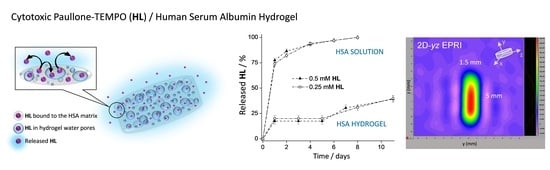
The Release of a Highly Cytotoxic Paullone Bearing a TEMPO Free Radical from the HSA Hydrogel: An EPR Spectroscopic Characterization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. The Binding and Release of HL from HSA
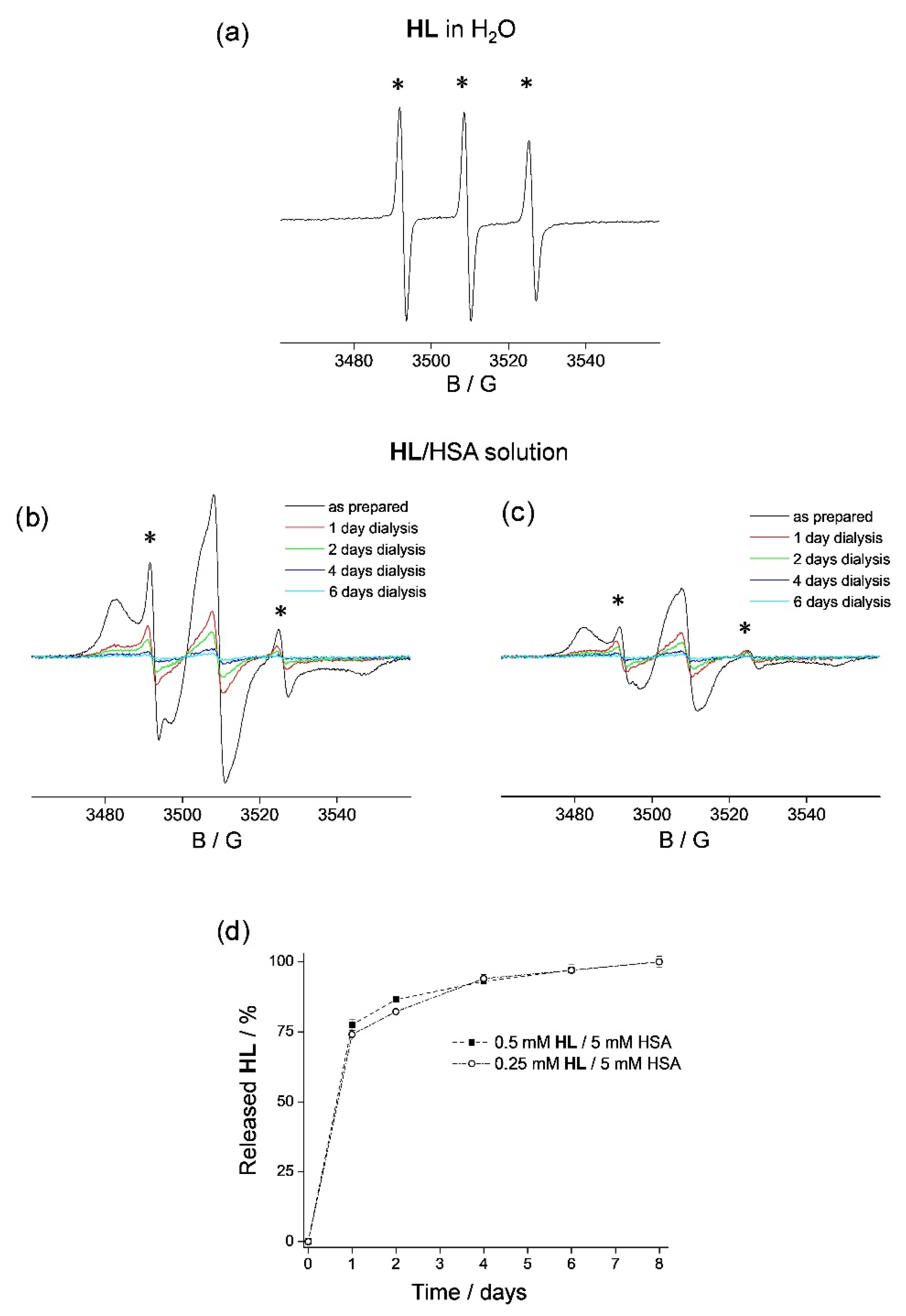
2.2.1. HL Release from HSA Solutions
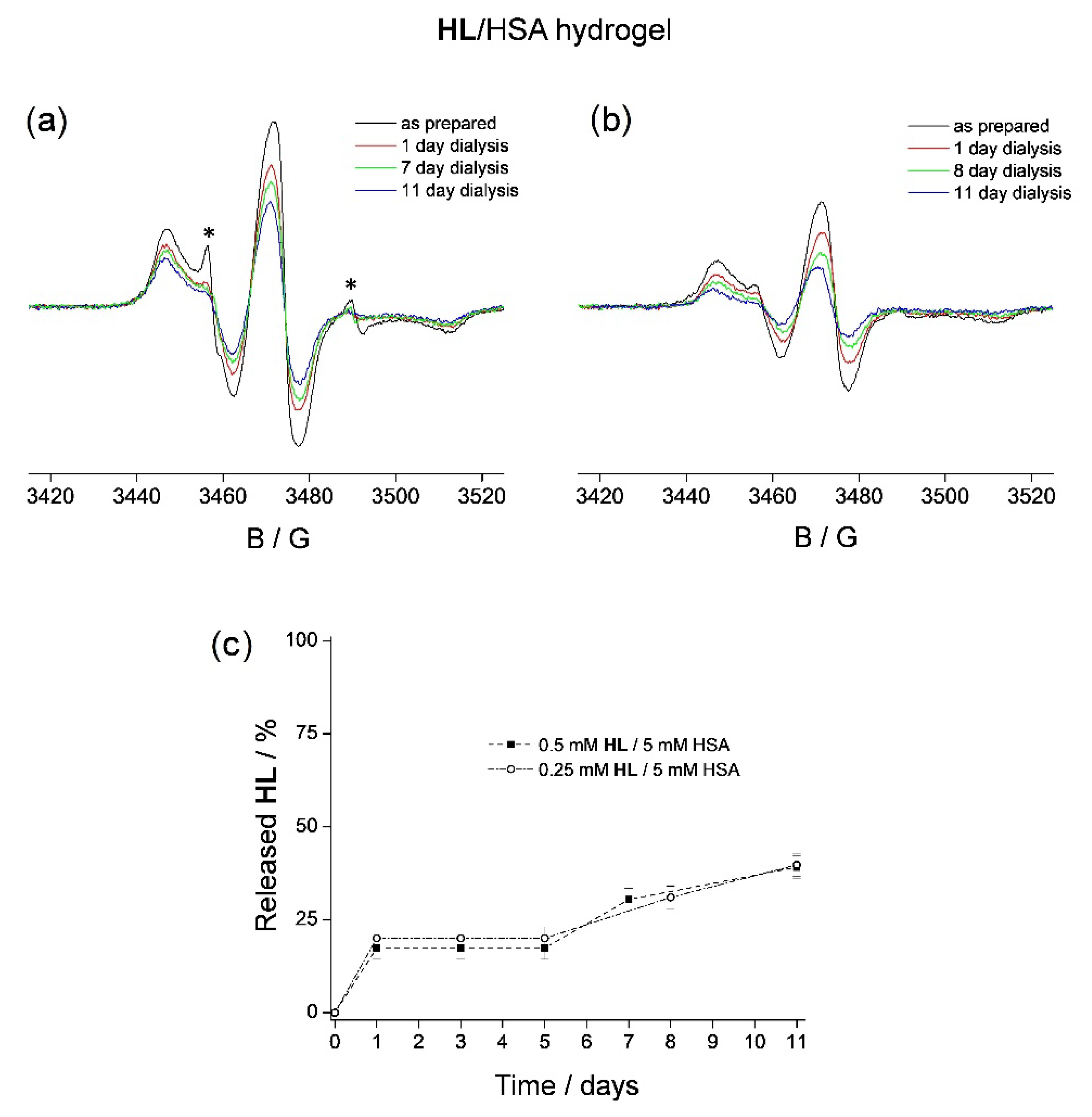
2.2.2. HL Release from HSA Hydrogels
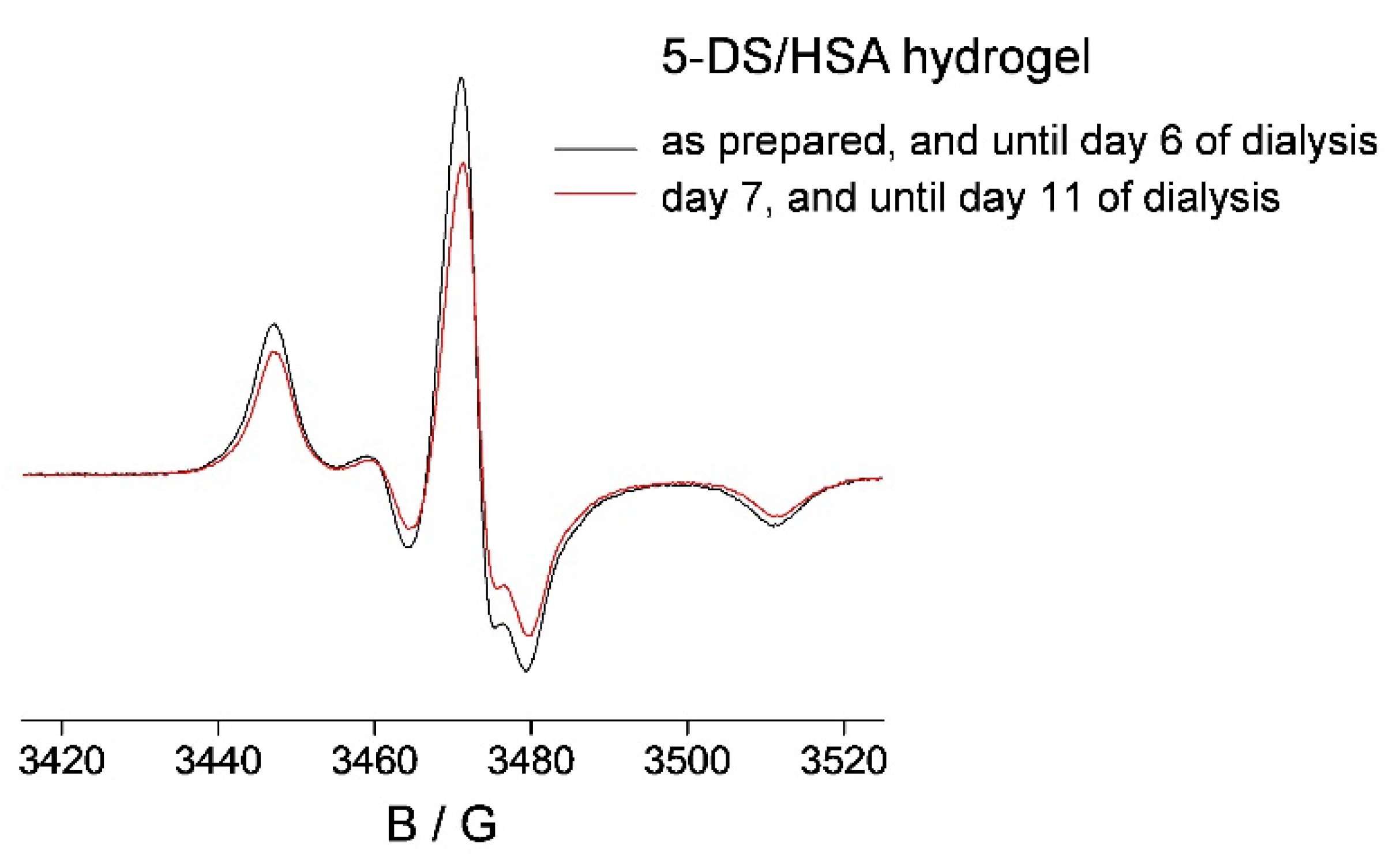
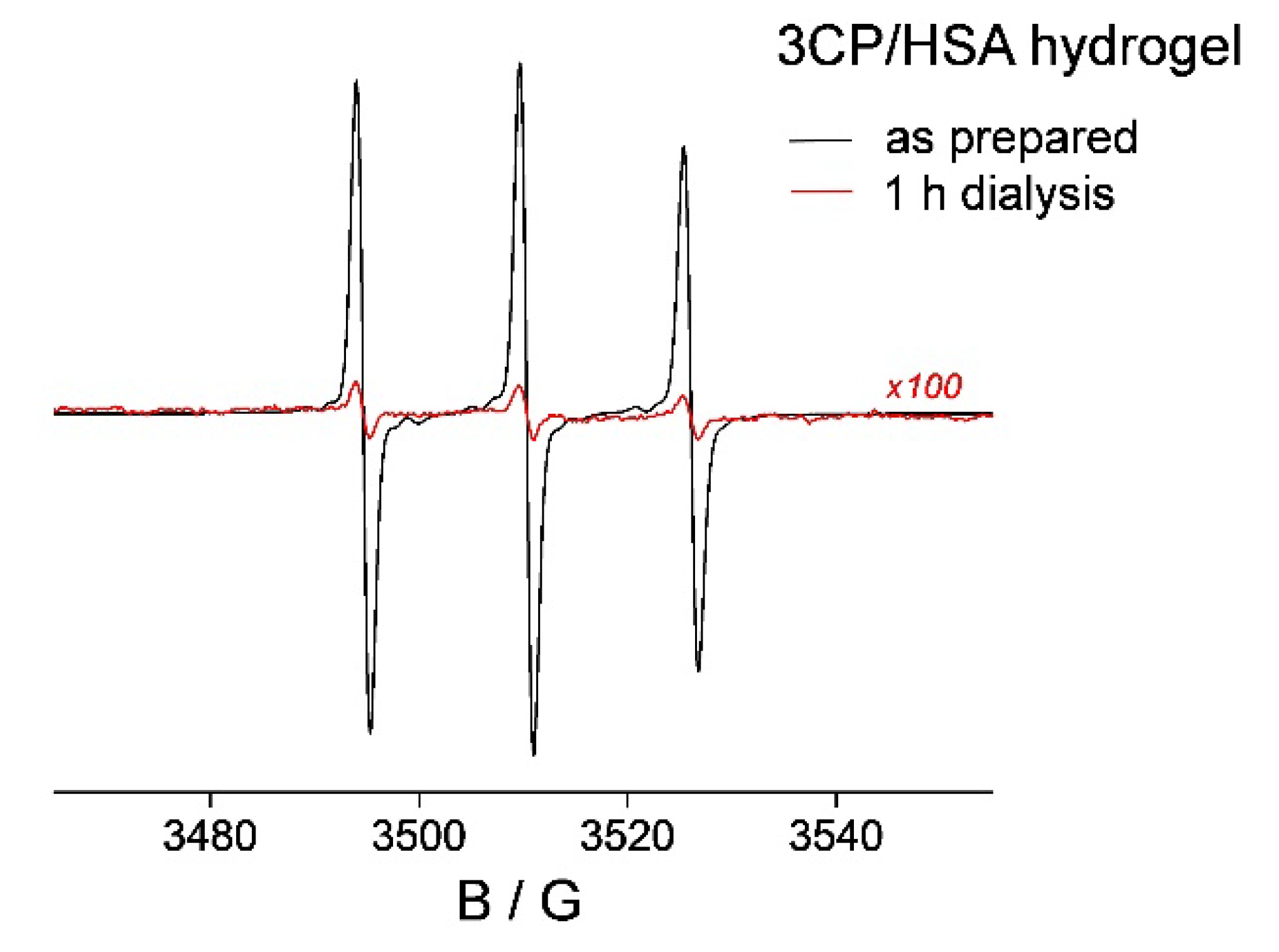
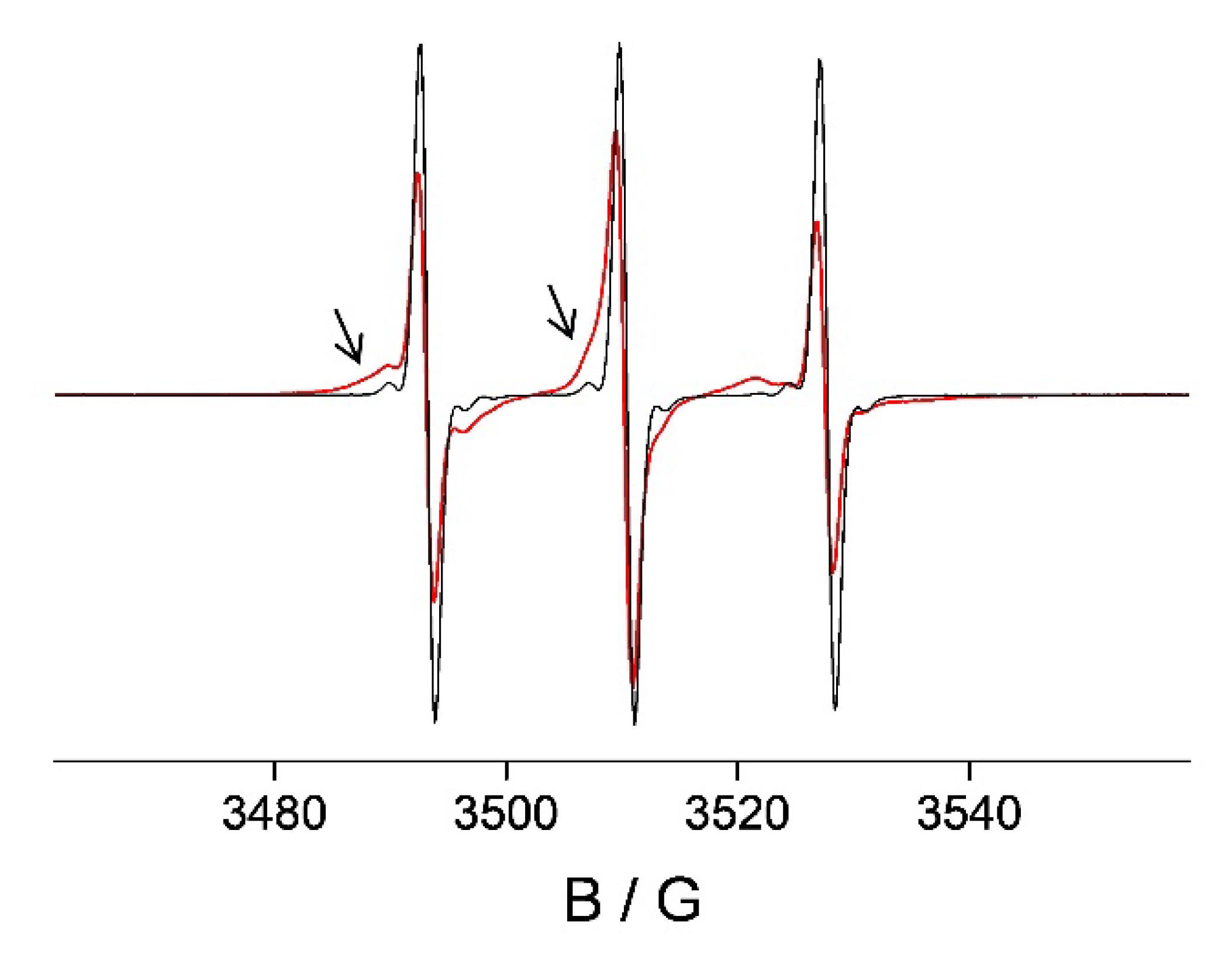
2.3. 5-DS/HSA and 3CP/HSA Spin-Labeled Hydrogels
2.4. In Vitro Cell Studies on HL and the HL/HSA Hydrogel: Cell Line and Culture Conditions and MTT Assay
2.5. Molecular Docking (MD)
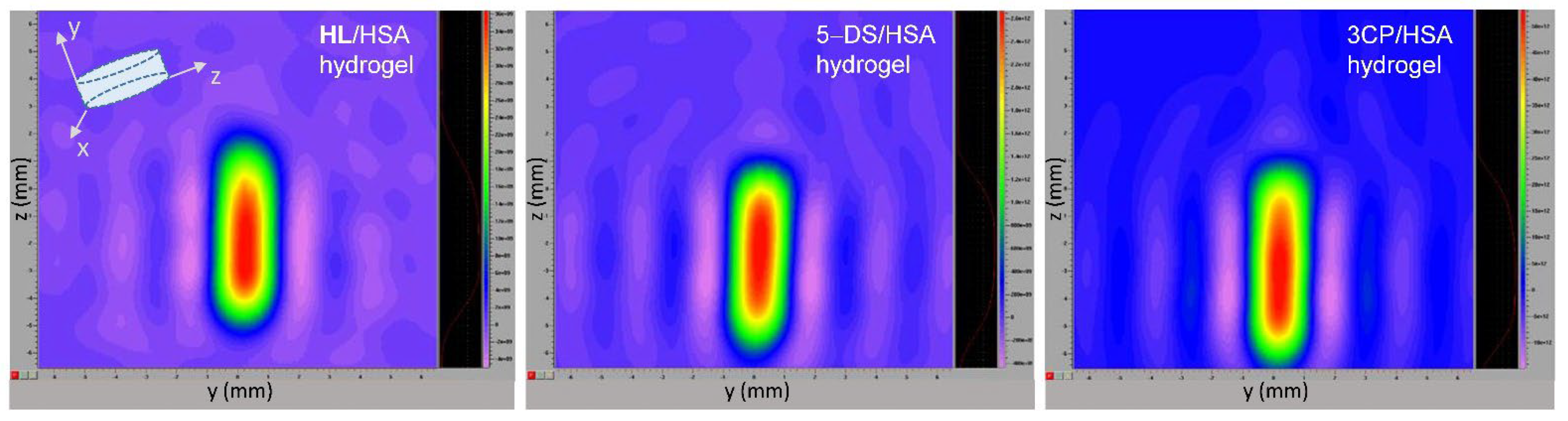
2.6. EPR Imaging of the Spin-Labeled Hydrogels
3. Results and Discussion
3.1. HL Binding to HSA
3.2. HL Binding to HSA Hydrogel Matrix
3.3. HSA Hydrogel Matrix Degradation
3.4. HL Diffusion from HSA Hydrogel Water Pores
3.5. In Vitro Cytotoxicity of HL and HL/HSA Hydrogel
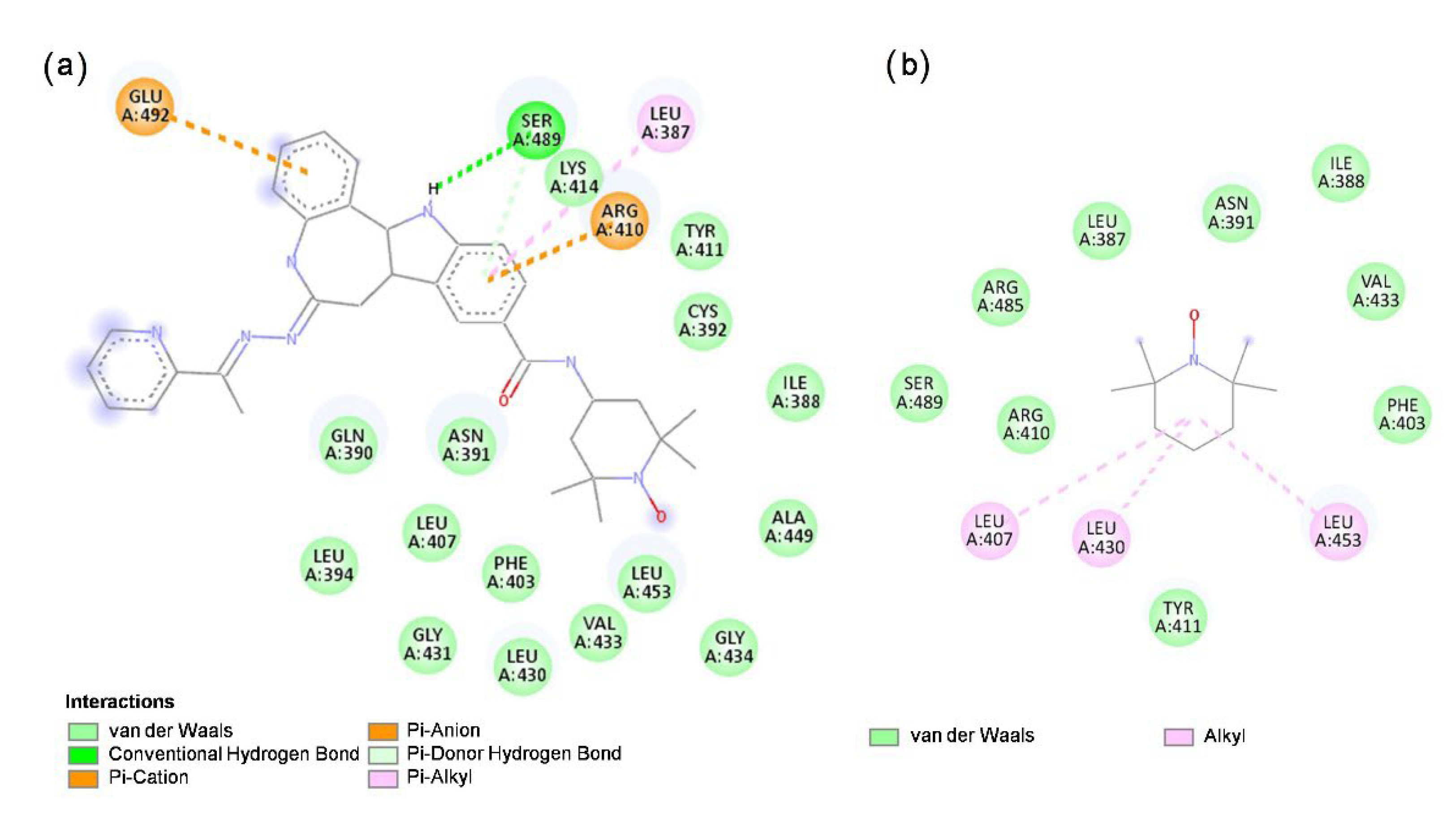
3.6. Molecular Docking of HL and TEMPO to HSA
3.7. EPR Imaging of Spin-Labeled Hydrogels
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Horn, G.; Moulton, K.; Oza, A.; Byler, S.; Kokolus, S.; Longacre, M. Cancer Development, Progression, and Therapy: An Epigenetic Overview. Int. J. Mol. Sci. 2013, 14, 21087–21113. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000; Chapter 15; ISBN 978-0-87893-106-4. [Google Scholar]
- Weller, D.; Vedsted, P.; Rubin, G.; Walter, F.M.; Emery, J.; Scott, S.; Campbell, C.; Andersen, R.S.; Hamilton, W.; Olesen, F.; et al. The Aarhus Statement: Improving Design and Reporting of Studies on Early Cancer Diagnosis. Br. J. Cancer 2012, 106, 1262–1267. [Google Scholar] [CrossRef]
- Fass, L. Imaging and Cancer: A Review. Mol. Oncol. 2008, 2, 115–152. [Google Scholar] [CrossRef] [PubMed]
- Beyer, T.; Bidaut, L.; Dickson, J.; Kachelriess, M.; Kiessling, F.; Leitgeb, R.; Ma, J.; Shiyam Sundar, L.K.; Theek, B.; Mawlawi, O. What Scans We Will Read: Imaging Instrumentation Trends in Clinical Oncology. Cancer Imaging 2020, 20, 1–38. [Google Scholar] [CrossRef]
- Hussain, T.; Nguyen, Q.T. Molecular Imaging for Cancer Diagnosis and Surgery. Adv. Drug Deliv. Rev. 2014, 66, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Shahbazi-Gahrouei, D.; Khaniabadi, P.; Khaniabadi, B.; Shahbazi-Gahrouei, S. Medical Imaging Modalities Using Nanoprobes for Cancer Diagnosis: A Literature Review on Recent Findings. J. Res. Med. Sci. 2019, 24, 38. [Google Scholar] [CrossRef]
- Jadvar, H.; Colletti, P.M.; Delgado-Bolton, R.; Esposito, G.; Krause, B.J.; Iagaru, A.H.; Nadel, H.; Quinn, D.I.; Rohren, E.; Subramaniam, R.M.; et al. Appropriate Use Criteria for 18 F-FDG PET/CT in Restaging and Treatment Response Assessment of Malignant Disease. J. Nucl. Med. 2017, 58, 2026–2037. [Google Scholar] [CrossRef] [Green Version]
- Hope, T.A.; Kassam, Z.; Loening, A.; McNamara, M.M.; Paspulati, R. The Use of PET/MRI for Imaging Rectal Cancer. Abdom. Radiol. 2019, 44, 3559–3568. [Google Scholar] [CrossRef]
