Abstract
Ritodrine, a β2-adrenergic receptor agonist, is among most commonly prescribed tocolytic agents. This study aimed to evaluate the associations of single nucleotide polymorphisms in GNAS, RGS2, and RGS5 with the risk of ritodrine-induced adverse events (AEs) and develop a risk scoring system to identify high-risk patients. This is the prospective cohort study conducted at the Ewha Woman’s University Mokdong Hospital between January 2010 and October 2016. Pregnant women were included if they were treated with ritodrine for preterm labor with regular uterine contractions (at least 3 every 10 min) and cervical dilation. A total of 6, 3, and 5 single nucleotide polymorphisms (SNPs) of GNAS, RGS2, and RGS5 genes were genotyped and compared in patients with and without ritodrine-induced AEs. A total of 163 patients were included in this study. After adjusting confounders, GNAS rs3730168 (per-allele odds ratio (OR): 2.1; 95% confidence interval (95% CI): 1.0–4.3) and RGS2 rs1152746 (per-allele OR: 2.6, 95% CI: 1.1–6.5) were significantly associated with ritodrine-induced AEs. According to the constructed risk scoring models, patients with 0, 1, 2, 3, 4, and 5 points showed 0%, 13%, 19%, 31%, 46%, and 100% risks of AEs. This study suggested that GNAS and RGS2 polymorphisms could affect the risk of AEs in patients treated with ritodrine.
1. Introduction
Preterm birth is defined as delivery before 37 completed weeks of gestation [1]. Since it is one of the major causes of neonatal morbidity and mortality, it has a critical socioeconomic impact on healthcare systems [2]. According to the World Health Organization, about 15 million babies are born premature every year, and 1 million children die because of the complications associated with preterm birth [3]. The main cause of preterm labor is not well established; however, the etiology is considered complex and multifactorial. Many factors have been reported to be associated with increased risk of preterm birth, such as low socioeconomic status, maternal age, multiple gestations, stress, smoking, alcohol or substance abuse, inflammation, and intrauterine infection [4,5].
Various tocolytic agents are used to prevent preterm birth by relaxing uterine contraction via different mechanisms [6]. They include β2-adrenergic receptor agonists, oxytocin antagonists, calcium channel blockers, non-steroidal anti-inflammatory drugs, and magnesium sulfate [7,8]. Among them, ritodrine, a β2-adrenergic receptor agonist, is among the most commonly prescribed tocolytic agents, particularly in Asia and Europe [9,10]. However, due to safety issues, ritodrine has been withdrawn from the market in several countries, including the United States [11]. The adverse events (AEs) of ritodrine include tachycardia, palpitation, tremor, chest pain, and pulmonary edema [6,12]. These cardiopulmonary AEs can be explained by the stimulation of beta-adrenergic receptors by ritodrine with a lack of uterine selectivity [13].
For the safe use of ritodrine, it is important to identify the risk factors for AEs. For example, Shigemi et al. demonstrated that advanced maternal age, obesity, and long treatment duration were significant risk factors for ritodrine-induced AEs [14]. Along with clinical factors, several genetic polymorphisms have been reported to increase ritodrine-induced AEs [15,16,17]. Chung et al. showed that ADRB2 gene polymorphisms were related to ritodrine-induced AEs [15]. Two next-generation sequencing studies also identified SERPINA7, ADRA1A, and CPT2 genes as risk factors for ritodrine-induced AEs [16,17].
As the beta-adrenergic receptors are G protein-coupled receptor (GPCR) proteins, several proteins in the GPCR signaling pathway can affect ritodrine responses [18]. As β2-adrenergic receptors are coupled with stimulatory G (Gs) proteins, and regulator of G protein signaling (RGS) proteins inactivate the signals by increasing GTP hydrolysis [19], we herein focused on Gs α subunit gene (GNAS) and two RGS genes (RGS2 and RGS5), which are reportedly involved in the cardiovascular system [20]. Therefore, this study aimed to evaluate the associations of single nucleotide polymorphisms (SNPs) in GNAS, RGS2, and RGS5 genes with the risk of ritodrine-induced AEs in Korean women with preterm labor.
