pH-Responsive Lipid Nanoparticles Achieve Efficient mRNA Transfection in Brain Capillary Endothelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of ssPalm-LNP Encapsulating GFP-Encoding mRNA
2.3. Preparation of Lipofectamine/GFP-Encoding mRNA Complex (LFN)
2.4. Flow Cytometry Analysis of Cellular Uptake and Gene Expression
2.5. Observation of Cellular Uptake and Gene Expression with Con-Focal Laser Scanning Microscopy
2.6. Evaluation of Cytotoxicity in mRNA Transfection Using ssPalm-LNP and LFN in hCMEC/D3 Cells
2.7. SWATH-MS Analysis in hCMEC/D3 Cells Treated with ssPalm LNP and LFN
3. Results
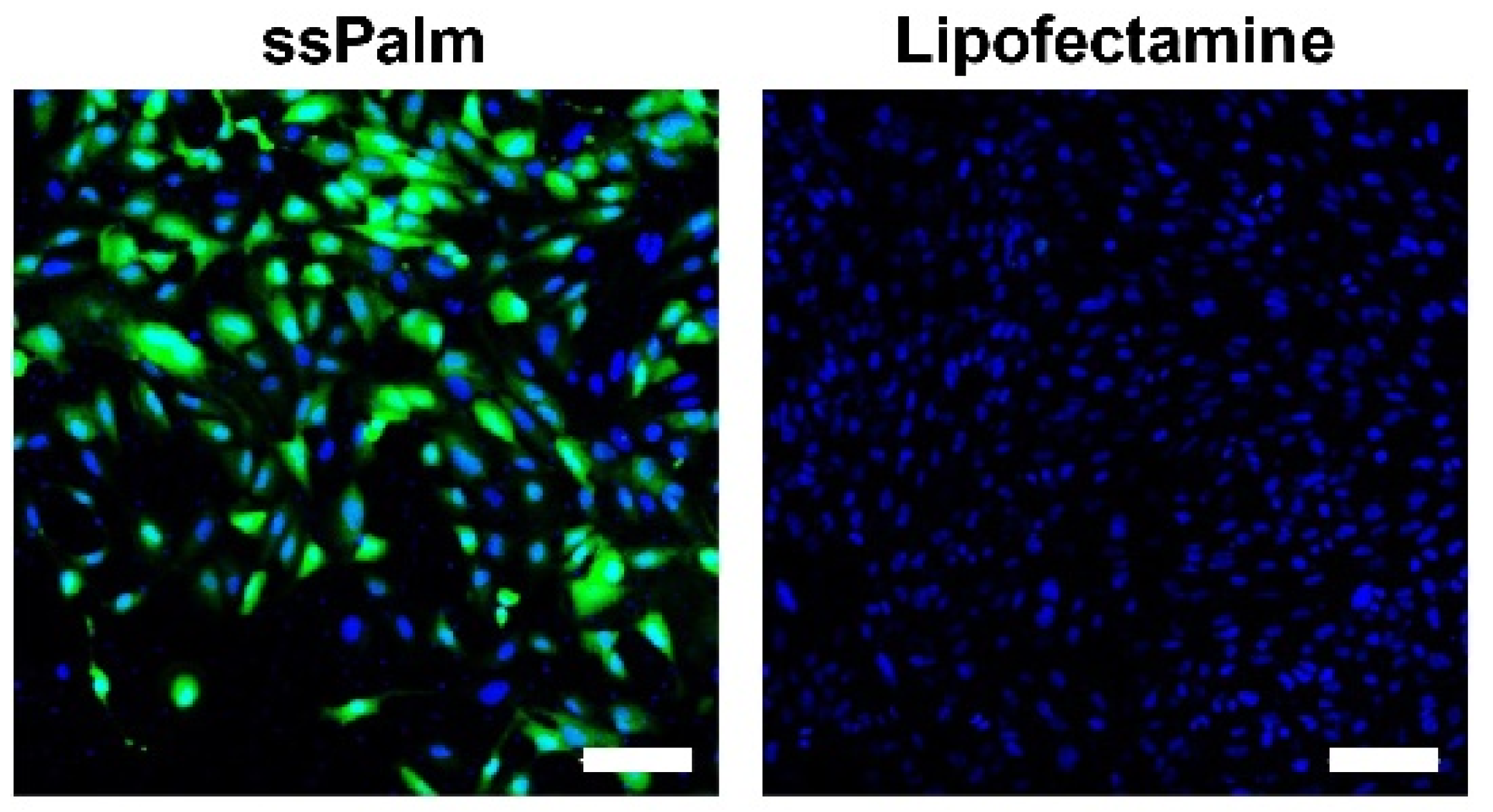
3.1. The mRNA Introduced by the ssPalm-LNP Is Homogeneously Translated in hCMEC/D3 Cells
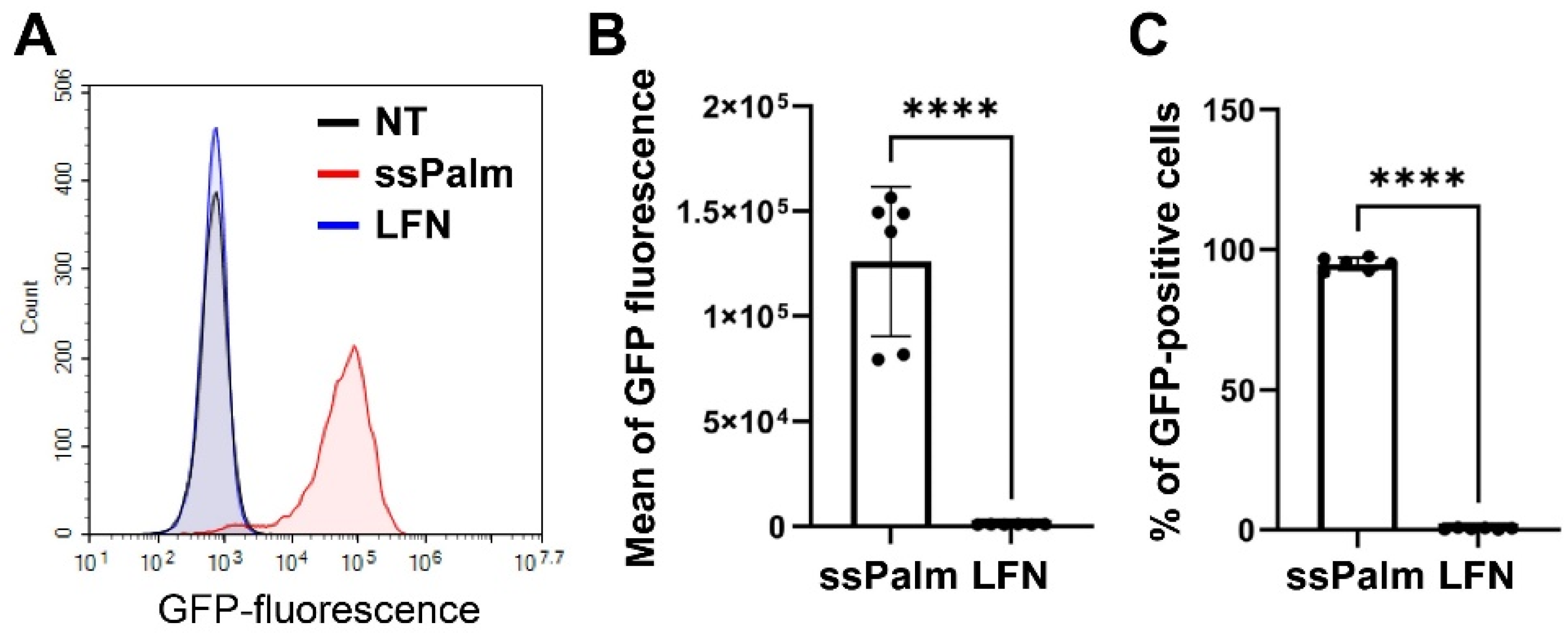
3.2. Higher Amount of the ssPalm-LNP Internalizes hCMEC/D3 Cells Than LFN
3.3. mRNA Transfection into hCMEC/D3 Cells by ssPalm-LNP Is Significantly Less Cytotoxic Than LFN
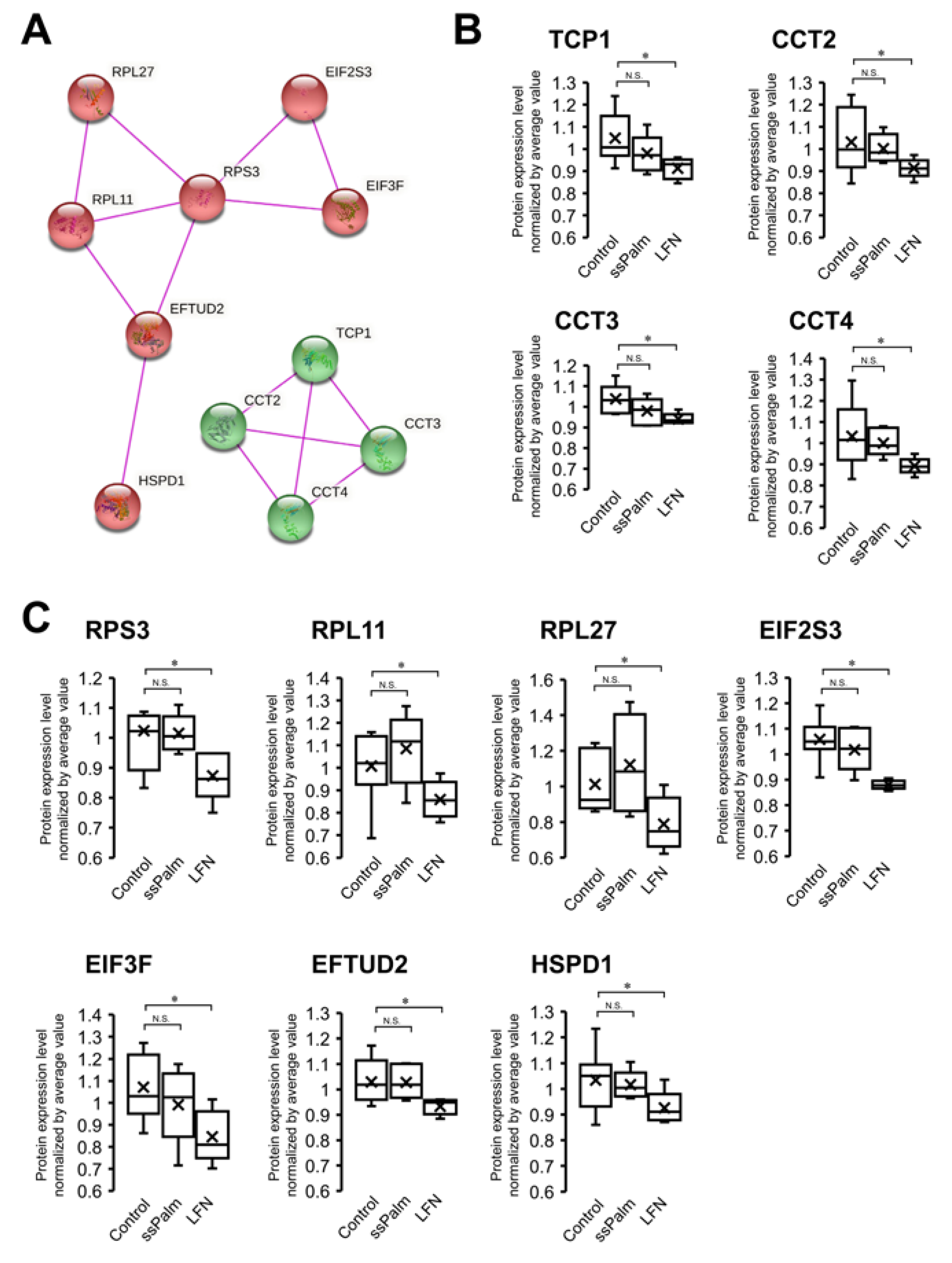
3.4. ssPalm LNP Has Significantly Fewer Adverse Effects than LFN
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohtsuki, S.; Ikeda, C.; Uchida, Y.; Sakamoto, Y.; Miller, F.; Glacial, F.; Decleves, X.; Scherrmann, J.M.; Couraud, P.O.; Kubo, Y.; et al. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Mol. Pharm. 2013, 10, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, T.; Sako, D.; Tega, Y.; Debori, Y.; Tomihara, Y.; Aoyama, K.; Kubo, Y.; Amano, N.; Deguchi, Y. Construction and Functional Evaluation of a Three-Dimensional Blood-Brain Barrier Model Equipped with Human Induced Pluripotent Stem Cell-Derived Brain Microvascular Endothelial Cells. Pharm. Res. 