An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
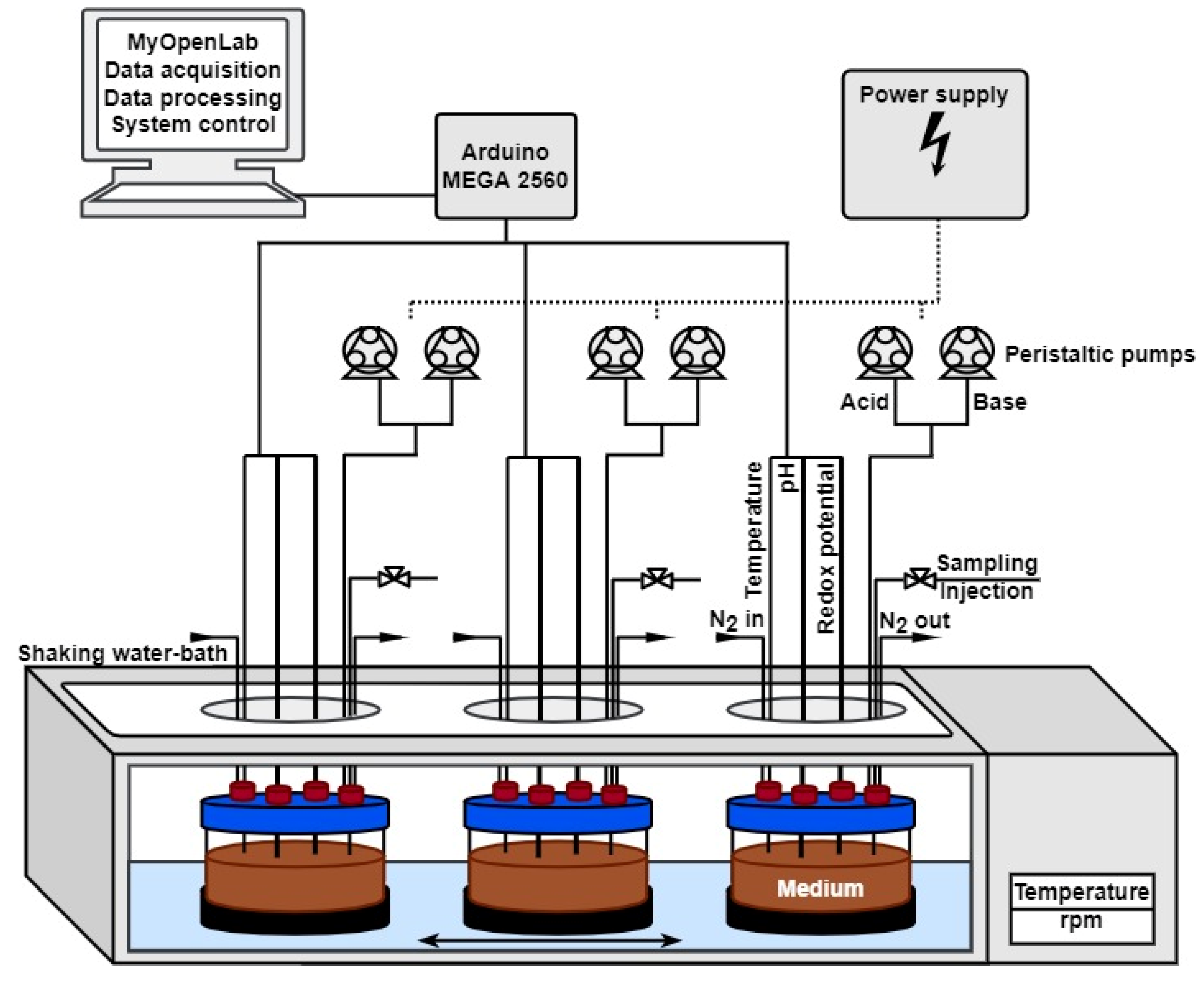
2.2.1. Dynamic Culture (MimiCol3)