2. Methods
2.1. Study Patients
This prospective cohort study was conducted at the Ewha Woman’s University Mokdong Hospital between January 2010 and October 2016. The study procedure was approved by the Ethics Committee of the Institutional Review Board (IRB number: 217-1-26) and performed in accordance with the ethical standards of the Declaration of Helsinki. All participants provided written informed consent before enrollment.
Eligible patients were pregnant adults (≥18 years old) treated with ritodrine for preterm labor with regular uterine contractions (at least 3 every 10 min) and cervical dilation. Patients were excluded if they were treated with ritodrine for the McDonald procedure, had severe conditions requiring urgent delivery or surgery (e.g., fetal distress, pre-eclampsia, placenta abruption, spontaneous premature membrane, or spontaneous rupture oligohydramnios), or had comorbidities of cardiovascular disease, asthma, and hyperthyroidism.
Ritodrine (Lavopa®; JW Pharmaceutical, Seoul, Korea) was administered by intravenous infusion. The initial dose was 0.05 mg/min, which was increased every 10 min by 0.05 mg/min until adequate response was achieved. Intravenous treatment was discontinued during uterine quiescence. Thereafter, maintenance therapy was continued via an infusion of 0.05 mg/min for 12–48 h.
Data collection was performed by reviewing paper-based and electronic medical records. The following variables were included for data collection: maternal age, gestational age, weight, height, maximum infusion rate, and ritodrine-induced AEs. The presence of ritodrine-induced AEs was defined as dose reduction of ritodrine or change of ritodrine to other medications because of tachycardia (heart rate ≥ 100 bpm), shortness of breath, palpitation, dyspnea, or pulmonary edema.
2.2. Genotyping Methods
Genomic DNA was extracted from whole blood samples using QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). The following single-nucleotide polymorphisms (SNPs) were genotyped using SNaPshot Multiplex Kit (Applied Biosystems, Foster City, CA, USA): GNAS (rs12625436 (NC_000020.11: g.58870158G>A), rs13831 (NC_000020.11: g.58900136A>G), rs6128461 (NC_000020.11: g.58902035T>C), rs7121 (NC_000020.11: g.58903752C>T), rs3730168 (NC_000020.11: g.58903884G>A), and rs6026593 (NC_000020.11: g.58904078A>G)), RGS2 (rs1856840 (NC_000001.11: g.192842157T>C), rs4606 (NC_000001.11: g.192812042C>G), and rs1152746 (NC_000001.11: g.192827775C>T)), and RGS5 (rs3806366 (NC_000001.11: g.163145531A>G), rs2815276 (NC_000001.11: g.163155478A>G), rs2662776 (NC_000001.11: g.163195239A>G), rs1509018 (NC_000001.11: g.163218794G>C), and rs6698367 (NC_000001.11: g.163226647C>T)). All procedures were performed according to the manufacturer’s instructions.
2.3. Statistical Analyses
Chi-squared test or Fisher’s exact test was performed to compare patients with and without ritodrine-induced AEs. An additive model was applied to analyze the genetic effects on ritodrine-induced AEs. Haplotype analysis was performed using Plink [21]. To identify independent predictors, multivariable logistic regression analysis was performed using factors with p-value < 0.05 in the univariate analysis in addition to age and gestational age. Model discrimination was evaluated by area under the receiver operating curve (AUROC) analysis. A risk scoring system was developed based on the multivariable logistic regression model. To calculate the risk score, each coefficient from the multivariable logistic regression model was divided by the smallest one and rounded to the nearest integer. Statistical analysis was performed using SPSS version 20 (IBM, Chicago, IL, USA). A p-value of <0.05 was considered statistically significant.
