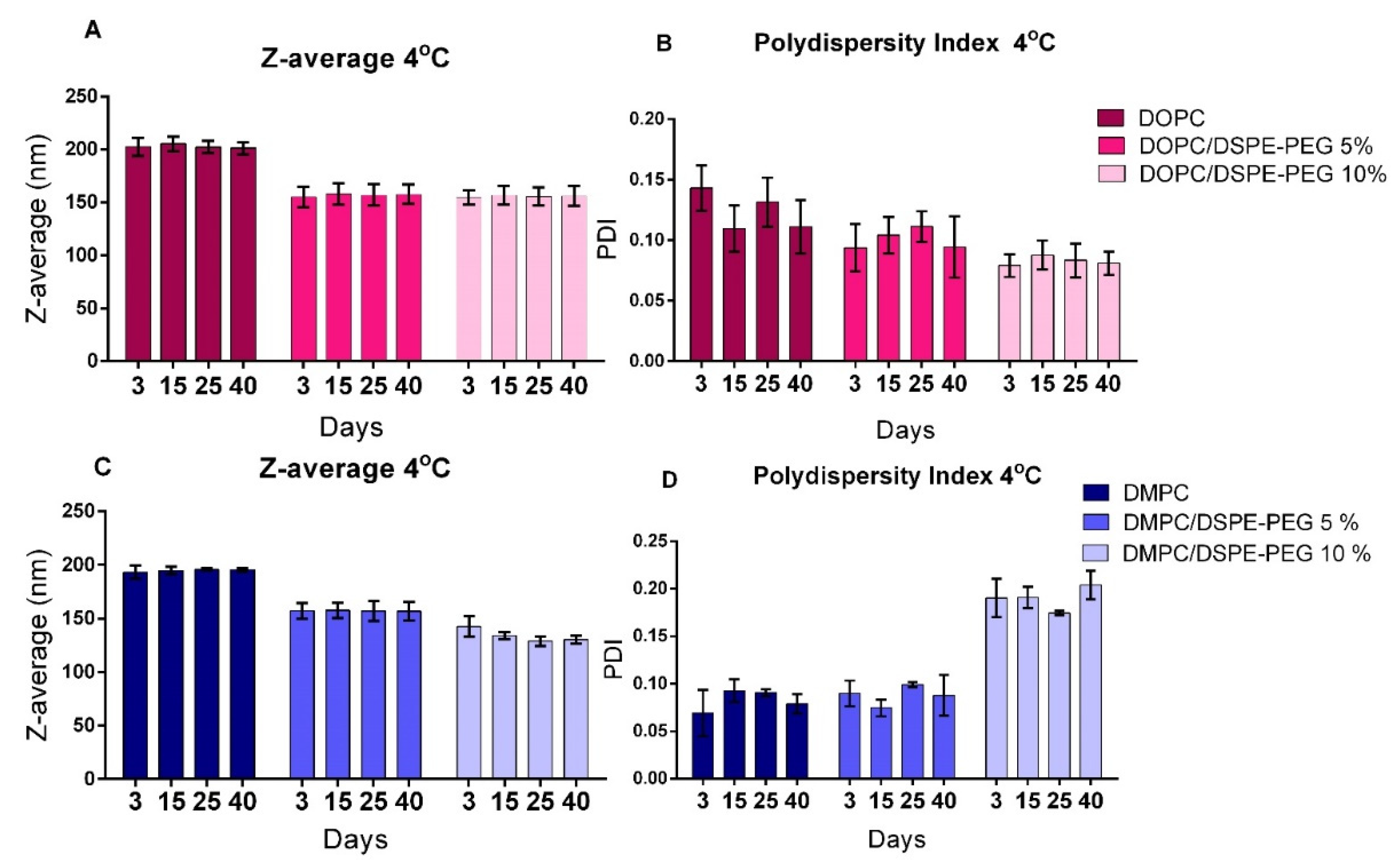
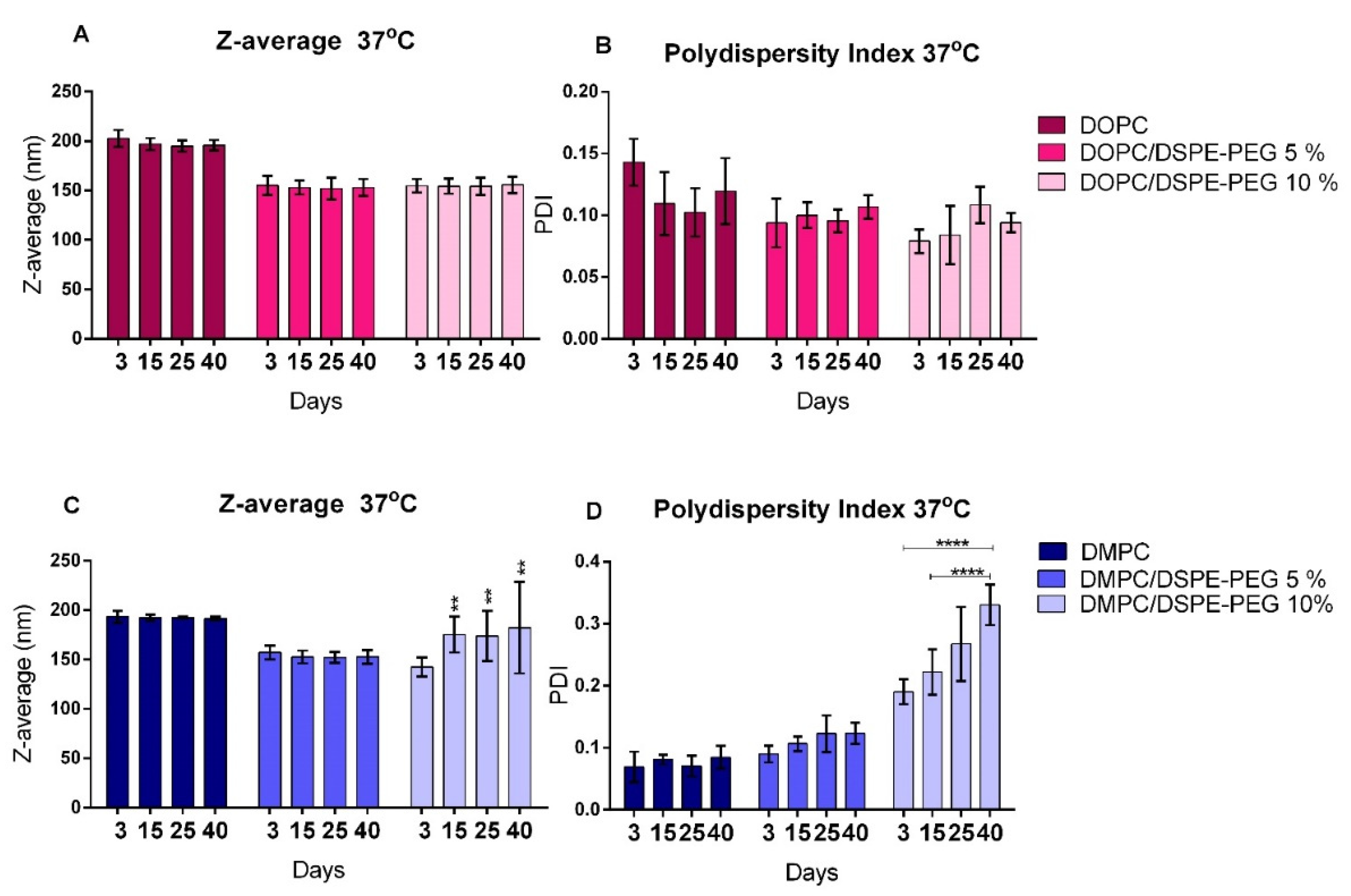

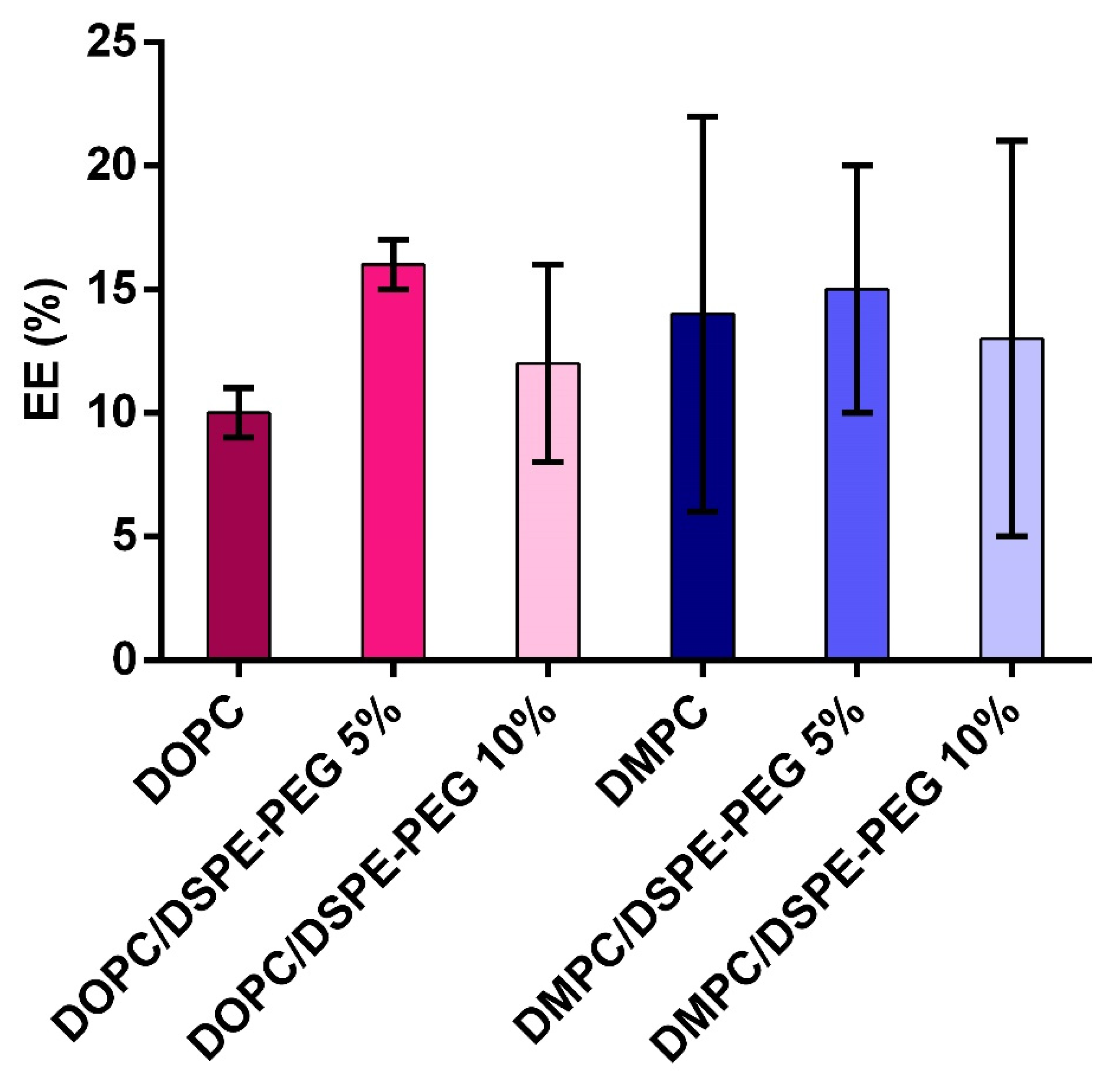
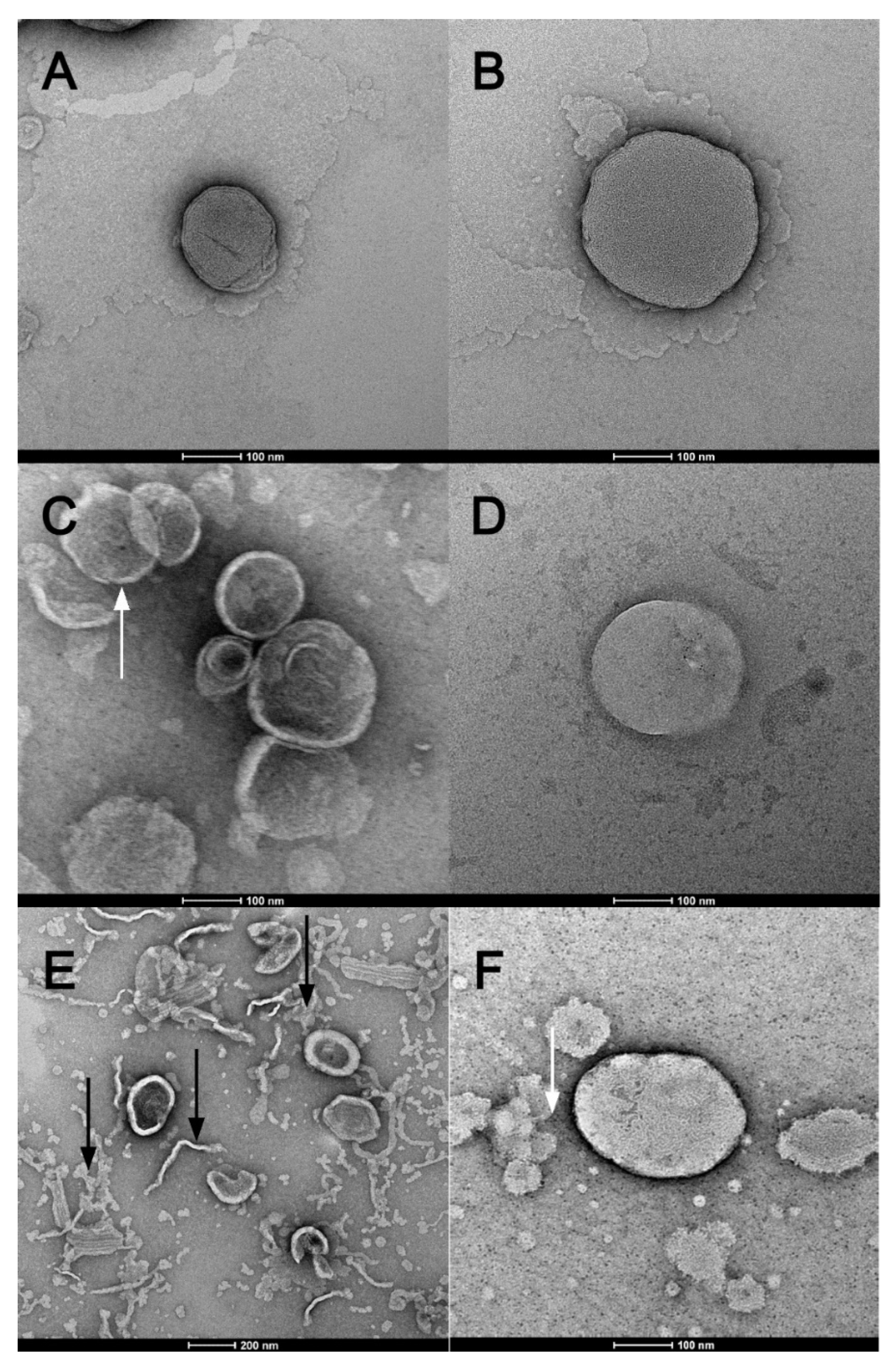
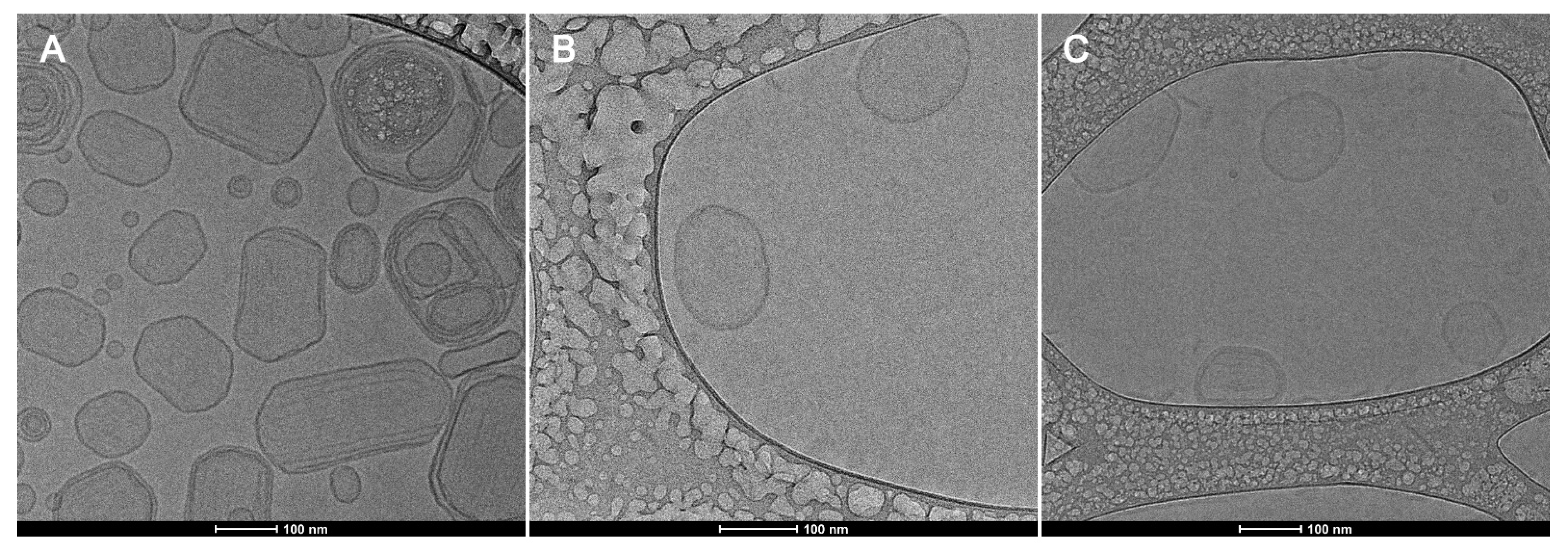
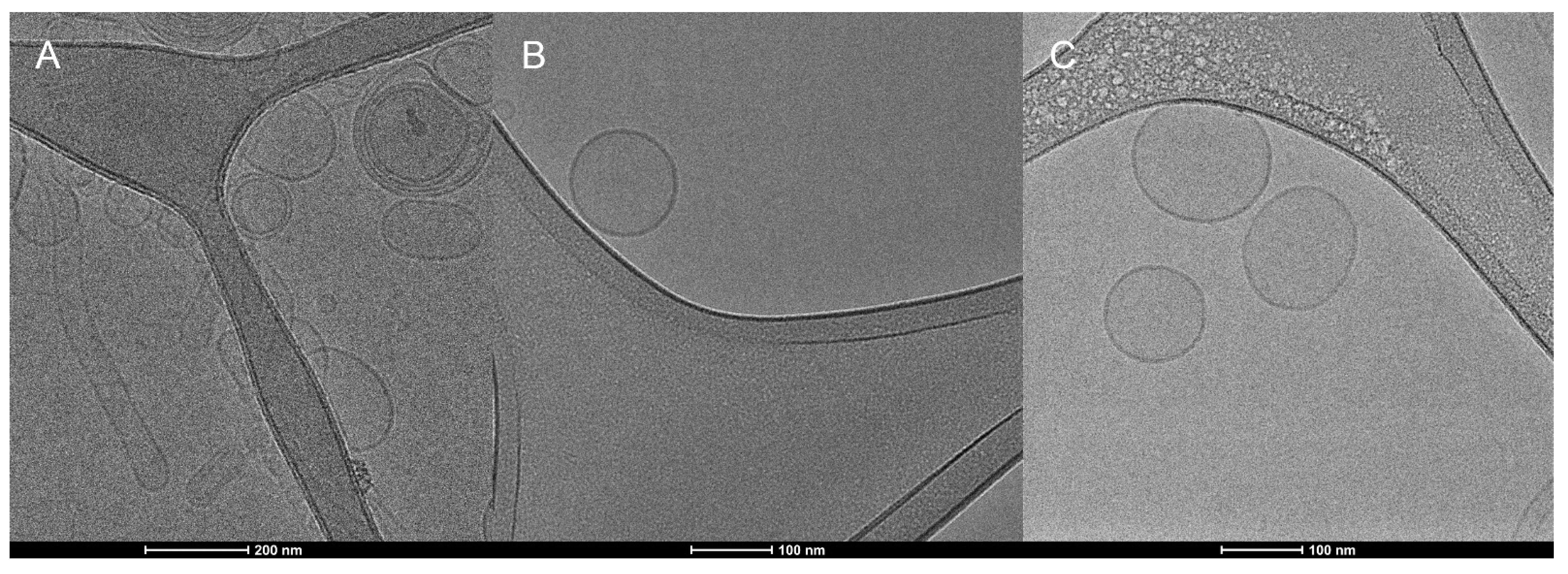

| Formulation | Z-Average (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| DOPC | 202 ± 9 | 0.143 ± 0.019 | −3.57 |
| DOPC/DSPE-PEG 5% | 155 ± 10 | 0.084 ± 0.014 | −3.69 |