2022, 39, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, A.; Thomsen, L.B.; Thomsen, M.S.; Lichota, J.; Fazakas, C.; Krizbai, I.; Moos, T. Transfection of brain capillary endothelial cells in primary culture with defined blood-brain barrier properties. Fluids Barriers CNS 2015, 12, 19. [Google Scholar] [CrossRef]
- Huber, O.; Brunner, A.; Maier, P.; Kaufmann, R.; Couraud, P.O.; Cremer, C.; Fricker, G. Localization microscopy (SPDM) reveals clustered formations of P-glycoprotein in a human blood-brain barrier model. PLoS ONE 2012, 7, e44776. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, Y.; Guo, M.; Wang, X.; Li, J.; Wang, Y.; Ye, L.; Dai, M.; Zhou, L.; Persidsky, Y.; Ho, W. TLR3 activation efficiency by high or low molecular mass poly I:C. Innate Immun. 2013, 19, 184–192. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, C.; Wang, Z.; Shen, J.; Xiang, P.; Chen, X.; Nan, J.; Lin, Y. Lipofectamine 2000/siRNA complexes cause endoplasmic reticulum unfolded protein response in human endothelial cells. J. Cell. Physiol. 2019, 234, 21166–21181. [Google Scholar] [CrossRef]
- Tanaka, H.; Sakurai, Y.; Anindita, J.; Akita, H. Development of lipid-like materials for RNA delivery based on intracellular environment-responsive membrane destabilization and spontaneous collapse. Adv. Drug Deliv. Rev. 2020, 154–155, 210–226. [Google Scholar] [CrossRef]
- Tanaka, H.; Miyama, R.; Sakurai, Y.; Tamagawa, S.; Nakai, Y.; Tange, K.; Yoshioka, H.; Akita, H. Improvement of mRNA Delivery Efficiency to a T Cell Line by Modulating PEG-Lipid Content and Phospholipid Components of Lipid Nanoparticles. Pharmaceutics 2021, 13, 2097. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 2012, 11, O111.016717. [Google Scholar] [CrossRef]