3. Results
Of 236 enrolled patients, 73 patients were excluded by the following reasons: 38 patients did not have enough samples, 15 patients were in a serious condition before arriving to the hospital, 10 patients received ritodrine for the McDonald procedure, and 10 patients had underlying cardiovascular diseases. Accordingly, a total of 163 patients were included in this study. The mean maternal age and gestational age were 31 years (standard deviation (SD): 4 years) and 30 weeks (SD: 4 weeks), respectively. The mean weight and height were 63 kg (SD: 8 kg) and 161 cm (SD: 5 cm), respectively. Table 1 shows the association between baseline characteristics and ritodrine-induced AEs. The height of the patient was statistically significantly associated with AEs, and patients with a height of <160 cm were at a higher risk of developing AEs than others. Similarly, patients weighing <60 kg showed a trend toward higher risk of AEs; however, the association did not reach statistical significance.

Table 1.
Effects of demographic characteristics on ritodrine-induced adverse events.
The genetic effects on ritodrine-induced AEs are evaluated in Table 2. Among the studied SNPs, rs7121 and rs3730168 of GNAS and rs1152746 of RGS2 were significantly associated with AEs. The odds ratios (ORs) per variant allele for rs7121, rs3730168, and rs1152746 were 0.60 (95% confidence interval (CI): 0.36–0.99), 2.18 (95% CI: 1.62–3.79), and 2.39 (95% CI: 1.05–5.41), respectively. As two SNPs of GNAS (rs7121 and rs3730168) showed significant associations with AEs, we constructed haplotypes of them: H1 (C-G in rs7121-rs3730168; 17.6%), H2 (C-A; 26.9%), H3 (T-G; 53.3%), and H4 (T-A; 2.2%). The most frequent haplotype was H3, containing major alleles at every locus, followed by H2, containing risk alleles. The OR per H2 allele was 1.88 (95% CI: 1.08–3.28).

Table 2.
Effects of genotypes on ritodrine-induced adverse events.
Table 3 represents the results of the multivariable logistic regression analysis using age, gestational age, and factors identified as significant (p-value < 0.05) in the univariate analysis. After adjusting for confounders, height, GNAS rs3730168, and RGS2 rs1152746 were identified as significant factors related to ritodrine-induced AEs. Patients with a height of <160 cm had a 2.41-fold (95% CI: 1.13–5.12) increased risk of AEs than the others. GNAS rs3730168 was associated with a higher risk of AEs (per-allele OR: 2.10, 95% CI: 1.03–4.30). RGS2 rs1152746 was also associated with a higher risk of AEs (per-allele OR: 2.63; 95% CI: 1.07–6.49). The AUROC for predicted probability by multivariable logistic regression was 0.709 (95% CI: 0.619–0.800, p-value < 0.001; Figure 1).

Table 3.
Multivariable analysis and risk scoring system for ritodrine-induced adverse events.
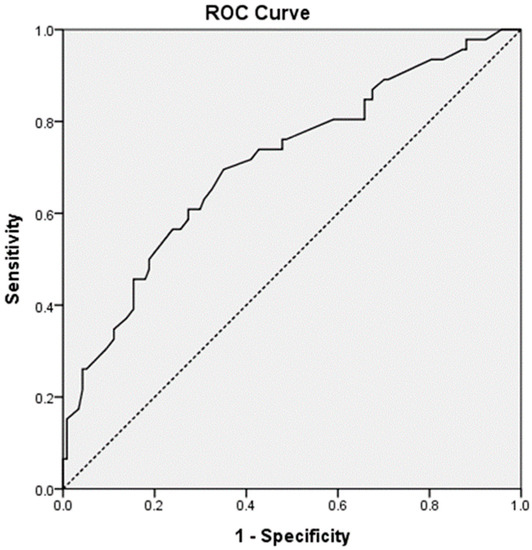
Figure 1.
The area under the receiver operating curve for ritodrine-induced adverse events.
Based on the multivariable analysis model, we constructed a risk scoring system to predict ritodrine-induced AEs. We assigned one point to each factor: height < 160 cm and number of variant alleles (each from 0 to 2) in rs3730168 and rs1152746. Thus, the total risk score range was 0 to 5. According to the constructed risk scoring system, patients with 0, 1, 2, 3, 4, and 5 points were at 0%, 13%, 19%, 31%, 46%, and 100% risks of developing AEs, respectively (Table 4). The risks predicted using the logistic regression curve are shown in Figure 2; the predicted risks of AEs for patients with 0, 1, 2, 3, 4, and 5 points were 4%, 9%, 18%, 32%, 51%, and 69%, respectively.