- Marin, J.F.G.; Nunes, R.F.; Coutinho, A.M.; Zaniboni, E.C.; Costa, L.B.; Barbosa, F.G.; Queiroz, M.A.; Cerri, G.G.; Buchpiguel, C.A. Theranostics in Nuclear Medicine: Emerging and Re-Emerging Integrated Imaging and Therapies in the Era of Precision Oncology. Radiographics 2020, 40, 1715–1740. [Google Scholar] [CrossRef]
- Chen, X.; Wong, S.T.C. Cancer Theranostics: An Introduction. In Cancer Theranostics; Chen, X., Wong, S.T.C., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 3–8. ISBN 978-0-12-407722-5. [Google Scholar]
- Jain, N.K.; Chathoth, B.M.; Bhaskar, V.S.; Meena, H.; Prasad, R.; Srivastava, R. Nanoengineered Photoactive Theranostic Agents for Cancer. Nanophotonics 2021, 10, 2973–2997. [Google Scholar] [CrossRef]
- Abbas, Z.; Rehman, S. An Overview of Cancer Treatment Modalities. In Neoplasm; Shahzad, H.N., Ed.; IntechOpen: London, UK, 2018; ISBN 978-1-78923-778-8. [Google Scholar]
- Dickens, E. Principles of Cancer Treatment by Chemotherapy. Surgery 2017, 36, 134–138. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The Side Effects of Platinum-Based Chemotherapy Drugs: A Review for Chemists. Dalt. Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Meegan, M.J.; O’Boyle, N.M. Special Issue “Anticancer Drugs”. Pharmaceuticals 2019, 12, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbugua, S.N.; Njenga, L.W.; Odhiambo, R.A.; Wandiga, S.O.; Onani, M.O. Beyond DNA-Targeting in Cancer Chemotherapy. Emerging Frontiers—A Review. Curr. Top. Med. Chem. 2020, 21, 28–47. [Google Scholar] [CrossRef]
- Senapati, S.; Mahanta, A.K.; Kumar, S.; Maiti, P. Controlled Drug Delivery Vehicles for Cancer Treatment and Their Performance. Signal Transduct. Target. Ther. 2018, 3, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Yin, X.; Yin, X.; Chen, A.; Zhao, L.; Zhang, G.; Liao, W.; Huang, X.; Li, J.; Zhang, C.Y. Dual ph/Redox-Responsive Mixed Polymeric Micelles for Anticancer Drug Delivery and Controlled Release. Pharmaceutics 2019, 11, 176. [Google Scholar] [CrossRef] [Green Version]
- Dicheva, B.M.; Ten Hagen, T.L.M.; Schipper, D.; Seynhaeve, A.L.B.; Van Rhoon, G.C.; Eggermont, A.M.M.; Koning, G.A. Targeted and Heat-Triggered Doxorubicin Delivery to Tumors by Dual Targeted Cationic Thermosensitive Liposomes. J. Control. Release 2014, 195, 37–48. [Google Scholar] [CrossRef]
- Guo, Z.; Sui, J.; Ma, M.; Hu, J.; Sun, Y.; Yang, L.; Fan, Y.; Zhang, X. pH-Responsive Charge Switchable PEGylated ε-poly-l-lysine Polymeric Nanoparticles-Assisted Combination Therapy for Improving Breast Cancer Treatment. J. Control. Release 2020, 326, 350–364. [Google Scholar] [CrossRef]
- Bastiancich, C.; Danhier, P.; Préat, V.; Danhier, F. Anticancer Drug-Loaded Hydrogels as Drug Delivery Systems for the Local Treatment of Glioblastoma. J. Control. Release 2016, 243, 29–42. [Google Scholar] [CrossRef]