- Shimomura, K.; Okura, T.; Kato, S.; Couraud, P.O.; Schermann, J.M.; Terasaki, T.; Deguchi, Y. Functional expression of a proton-coupled organic cation (H+/OC) antiporter in human brain capillary endothelial cell line hCMEC/D3, a human blood-brain barrier model. Fluids Barriers CNS 2013, 10, 8. [Google Scholar] [CrossRef]
- Weksler, B.B.; Subileau, E.A.; Perriere, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P.; et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef]
- Tanaka, H.; Nakatani, T.; Furihata, T.; Tange, K.; Nakai, Y.; Yoshioka, H.; Harashima, H.; Akita, H. In Vivo Introduction of mRNA Encapsulated in Lipid Nanoparticles to Brain Neuronal Cells and Astrocytes via Intracerebroventricular Administration. Mol. Pharm. 2018, 15, 2060–2067. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Takahashi, T.; Konishi, M.; Takata, N.; Gomi, M.; Shirane, D.; Miyama, R.; Hagiwara, S.; Yamasaki, Y.; Sakurai, Y.; et al. Self-Degradable Lipid-Like Materials Based on “Hydrolysis accelerated by the intra-Particle Enrichment of Reactant (HyPER)” for Messenger RNA Delivery. Adv. Funct. Mater. 2020, 30, 1910575. [Google Scholar] [CrossRef]
- Jeffs, L.B.; Palmer, L.R.; Ambegia, E.G.; Giesbrecht, C.; Ewanick, S.; MacLachlan, I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 2005, 22, 362–372. [Google Scholar] [CrossRef]
- Sato, R.; Ohmori, K.; Umetsu, M.; Takao, M.; Tano, M.; Grant, G.; Porter, B.; Bet, A.; Terasaki, T.; Uchida, Y. An Atlas of the Quantitative Protein Expression of Anti-Epileptic-Drug Transporters, Metabolizing Enzymes and Tight Junctions at the Blood-Brain Barrier in Epileptic Patients. Pharmaceutics 2021, 13, 2122. [Google Scholar] [CrossRef]