Table 4.
Observed risks of ritodrine-induced adverse events according to risk scores.
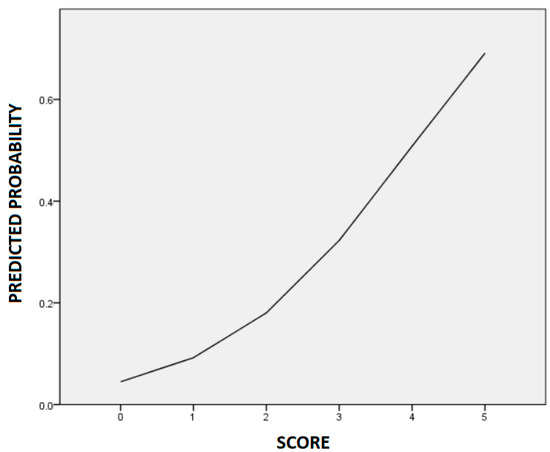
Figure 2.
The logistic regression curve of the predicted probability of ritodrine-induced adverse events.
4. Discussion
This study showed that GNAS rs3730168 (per-allele OR: 2.10; 95% CI: 1.03–4.30) and RGS2 rs1152746 (per-allele OR: 2.63; 95% CI: 1.07–6.49) along with height were significantly associated with ritodrine-induced AEs after adjustment for confounders. According to the risk scoring system, patients with higher scores were expected to have higher risks of AEs.
β2-adrenergic agonists have been reported to induce cardiovascular AEs, such as tachycardia, by stimulating β2-adrenergic receptors [22]. In the human heart, β1- and β2-adrenergic receptors are expressed in a ratio of 7:3 and have inotropic and chronotropic effects [23]. Both receptors are coupled to the Gs protein, which stimulates adenylyl cyclase to increase the level of cAMP [24]. It caused cAMP-mediated protein kinase A activation and calcium-induced cardiac contraction, thereby increasing the cardiac output and causing ventricular wall motion [25]. According to studies of mice models overexpressing the Gs α subunit, Gs α overexpression was associated with increased receptor sensitivity and enhanced GPCR signaling pathway, including adenylyl cyclase and L-type calcium channels, thereby leading to increased cardiac contractility and heart rate [26,27,28].
RGS proteins are multi-functional GTPase-activating proteins that accelerate the GTPase activity of the G protein α subunit and inhibit GPCR-induced signaling [29,30]. Among several subtypes, RGS2 and RGS5 are predominantly expressed in the cardiovascular system and are known as negative regulators of the GPCR signaling pathway [31]. Several in vivo studies have demonstrated the association of RGS2 inhibition with atrial tachycardia, hypertension, and cardiac hypertrophy [32,33,34]. RGS2 deficiency is reported to prolong vasoconstrictor signaling via AT1R and P2Y and increase internal calcium release via ryanodine receptors [35,36]. Similarly, loss of RGS5 has been reported to be associated with tachyarrhythmia and cardiac hypertrophy in mice [37,38]. Accordingly, genetic variants of GNAS and RGS-related genes are hypothesized to affect cardiovascular AEs associated with ritodrine.
The findings of this study show that rs3730168 of GNAS and rs1152746 of RGS2 are significantly associated with ritodrine-induced AEs after adjustment for confounders. Rs3730168 and rs1152746 are located in the 3’-untranslated region (3’-UTR) of GNAS and intronic region of RGS2, respectively. Although 3’-UTR and intron are non-coding parts of mRNA, they are known to regulate gene expression. 3’-UTR SNPs may regulate gene expression via degradation, translation, and localization of mRNAs [39], whereas intronic SNPs could affect mRNA stability and alternative splicing [40]. According to the eQTL analysis by GTEx [41], rs3730168 was identified as a significant expression quantitative trait locus, indicating that the variant allele had higher GNAS expression in whole blood cells (p-value = 8.2 × 10–6). Conversely, rs1152746 showed significant eQTL signals in different tissues, mainly with lower expression in the variant alleles. This finding was in agreement with previous reports that suggested that the upregulation of GNAS and downregulation of RGS2 may increase the cardiovascular risk.