- Cirillo, G.; Spizzirri, U.G.; Curcio, M.; Nicoletta, F.P.; Iemma, F. Injectable Hydrogels for Cancer Therapy over the Last Decade. Pharmaceutics 2019, 11, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, Q.; Jiao, Y.; Yu, X. Hydrogels for Biomedical Applications: Their Characteristics and the Mechanisms behind Them. Gels 2017, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, E.M. Hydrogel: Preparation, Characterization, and Applications: A Review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trombino, S.; Servidio, C.; Curcio, F.; Cassano, R. Strategies for Hyaluronic Acid-Based Hydrogel Design in Drug Delivery. Pharmaceutics 2019, 11, 407. [Google Scholar] [CrossRef] [Green Version]
- Ciolacu, D.E.; Nicu, R.; Ciolacu, F. Cellulose-Based Hydrogels as Sustained Drug-Delivery Systems. Materials 2020, 13, 5270. [Google Scholar] [CrossRef]
- Buwalda, S.J.; Boere, K.W.M.; Dijkstra, P.J.; Feijen, J.; Vermonden, T.; Hennink, W.E. Hydrogels in a Historical Perspective: From Simple Networks to Smart Materials. J. Control. Release 2014, 190, 254–273. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y.S.; Pang, B.; Hyun, D.C.; Yang, M.; Xia, Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem.-Int. Ed. 2014, 53, 12320–12364. [Google Scholar] [CrossRef]
- Gyles, D.A.; Castro, L.D.; Silva, J.O.C.; Ribeiro-Costa, R.M. A Review of the Designs and Prominent Biomedical Advances of Natural and Synthetic Hydrogel Formulations. Eur. Polym. J. 2017, 88, 373–392. [Google Scholar] [CrossRef]
- Ahsan, A.; Farooq, M.A.; Parveen, A. Thermosensitive Chitosan-Based Injectable Hydrogel as an Efficient Anticancer Drug Carrier. ACS Omega 2020, 5, 20450–20460. [Google Scholar] [CrossRef]
- Reig-Vano, B.; Tylkowski, B.; Montané, X.; Giamberini, M. Alginate-Based Hydrogels for Cancer Therapy and Research. Int. J. Biol. Macromol. 2021, 170, 424–436. [Google Scholar] [CrossRef]
- Niu, X.; Zhang, Z.; Zhong, Y. Hydrogel Loaded with Self-Assembled Dextran Sulfate-Doxorubicin Complexes as a Delivery System for Chemotherapy. Mater. Sci. Eng. C 2017, 77, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Tabata, Y.; Kariya, M.; Suzuki, A.; Mandai, M.; Nanbu, K.; Takakura, K.; Fujii, S. In Vivo Anti-Tumor Effect through the Controlled Release of Cisplatin from Biodegradable Gelatin Hydrogel. J. Control. Release 2003, 92, 301–313. [Google Scholar] [CrossRef]
- Upadhyay, A.; Kandi, R.; Rao, C.P. Injectable, Self-Healing, and Stress Sustainable Hydrogel of BSA as a Functional Biocompatible Material for Controlled Drug Delivery in Cancer Cells. ACS Sustain. Chem. Eng. 2018, 6, 3321–3330. [Google Scholar] [CrossRef]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the Mysteries of Serum Albumin-More Than Just a Serum Protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoogenboezem, E.N.; Duvall, C.L. Harnessing Albumin as a Carrier for Cancer Therapies. Adv. Drug Deliv. Rev. 2018, 130, 73–89. [Google Scholar] [CrossRef]
- Doweiko, J.P.; Nompleggi, D.J. Reviews: Role of Albumin in Human Physiology and Pathophysiology. J. Parenter. Enter. Nutr. 1991, 15, 207–211. [Google Scholar] [CrossRef]
- Evans, T.W. Review Article: Albumin as a Drug-Biological Effects of Albumin Unrelated to Oncotic Pressure. Aliment. Pharmacol. Ther. 2002, 16, 6–11. [Google Scholar] [CrossRef]