- Tezuka, K.; Suzuki, M.; Sato, R.; Kawarada, S.; Terasaki, T.; Uchida, Y. Activation of Annexin A2 signaling at the blood-brain barrier in a mouse model of multiple sclerosis. J. Neurochem. 2022, 160, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Sasaki, H.; Terasaki, T. Establishment and validation of highly accurate formalin-fixed paraffin-embedded quantitative proteomics by heat-compatible pressure cycling technology using phase-transfer surfactant and SWATH-MS. Sci. Rep. 2020, 10, 11271. [Google Scholar] [CrossRef]
- Uchida, Y.; Higuchi, T.; Shirota, M.; Kagami, S.; Saigusa, D.; Koshiba, S.; Yasuda, J.; Tamiya, G.; Kuriyama, S.; Kinoshita, K.; et al. Identification and Validation of Combination Plasma Biomarker of Afamin, Fibronectin and Sex Hormone-Binding Globulin to Predict Pre-eclampsia. Biol. Pharm. Bull. 2021, 44, 804–815. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Sakurai, Y.; Kato, A.; Hida, Y.; Hamada, J.; Maishi, N.; Hida, K.; Harashima, H. Synergistic Enhancement of Cellular Uptake with CD44-Expressing Malignant Pleural Mesothelioma by Combining Cationic Liposome and Hyaluronic Acid-Lipid Conjugate. J. Pharm. Sci. 2019, 108, 3218–3224. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Gao, Y.; Zhu, J.; Wientjes, M.G.; Au, J.L. Relationships between liposome properties, cell membrane binding, intracellular processing, and intracellular bioavailability. AAPS J. 2011, 13, 585–597. [Google Scholar] [CrossRef]