Among the baseline characteristics, height was the only factor associated with ritodrine-induced AEs. As height was correlated with weight (r = 0.366, p-value < 0.001) in the study, this can be expected to have high drug concentrations in the smaller patients, thereby leading to AEs. As this was a single-center cohort study, the possibility of biased sampling could not be excluded. Further research is required to confirm these associations.
5. Limitation
This study has several limitations. First, this was a single-center study and included only Asian women, and thus, the findings should be validated in a multicenter study with a larger sample and multiple ethnicities. In addition, we did not perform multiple test corrections to avoid the possible loss of the true positives.
6. Conclusions
This is the first study evaluating the genetic effects of GNAS, RGS2, and RGS5 genes on ritodrine-induced AEs in preterm labor women. GNAS rs3730168 and RGS2 rs1152746 were identified as risk variants for ritodrine-induced AEs. In addition, we constructed a risk scoring system for predicting ritodrine-induced AEs by incorporating clinical and genetic factors. This tool can be used to identify high-risk patients requiring more caution during ritodrine treatment.
Author Contributions
Conceptualization, E.-J.J., Y.-J.K., H.-S.H., J.Y. and H.-S.G.; Formal analysis, E.-J.J. and J.Y.; Funding acquisition, H.-S.G.; Methodology, E.-J.J., Y.-J.K., H.-S.H., J.Y. and H.-S.G.; Resources, Y.-J.K. and H.-S.H.; Writing—original draft, E.-J.J., J.Y. and H.-S.G.; Writing—review & editing, J.Y. and H.-S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant (No. NRF-2010-0022544) funded by the Korea Government (MEST) and the Korea Health Industry Development Institute (KHIDI) (No. HI14C0306) funded by the Ministry of Health and Welfare.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the Institutional Review Board (IRB number: 217-1-26).
Informed Consent Statement
All participants provided written informed consent before enrollment.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Romero, R.; Dey, S.K.; Fisher, S.J. Preterm labor: One syndrome, many causes. Science 2014, 345, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Waitzman, N.J.; Jalali, A.; Grosse, S.D. Preterm birth lifetime costs in the United States in 2016: An update. Semin. Perinatol. 2021, 45, 151390. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Preterm Birth: Fact Sheet 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/preterm-birth (accessed on 1 August 2021).
- Li, C.; Cao, M.; Zhou, X. Role of epigenetics in parturition and preterm birth. Biol. Rev. Camb. Philos. Soc. 2022, 97, 851–873. [Google Scholar] [CrossRef] [PubMed]
- Gilman-Sachs, A.; Dambaeva, S.; Salazar Garcia, M.D.; Hussein, Y.; Kwak-Kim, J.; Beaman, K. Inflammation induced preterm labor and birth. J. Reprod. Immunol. 2018, 129, 53–58. [Google Scholar] [CrossRef]
- Garfield, L.; Chin, E. Pharmacology for Preterm Labor. J. Perinat. Neonatal Nurs. 2020, 34, 155–161. [Google Scholar] [CrossRef]
- Patel, S.S.; Ludmir, J. Drugs for the Treatment and Prevention of Preterm Labor. Clin. Perinatol. 2019, 46, 159–172. [Google Scholar] [CrossRef]
- Haas, D.M.; Caldwell, D.M.; Kirkpatrick, P.; McIntosh, J.J.; Welton, N.J. Tocolytic therapy for preterm delivery: Systematic review and network meta-analysis. BMJ 2012, 345, e6226. [Google Scholar] [CrossRef]