- Roche, M.; Rondeau, P.; Singh, N.R.; Tarnus, E.; Bourdon, E. The Antioxidant Properties of Serum Albumin. FEBS Lett. 2008, 582, 1783–1787. [Google Scholar] [CrossRef]
- Utariani, A.; Rahardjo, E.; Perdanakusuma, D.S. Effects of Albumin Infusion on Serum Levels of Albumin, Proinflammatory Cytokines (TNF- α, IL-1, and IL-6), CRP, and MMP-8; Tissue Expression of EGRF, ERK1, ERK2, TGF- β, Collagen, and MMP-8; and Wound Healing in Sprague Dawley Rats. Int. J. Inflam. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- He, X.M.; Carter, D.C. Atomic Structure and Chemistry of Human Serum Albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal Structure of Human Serum Albumin Complexed with Fatty Acid Reveals an Asymmetric Distribution of Binding Sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Kratz, F. Albumin as a Drug Carrier: Design of Prodrugs, Drug Conjugates and Nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Ascoli, G.A.; Domenici, E.; Bertucci, C. Drug Binding to Human Serum Albumin: Abridged Review of Results Obtained with High-Performance Liquid Chromatography and Circular Dichroism. Chirality 2006, 18, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Sanaeifar, N.; Mäder, K.; Hinderberger, D. Molecular-Level Release of Coumarin-3-Carboxylic Acid and Warfarin-Derivatives from BSA-Based Hydrogels. Pharmaceutics 2021, 13, 1661. [Google Scholar] [CrossRef] [PubMed]
- Merlot, A.M.; Kalinowski, D.S.; Kovacevic, Z.; Jansson, P.J.; Lane, D.J.; Huang, M.L.-H.; Sahni, S.; Richardson, D.R. Making a Case for Albumin—A Highly Promising Drug-Delivery System. Future Med. Chem. 2015, 7, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, F.; Dinarvand, R.; Ghahremani, M.H.; Amini, M.; Rouhani, H.; Sepehri, N.; Ostad, S.N.; Atyabi, F. Docetaxel–Albumin Conjugates: Preparation, In Vitro Evaluation and Biodistribution Studies. J. Pharm. Sci. 2009, 98, 2718–2730. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Jiang, S.; Zhang, X.; Fu, Y.; Liu, Z. Preparation, Characterization and in Vitro Activity of a Docetaxel–Albumin Conjugate. Bioorg. Chem. 2019, 83, 154–160. [Google Scholar] [CrossRef]
- Melder, R.J.; Osborn, B.L.; Riccobene, T.; Kanakaraj, P.; Wei, P.; Chen, G.; Stolow, D.; Halpern, W.G.; Migone, T.S.; Wang, Q.; et al. Pharmacokinetics and In Vitro and In Vivo Anti-Tumor Response of an Interleukin-2-Human Serum Albumin Fusion Protein in Mice. Cancer Immunol. Immunother. 2005, 54, 535–5475. [Google Scholar] [CrossRef]
- Li, R.; Yang, H.; Jia, D.; Nie, Q.; Cai, H.; Fan, Q.; Wan, L.; Li, L.; Lu, X. Fusion to an Albumin-Binding Domain with a High Affinity for Albumin Extends the Circulatory Half-Life and Enhances the In Vivo Antitumor Effects of Human TRAIL. J. Control. Release 2016, 228, 96–106. [Google Scholar] [CrossRef]
- Kratz, F.; Müller-Driver, R.; Hofmann, I.; Drevs, J.; Unger, C. A Novel Macromolecular Prodrug Concept Exploiting Endogenous Serum Albumin as a Drug Carrier for Cancer Chemotherapy. J. Med. Chem. 2000, 43, 1253–1256. [Google Scholar] [CrossRef]