- Mounkes, L.C.; Zhong, W.; Cipres-Palacin, G.; Heath, T.D.; Debs, R.J. Proteoglycans mediate cationic liposome-DNA complex-based gene delivery in vitro and in vivo. J. Biol. Chem. 1998, 273, 26164–26170. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Querbes, W.; De, S.; Qin, J.; Frank-Kamenetsky, M.; Jayaprakash, K.N.; Jayaraman, M.; Rajeev, K.G.; Cantley, W.L.; Dorkin, J.R.; et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010, 18, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Hatakeyama, H.; Sakurai, Y.; Hyodo, M.; Akita, H.; Harashima, H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J. Control. Release 2012, 163, 267–276. [Google Scholar] [CrossRef]
- Akita, H.; Noguchi, Y.; Hatakeyama, H.; Sato, Y.; Tange, K.; Nakai, Y.; Harashima, H. Molecular Tuning of a Vitamin E-Scaffold pH-Sensitive and Reductive Cleavable Lipid-like Material for Accelerated In Vivo Hepatic siRNA Delivery. ACS Biomater. Sci. Eng. 2015, 1, 834–844. [Google Scholar] [CrossRef]
- Ribecco-Lutkiewicz, M.; Sodja, C.; Haukenfrers, J.; Haqqani, A.S.; Ly, D.; Zachar, P.; Baumann, E.; Ball, M.; Huang, J.; Rukhlova, M.; et al. A novel human induced pluripotent stem cell blood-brain barrier model: Applicability to study antibody-triggered receptor-mediated transcytosis. Sci. Rep. 2018, 8, 1873. [Google Scholar] [CrossRef] [PubMed]
- Molino, Y.; David, M.; Varini, K.; Jabes, F.; Gaudin, N.; Fortoul, A.; Bakloul, K.; Masse, M.; Bernard, A.; Drobecq, L.; et al. Use of LDL receptor-targeting peptide vectors for in vitro and in vivo cargo transport across the blood-brain barrier. FASEB J. 2017, 31, 1807–1827. [Google Scholar] [CrossRef]
- Hanafy, A.S.; Dietrich, D.; Fricker, G.; Lamprecht, A. Blood-brain barrier models: Rationale for selection. Adv. Drug Deliv. Rev. 2021, 176, 113859. [Google Scholar] [CrossRef]