- Lamont, C.D.; Jørgensen, J.S.; Lamont, R.F. The safety of tocolytics used for the inhibition of preterm labour. Expert Opin. Drug Saf. 2016, 15, 1163–1173. [Google Scholar] [CrossRef]
- Sameshima, H.; Saito, S.; Matsuda, Y.; Kamitomo, M.; Makino, S.; Ohashi, M.; Kino, E.; Kanayama, N.; Takeda, S. Annual Report of the Perinatology Committee, Japan Society of Obstetrics and Gynecology, 2016: Overall report on a comprehensive retrospective study of obstetric management of preterm labor and preterm premature rupture of the membranes. J. Obstet. Gynaecol. Res. 2018, 44, 5–12. [Google Scholar] [CrossRef]
- Lam, F.; Gill, P. Beta-Agonist tocolytic therapy. Obstet. Gynecol. Clin. N. Am. 2005, 32, 457–484. [Google Scholar] [CrossRef]
- Canadian Preterm Labor Investigators Group. Treatment of preterm labor with the beta-adrenergic agonist ritodrine. N. Engl. J. Med. 1992, 327, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Modzelewska, B. Beta-adrenoceptors in obstetrics and gynecology. Dev. Period Med. 2016, 20, 93–98. [Google Scholar] [PubMed]
- Shigemi, D.; Aso, S.; Yasunaga, H. Inappropriate use of ritodrine hydrochloride for threatened preterm birth in Japan: A retrospective cohort study using a national inpatient database. BMC Prgenancy Childbirth 2019, 19, 204. [Google Scholar] [CrossRef]
- Chung, J.E.; Choi, S.A.; Hwang, H.S.; Park, J.Y.; Lee, K.E.; Yee, J.; Kim, Y.J.; Gwak, H.S. Association between B2-adrenergic receptor gene polymorphisms and adverse events of ritodrine in the treatment of preterm labor: A prospective observational study. BMC Genet. 2017, 18, 96. [Google Scholar] [CrossRef]
- Seo, H.; Kwon, E.J.; You, Y.A.; Park, Y.; Min, B.J.; Yoo, K.; Hwang, H.S.; Kim, J.H.; Kim, Y.J. Deleterious genetic variants in ciliopathy genes increase risk of ritodrine-induced cardiac and pulmonary side effects. BMC Med. Genom. 2018, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Park, Y.; Kim, Y.J.; Hwang, H.S.; Seo, H.; Min, B.J.; Lee, K.H.; Kim, S.Y.; Jung, Y.M.; Lee, S.; et al. Identifying genetic variants associated with ritodrine-induced pulmonary edema. PLoS ONE. 2020, 15, e0241215. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.D.; Cole, D.E.; Jose, P.A.; Chidiac, P. G protein-coupled receptor accessory proteins and signaling: Pharmacogenomic insights. Methods Mol. Biol. 2014, 1175, 121–152. [Google Scholar]
- Masuho, I.; Balaji, S.; Muntean, B.S.; Skamangas, N.K.; Chavali, S.; Tesmer, J.J.G.; Babu, M.M.; Martemyanov, K.A. A Global Map of G protein Signaling Regulation by RGS Proteins. Cell 2020, 183, 503–521.e19. [Google Scholar] [CrossRef]
- Hao, J.; Michalek, C.; Zhang, W.; Zhu, M.; Xu, X.; Mende, U. Regulation of cardiomyocyte signaling by RGS proteins: Differential selectivity towards G proteins and susceptibility to regulation. J. Mol. Cell Cardiol. 2006, 41, 51–61. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Hall, J.A.; Petch, M.C.; Brown, M.J. Intracoronary injections of salbutamol demonstrate the presence of functional beta 2-adrenoceptors in the human heart. Circ. Res. 1989, 65, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Brodde, O.E.; Michel, M.C. Adrenergic and muscarinic receptors in the human heart. Pharmacol. Rev. 1999, 51, 651–690. [Google Scholar] [PubMed]
- Brodde, O.E. Beta 1- and beta 2-adrenoceptors in the human heart: Properties, function, and alterations in chronic heart failure. Pharmacol. Rev. 1991, 43, 203–242. [Google Scholar] [PubMed]