- Kratz, F.; Warnecke, A.; Scheuermann, K.; Stockmar, C.; Schwab, J.; Lazar, P.; Drückes, P.; Esser, N.; Drevs, J.; Rognan, D.; et al. Probing the Cysteine-34 Position of Endogenous Serum Albumin with Thiol-Binding Doxorubicin Derivatives. Improved Efficacy of an Acid-Sensitive Doxorubicin Derivative with Specific Albumin-Binding Properties Compared to That of the Parent Compound. J. Med. Chem. 2002, 45, 5523–5533. [Google Scholar] [CrossRef] [PubMed]
- Kratz, F.; Mansour, A.; Soltau, J.; Warnecke, A.; Fichtner, I.; Unger, C.; Drevs, J. Development of Albumin-Binding Doxorubicin pro-Drugs That Are Cleaved by Prostate-Specific Antigen. Arch. Pharm. (Weinheim) 2005, 338, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Schmid, B.; Chung, D.E.; Warnecke, A.; Fichtner, I.; Kratz, F. Albumin-Binding Prodrugs of Camptothecin and Doxorubicin with an Ala-Leu-Ala-Leu-Linker That Are Cleaved by Cathepsin B: Synthesis and Antitumor Efficacy. Bioconjug. Chem. 2007, 18, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, A.; Kratz, F. Maleimide-Oligo(Ethylene Glycol) Derivatives of Camptothecin as Albumin-Binding Prodrugs: Synthesis and Antitumor Efficacy. Bioconjug. Chem. 2003, 14, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Warnecke, A.; Fichtner, I.; Garmann, D.; Jaehde, U.; Kratz, F. Synthesis and Biological Activity of Water-Soluble Maleimide Derivatives of the Anticancer Drug Carboplatin Designed as Albumin-Binding Prodrugs. Bioconjug. Chem. 2004, 15, 1349–1359. [Google Scholar] [CrossRef]
- Mayr, J.; Heffeter, P.; Groza, D.; Galvez, L.; Koellensperger, G.; Roller, A.; Alte, B.; Haider, M.; Berger, W.; Kowol, C.R.; et al. An Albumin-Based Tumor-Targeted Oxaliplatin Prodrug with Distinctly Improved Anticancer Activity In Vivo. Chem. Sci. 2017, 8, 2241–2250. [Google Scholar] [CrossRef] [Green Version]
- Stepanenko, I.N.; Casini, A.; Edafe, F.; Novak, M.S.; Arion, V.B.; Dyson, P.J.; Jakupec, M.A.; Keppler, B.K. Conjugation of Organoruthenium(II) 3-(1H-Benzimidazol-2-Yl)pyrazolo [3,4-b]pyridines and Indolo [3,2-d]benzazepines to Recombinant Human Serum Albumin: A Strategy To Enhance Cytotoxicity in Cancer Cells. Inorg. Chem. 2011, 50, 12669–12679. [Google Scholar] [CrossRef]
- Desai, N. Nanoparticle Albumin-Bound Paclitaxel (Abraxane®). In Albumin in Medicine: Pathological and Clinical Applications; Otagiri, M., Giam Chuang, V.T., Eds.; Springer: London, UK, 2016; pp. 101–119. ISBN 978-981-10-2116-9. [Google Scholar]
- Dobrov, A.; Göschl, S.; Jakupec, M.A.; Popović-Bijelić, A.; Gräslund, A.; Rapta, P.; Arion, V.B. A Highly Cytotoxic Modified Paullone Ligand Bearing a TEMPO Free-Radical Unit and Its Copper(II) Complex as Potential hR2 RNR Inhibitors. Chem. Commun. 2013, 49, 10007. [Google Scholar] [CrossRef] [Green Version]
- Hauenschild, T.; Reichenwallner, J.; Enkelmann, V.; Hinderberger, D. Characterizing Active Pharmaceutical Ingredient Binding to Human Serum Albumin by Spin-Labeling and EPR Spectroscopy. Chem.-A Eur. J. 2016, 22, 12825–12838. [Google Scholar] [CrossRef]