| DOPC/DSPE-PEG 10% | 154 ± 7 | 0.079 ± 0.009 | −5.89 |
| DMPC | 193 ± 6 | 0.069 ± 0.024 | −4.55 |
| DMPC/DSPE-PEG 5% | 157 ± 7 | 0.090 ± 0.014 | −3.73 |
| DMPC/DSPE-PEG 10% | 142 ± 10 | 0.190 ± 0.020 | −2.57 |
| Formulations | Average Size (nm) | Bilayer Thickness Average (nm) |
|---|---|---|
| DOPC | 114 | 4.7 |
| DOPC/DSPE-PEG 5% | 140 | 6.0 |
| DMPC | 157 | 4.9 |
| DMPC/DSPE-PEG 10% | 126 | 5.8 |
| ASNase-DOPC/DSPE-PEG 5% | 148 | 5.8 |
| ASNase-DMPC/DSPE-PEG 10% | 145 | 6.6 |
| Formulations | IC50 (U/mL) |
|---|---|
| ASNase | 0.000376 ± 0.000027 |
| ASNase-DMPC | 0.000548 ± 0.000044 |
| ASNase-DMPC/DSPE-PEG 10% | 0.000267 ± 0.000029 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza Guimarães, M.; Cachumba, J.J.M.; Bueno, C.Z.; Torres-Obreque, K.M.; Lara, G