- Ali, D.C.; Naveed, M.; Gordon, A.; Majeed, F.; Saeed, M.; Ogbuke, M.I.; Atif, M.; Zubair, H.M.; Changxing, L. B-adrenergic receptor, an essential target in cardiovascular diseases. Heart Fail. Rev. 2020, 25, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, C.; Ishikawa, Y.; Wight, D.C.; Mahdavi, V.; Nadal-Ginard, B.; Wagner, T.E.; Vatner, C.J. Overexpression of Gs alpha protein in the hearts of transgenic mice. J. Clin. Investig. 1995, 95, 1676–1683. [Google Scholar] [CrossRef]
- Iwase, M.; Uechi, M.; Vatner, D.E.; Asai, K.; Shannon, R.P.; Kudej, R.K.; Wagner, T.E.; Wight, D.C.; Patrick, T.A.; Ishikawa, Y.; et al. Cardiomyopathy induced by cardiac Gs alpha overexpression. Am. J. Physiol. 1997, 272, H585–H589. [Google Scholar] [CrossRef]
- Iwase, M.; Bishop, S.P.; Uechi, M.; Vatner, D.E.; Shannon, R.P.; Kudej, R.K.; Wight, D.C.; Wagner, T.E.; Ishikawa, Y.; Homcy, C.J.; et al. Adverse effects of chronic endogenous sympathetic drive induced by cardiac GS alpha overexpression. Circ. Res. 1996, 78, 517–524. [Google Scholar] [CrossRef]
- Hollinger, S.; Hepler, J.R. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol. Rev. 2002, 54, 527–559. [Google Scholar] [CrossRef]
- Wieland, T.; Mittmann, C. Regulators of G-protein signalling: Multifunctional proteins with impact on signalling in the cardiovascular system. Pharmacol. Ther. 2003, 97, 95–115. [Google Scholar] [CrossRef]
- Tsang, S.; Woo, A.Y.; Zhu, W.; Xiao, R.P. Deregulation of RGS2 in cardiovascular diseases. Front. Biosci. 2010, 2, 547–557. [Google Scholar]
- Tuomi, J.M.; Chidiac, P.; Jones, D.L. Evidence for enhanced M3 muscarinic receptor function and sensitivity to atrial arrhythmia in the RGS2-deficient mouse. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H554–H561. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heximer, S.P.; Knutsen, R.H.; Sun, X.; Kaltenbronn, K.M.; Rhee, M.H.; Peng, N.; Oliveira-dos-Santos, A.; Penninger, J.M.; Muslin, A.J.; Steinberg, T.H.; et al. Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J. Clin. Investig. 2003, 111, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Anger, T.; Su, J.; Hao, J.; Xu, X.; Zhu, M.; Gach, A.; Cui, L.; Liao, R.; Mende, U. Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy. J. Biol. Chem. 2006, 281, 5811–5820. [Google Scholar] [CrossRef] [PubMed]
- McNabb, H.J.; Zhang, Q.; Sjogren, B. Emerging Roles for Regulator of G protein Signaling 2 in (Patho)physiology. Mol. Pharmacol. 2020, 98, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Jie, L.; Owens, E.A.; Plante, L.A.; Fang, Z.; Rensing, D.T.; Moeller, K.D.; Osei-Owusu, P. RGS2 squelches vascular Gi/o and Gq signaling to modulate myogenic tone and promote uterine blood flow. Physiol. Rep. 2016, 4, e12692. [Google Scholar] [CrossRef]
- Qin, M.; Huang, H.; Wang, T.; Hu, H.; Liu, Y.; Gu, Y.; Cao, H.; Li, H.; Huang, C. Atrial tachyarrhythmia in Rgs5-null mice. PLoS ONE 2012, 7, e46856. [Google Scholar] [CrossRef]
- Perschbacher, K.J.; Deng, G.; Fisher, R.A.; Gibson-Corley, K.N.; Santillan, M.K.; Grobe, J.L. Regulators of G protein signaling in cardiovascular function during pregnancy. Physiol. Genom. 2018, 50, 590–604. [Google Scholar] [CrossRef]
- Mary, C. Regulation by 3’-Untranslated Regions. Annu. Rev. Genet. 2017, 51, 171–194. [Google Scholar] [CrossRef]
- Rose, A.B. Nuclear Pre-mRNA Processing in Plants; Springer: Berlin/Heidelberg, Germany, 2008; pp. 277–290. [Google Scholar]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T.; et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv. Biobank. 2015, 13, 311–319. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).