- Dömötör, O.; Rathgeb, A.; Kuhn, P.-S.; Popović-Bijelić, A.; Bačić, G.; Enyedy, E.A.; Arion, V.B. Investigation of the Binding of Cis/Trans-[MCl4(1H-Indazole)(NO)](−) (M = Ru, Os) Complexes to Human Serum Albumin. J. Inorg. Biochem. 2016, 159, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, J.; Tozer, T.N.; Sorby, D.L.; Tuck, L.D. Investigation of Drug-Albumin Interactions Using Spin-Labeled Bovine Serum Albumin. Mol. Pharmacol. 1975, 11, 133–143. [Google Scholar] [PubMed]
- Dömötör, O.; May, N.V.; Gál, G.T.; Spengler, G.; Dobrova, A.; Arion, V.B.; Enyedy, É.A. Solution Equilibrium Studies on Salicylidene Aminoguanidine Schiff Base Metal Complexes: Impact of the Hybridization with L-Proline on Stability, Redox Activity and Cytotoxicity. Molecules 2022, 27, 2044. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Prism Version 7.00 for Windows; Graph Pad Software: La Jolla, CA, USA, 2018; Available online: http://www.graphpad.com (accessed on 19 April 2022).
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacher, F.; Wittmann, C.; Nové, M.; Spengler, G.; Marć, M.A.; Enyedy, E.A.; Darvasiová, D.; Rapta, P.; Reiner, T.; Arion, V.B. Novel Latonduine Derived Proligands and Their Copper(II) Complexes Show Cytotoxicity in the Nanomolar Range in Human Colon Adenocarcinoma Cells and In Vitro Cancer Selectivity. Dalt. Trans. 2019, 48, 10464–10478. [Google Scholar] [CrossRef] [Green Version]
- Kocherginsky, N.; Swartz, H.M. Nitroxide Spin Labels. Reactions in Biology and Chemistry; CRC Press Inc.: Boca Raton, FL, USA, 1995; ISBN 978-0-8493-4204-2. [Google Scholar]
- Hagen, W.R. Biomolecular EPR Spectroscopy; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2009; ISBN 978-1-4200-5957-1. [Google Scholar]
- Marsh, D. Reaction Fields and Solvent Dependence of the EPR Parameters of Nitroxides: The Microenvironment of Spin Labels. J. Magn. Reson. 2008, 190, 60–67. [Google Scholar] [CrossRef]
- Vesković, A.; Nakarada, Đ.; Popović Bijelić, A. A Novel Methodology for Hydrogel Water Content Determination by EPR: The Basis for Real-Time Monitoring of Controlled Drug Release and Hydrogel Swelling and Degradation. Polym. Test. 2021, 98, 107187. [Google Scholar] [CrossRef]
- Murayama, K.; Tomida, M. Heat-Induced Secondary Structure and Conformation Change of Bovine Serum Albumin Investigated by Fourier Transform Infrared Spectroscopy. Biochemistry 2004, 43, 11526–11532. [Google Scholar] [CrossRef]
- Navarra, G.; Peres, C.; Contardi, M.; Picone, P.; San Biagio, P.L.; Di Carlo, M.; Giacomazza, D.; Militello, V. Heat- and pH-Induced BSA Conformational Changes, Hydrogel Formation and Application as 3D Cell Scaffold. Arch. Biochem. Biophys. 2016, 606, 134–142. [Google Scholar] [CrossRef]
- Berliner, L.J.; Reuben, J. (Eds.) Spin Labeling: Theory and Applications; Plenum Press: New York, NY, USA, 1989; Volume 8, Biological Magnetic Resonance; ISBN 978-1-4612-8060-6. [Google Scholar]
- van der Vusse, G.J. Albumin as Fatty Acid Transporter. Drug Metab. Pharmacokinet. 2009, 24, 300–307. [Google Scholar] [CrossRef]