.V.R.; Monteiro, G.; Barbosa, L.R.S.; Pessoa, A., Jr.; Rangel-Yagui, C.d.O. Peg-Grafted Liposomes for L-Asparaginase Encapsulation. Pharmaceutics 2022, 14, 1819. https://doi.org/10.3390/pharmaceutics14091819
de Souza Guimarães M, Cachumba JJM, Bueno CZ, Torres-Obreque KM, Lara GVR, Monteiro G, Barbosa LRS, Pessoa A Jr., Rangel-Yagui CdO. Peg-Grafted Liposomes for L-Asparaginase Encapsulation. Pharmaceutics. 2022; 14(9):1819. https://doi.org/10.3390/pharmaceutics14091819
Chicago/Turabian Stylede Souza Guimarães, Marina, Jorge Javier Muso Cachumba, Cecilia Zorzi Bueno, Karin Mariana Torres-Obreque, Grace Verónica Ruiz Lara, Gisele Monteiro, Leandro Ramos Souza Barbosa, Adalberto Pessoa, Jr., and Carlota de Oliveira Rangel-Yagui. 2022. "Peg-Grafted Liposomes for L-Asparaginase Encapsulation" Pharmaceutics 14, no. 9: 1819. https://doi.org/10.3390/pharmaceutics14091819
APA Stylede Souza Guimarães, M., Cachumba, J. J. M., Bueno, C. Z., Torres-Obreque, K. M., Lara, G. V. R., Monteiro, G., Barbosa, L. R. S., Pessoa, A., Jr., & Rangel-Yagui, C. d. O. (2022). Peg-Grafted Liposomes for L-Asparaginase Encapsulation. Pharmaceutics, 14(9), 1819. https://doi.org/10.3390/pharmaceutics14091819