- Jagtiani, E.; Yeolekar, M.; Naik, S.; Patravale, V. In vitro blood brain barrier models: An overview. J. Control. Release 2022, 343, 13–30. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakurai, Y.; Watanabe, H.; Nishio, K.; Hashimoto, K.; Harada, A.; Gomi, M.; Suzuki, M.; Oyama, R.; Handa, T.; Sato, R.; et al. pH-Responsive Lipid Nanoparticles Achieve Efficient mRNA Transfection in Brain Capillary Endothelial Cells. Pharmaceutics 2022, 14, 1560. https://doi.org/10.3390/pharmaceutics14081560
Sakurai Y, Watanabe H, Nishio K, Hashimoto K, Harada A, Gomi M, Suzuki M, Oyama R, Handa T, Sato R, et al. pH-Responsive Lipid Nanoparticles Achieve Efficient mRNA Transfection in Brain Capillary Endothelial Cells. Pharmaceutics. 2022; 14(8):1560. https://doi.org/10.3390/pharmaceutics14081560
Chicago/Turabian StyleSakurai, Yu, Himeka Watanabe, Kazuma Nishio, Kohei Hashimoto, Atsuki Harada, Masaki Gomi, Masayoshi Suzuki, Ryotaro Oyama, Takumi Handa, Risa Sato, and et al. 2022. "pH-Responsive Lipid Nanoparticles Achieve Efficient mRNA Transfection in Brain Capillary Endothelial Cells" Pharmaceutics 14, no. 8: 1560. https://doi.org/10.3390/pharmaceutics14081560
APA StyleSakurai, Y., Watanabe, H., Nishio, K., Hashimoto, K., Harada, A., Gomi, M., Suzuki, M., Oyama, R., Handa, T., Sato, R., Takeuchi, H., Taira, R., Tezuka, K., Tange, K., Nakai, Y., Akita, H., & Uchida, Y. (2022). pH-Responsive Lipid Nanoparticles Achieve Efficient mRNA Transfection in Brain Capillary Endothelial Cells. Pharmaceutics, 14(8), 1560. https://doi.org/10.3390/pharmaceutics14081560