- Petitpas, I.; Grüne, T.; Bhattacharya, A.A.; Curry, S. Crystal Structures of Human Serum Albumin Complexed with Monounsaturated and Polyunsaturated Fatty Acids. J. Mol. Biol. 2001, 314, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.A.; Grüne, T.; Curry, S. Crystallographic Analysis Reveals Common Modes of Binding of Medium and Long-Chain Fatty Acids to Human Serum Albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Pavićević, A.A.; Popović-Bijelić, A.D.; Mojović, M.D.; Šušnjar, S.V.; Bačić, G.G. Binding of Doxyl Stearic Spin Labels to Human Serum Albumin: An EPR Study. J. Phys. Chem. B 2014, 118, 10898–10905. [Google Scholar] [CrossRef] [PubMed]
- Cistola, D.P.; Small, D.M. Fatty Acid Distribution in Systems Modeling the Normal and Diabetic Human Circulation: A 13C Nuclear Magnetic Resonance Study. J. Clin. Invest. 1991, 87, 1431–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arabi, S.H.; Aghelnejad, B.; Schwieger, C.; Meister, A.; Kerth, A.; Hinderberger, D. Serum Albumin Hydrogels in Broad pH and Temperature Ranges: Characterization of Their Self-Assembled Structures and Nanoscopic and Macroscopic Properties. Biomater. Sci. 2018, 6, 478–492. [Google Scholar] [CrossRef] [Green Version]
- Sanaeifar, N.; Mäder, K.; Hinderberger, D. Nanoscopic Characterization of Stearic Acid Release from Bovine Serum Albumin Hydrogels. Macromol. Biosci. 2020, 20, 2000126. [Google Scholar] [CrossRef]
- Gayet, J.C.; Fortier, G. High Water Content BSA-PEG Hydrogel for Controlled Release Device: Evaluation of the Drug Release Properties. J. Control. Release 1996, 38, 177–184. [Google Scholar] [CrossRef]
- Peers, S.; Montembault, A.; Ladavière, C. Chitosan Hydrogels for Sustained Drug Delivery. J. Control. Release 2020, 326, 150–163. [Google Scholar] [CrossRef]
- Li, J.; Mooney, D.J. Designing Hydrogels for Controlled Drug Delivery. Nat. Rev. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef]
- Hoffman, A.S. Hydrogels for Biomedical Applications. Adv. Drug Deliv. Rev. 2012, 64, 18–23. [Google Scholar] [CrossRef]
- Stepanenko, I.; Babak, M.V.; Spengler, G.; Hammerstad, M.; Popovic-Bijelic, A.; Shova, S.; Büchel, G.E.; Darvasiova, D.; Rapta, P.; Arion, V.B. Coumarin-Based Triapine Derivatives and Their Copper(II) Complexes: Synthesis, Cytotoxicity and MR2 RNR Inhibition Activity. Biomolecules 2021, 11, 862. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, L.; Iacobazzi, R.M.; Quatrale, A.E.; Bergamini, C.; Denora, N.; Crupi, P.; Antonacci, D.; Mangia, A.; Simone, G.; Silvestris, N.; et al. Grape Seed Extracts Modify the Outcome of Oxaliplatin in Colon Cancer Cells by Interfering with Cellular Mechanisms of Drug Cytotoxicity. Oncotarget 2017, 8, 50845–50863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiebke, E.A.; Grieshop, N.A.; Loehrer, P.J.; Eckert, G.J.; Sidner, R.A. Antitumor Effects of 5-Fluorouracil on Human Colon Cancer Cell Lines: Antagonism by Levamisole. J. Surg. Res. 2003, 111, 63–69. [Google Scholar] [CrossRef]
- Jain, C.K.; Roychoudhury, S.; Majumder, H.K. Selective Killing of G2 Decatenation Checkpoint Defective Colon Cancer Cells by Catalytic Topoisomerase II Inhibitor. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1195–1204. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schütz, G.; Thierauch, K.-H.; Zopf, D. Regorafenib (BAY 73-4506): A New Oral Multikinase Inhibitor of Angiogenic, Stromal and Oncogenic Receptor Tyrosine Kinases with Potent Preclinical Antitumor Activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal Structure Analysis of Warfarin Binding to Human Serum Albumin. Anatomy of Drug Site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [Green Version]
- Vesković, A.; Nakarada, Đ.; Popović Bijelić, A. Spin-Labeled Hydrogels for Cell Viability Assessment by EPR. Free Rad. Biol. Med. 2021, 177, S76. [Google Scholar] [CrossRef]
- Swartz, H.M.; Boyer, S.; Gast, P.; Glockner, J.F.; Hu, H.; Liu, K.J.; Moussavi, M.; Norby, S.W.; Vahidi, N.; Walczak, T.; et al. Measurements of Pertinent Concentrations of Oxygen In Vivo. Magn. Reson. Med. 1991, 20, 333–339. [Google Scholar] [CrossRef]
- Berliner, L.J. (Ed.) In Vivo EPR (ESR): Theory and Applications; Kluwer Academic/Plenum Publishing Corp.: New York, NY, USA, 2003; Volume 18, Biological Magnetic Resonance; ISBN 978-1-4613-4906-8. [Google Scholar]
- Halpern, H.J.; Yu, C.; Peric, M.; Barth, E.; Grdina, D.J.; Teicher, B.A. Oxymetry Deep in Tissues with Low-Frequency Electron Paramagnetic Resonance. Proc. Natl. Acad. Sci. USA 1994, 91, 13047–13051. [Google Scholar] [CrossRef] [Green Version]
| Sample | Day | Observation | |
|---|---|---|---|
| 1:10 molar ratio | 1:20 molar ratio | ||
| HL/HSA solution | |||
| as prepared |
|
| |
| after dialysis in physiological saline | 1 |
|
|
| 8 |
|
| |
| HL/HSA hydrogel | |||
| as prepared |
|
| |
| after dialysis in physiological saline | 1 |
|
|
| 7 |
|
| |
| 8 |
|
| |
| 11 |
|
| |
| Ligand | Calculated Binding Affinity (kJ/mol) | |
|---|---|---|
| HSA | BSA | |
| HL | −33.89 | −30.54 |
| TEMPO | −25.52 | −17.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vesković, A.; Nakarada, Đ.; Vasiljević, O.; Dobrov, A.; Spengler, G.; Enyedy, É.A.; Arion, V.B.; Popović Bijelić, A. The Release of a Highly Cytotoxic Paullone Bearing a TEMPO Free Radical from the HSA Hydrogel: An EPR Spectroscopic Characterization. Pharmaceutics 2022, 14, 1174. https://doi.org/10.3390/pharmaceutics14061174
Vesković A, Nakarada Đ, Vasiljević O, Dobrov A, Spengler G, Enyedy ÉA, Arion VB, Popović Bijelić A. The Release of a Highly Cytotoxic Paullone Bearing a TEMPO Free Radical from the HSA Hydrogel: An EPR Spectroscopic Characterization. Pharmaceutics. 2022; 14(6):1174. https://doi.org/10.3390/pharmaceutics14061174
Chicago/Turabian StyleVesković, Ana, Đura Nakarada, Olga Vasiljević, Anatolie Dobrov, Gabriella Spengler, Éva A. Enyedy, Vladimir B. Arion, and Ana Popović Bijelić. 2022. "The Release of a Highly Cytotoxic Paullone Bearing a TEMPO Free Radical from the HSA Hydrogel: An EPR Spectroscopic Characterization" Pharmaceutics 14, no. 6: 1174. https://doi.org/10.3390/pharmaceutics14061174
APA StyleVesković, A., Nakarada, Đ., Vasiljević, O., Dobrov, A., Spengler, G., Enyedy, É. A., Arion, V. B., & Popović Bijelić, A. (2022). The Release of a Highly Cytotoxic Paullone Bearing a TEMPO Free Radical from the HSA Hydrogel: An EPR Spectroscopic Characterization. Pharmaceutics, 14(6), 1174. https://doi.org/10.3390/pharmaceutics14061174