2.2.2. Experimental Procedure
2.2.3. Determination of Bacterial Count by 16S rRNA Sequencing
3. Results
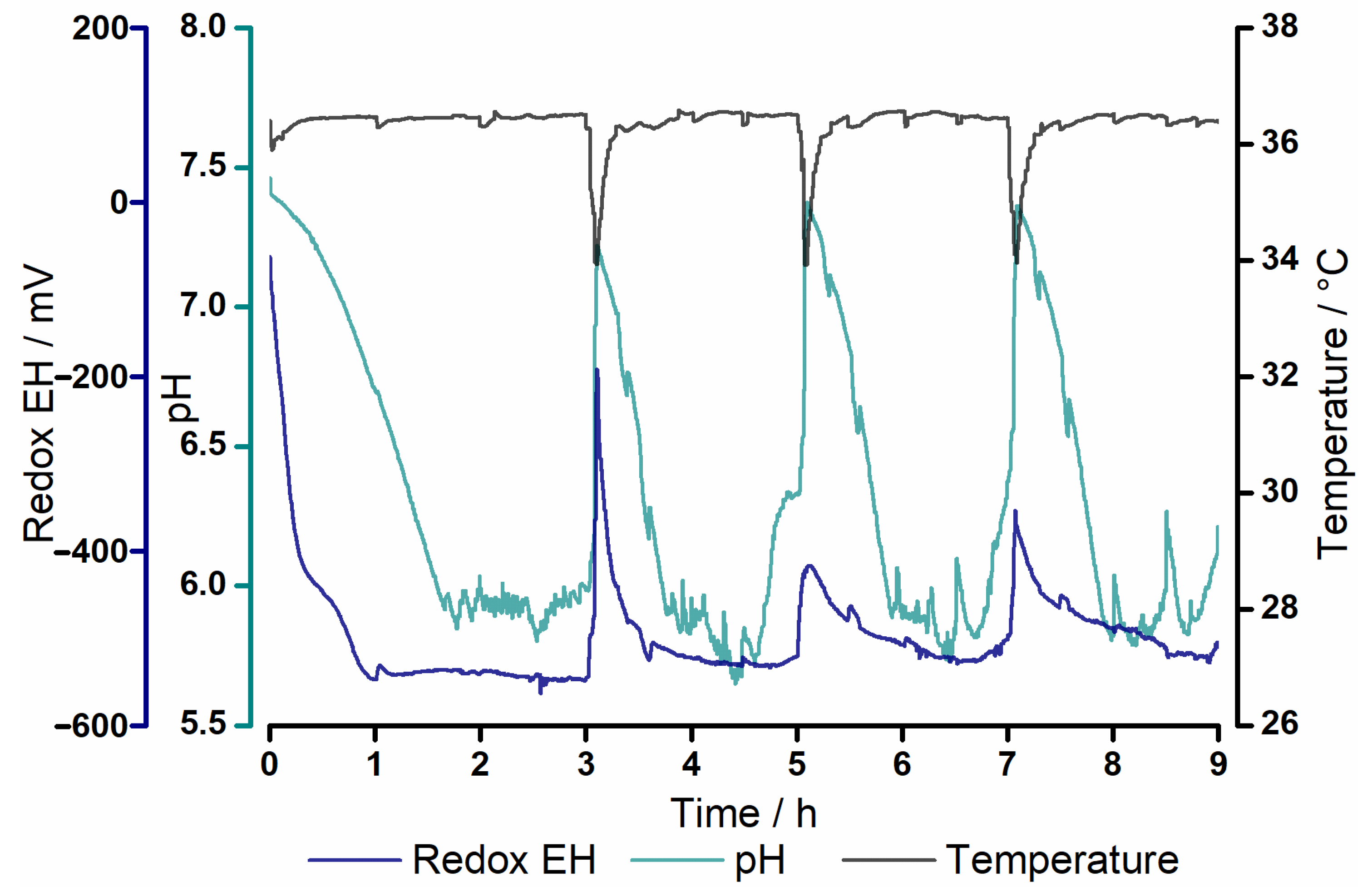
3.1. Process Parameters
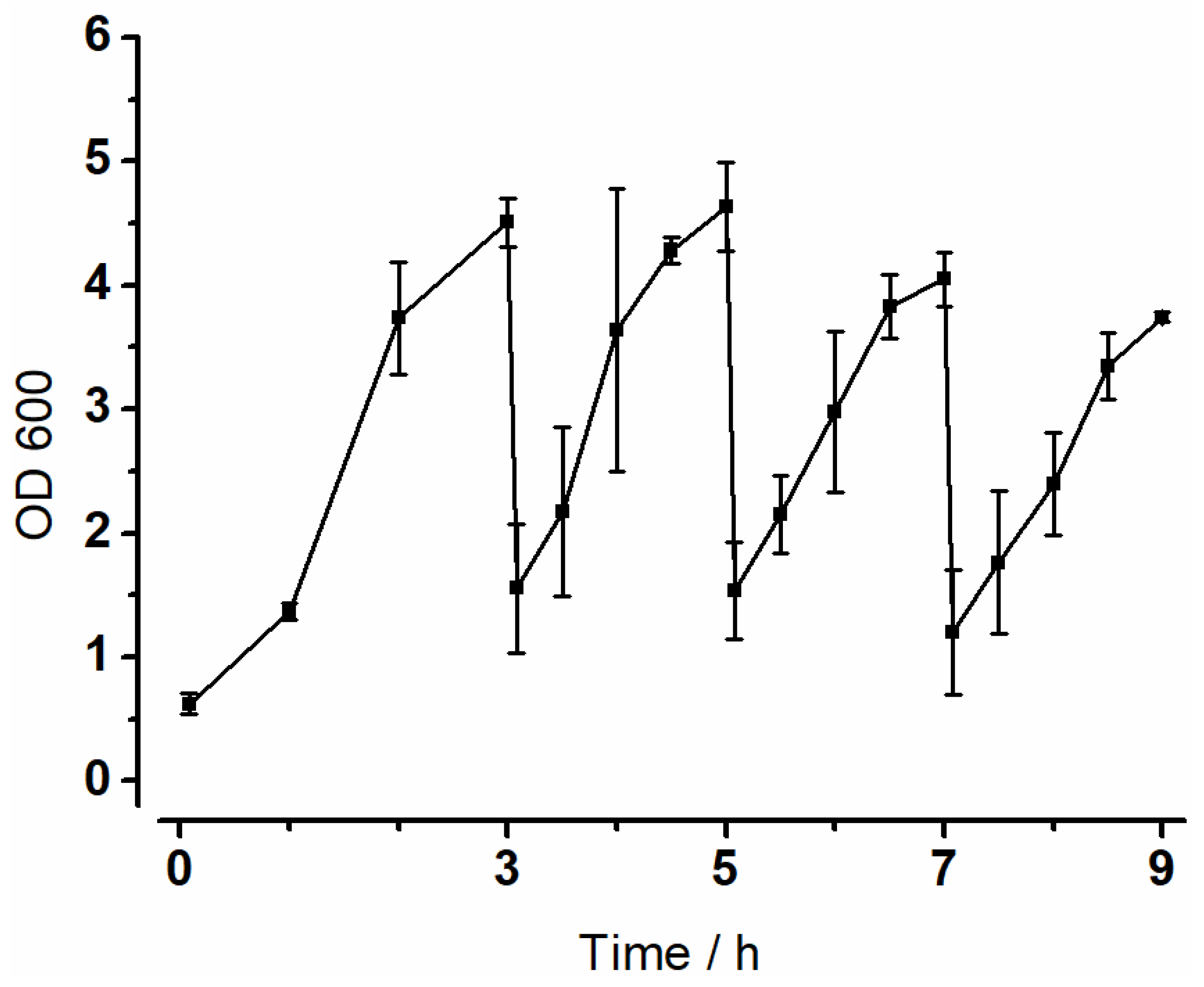
3.2. Optical Density (OD600)
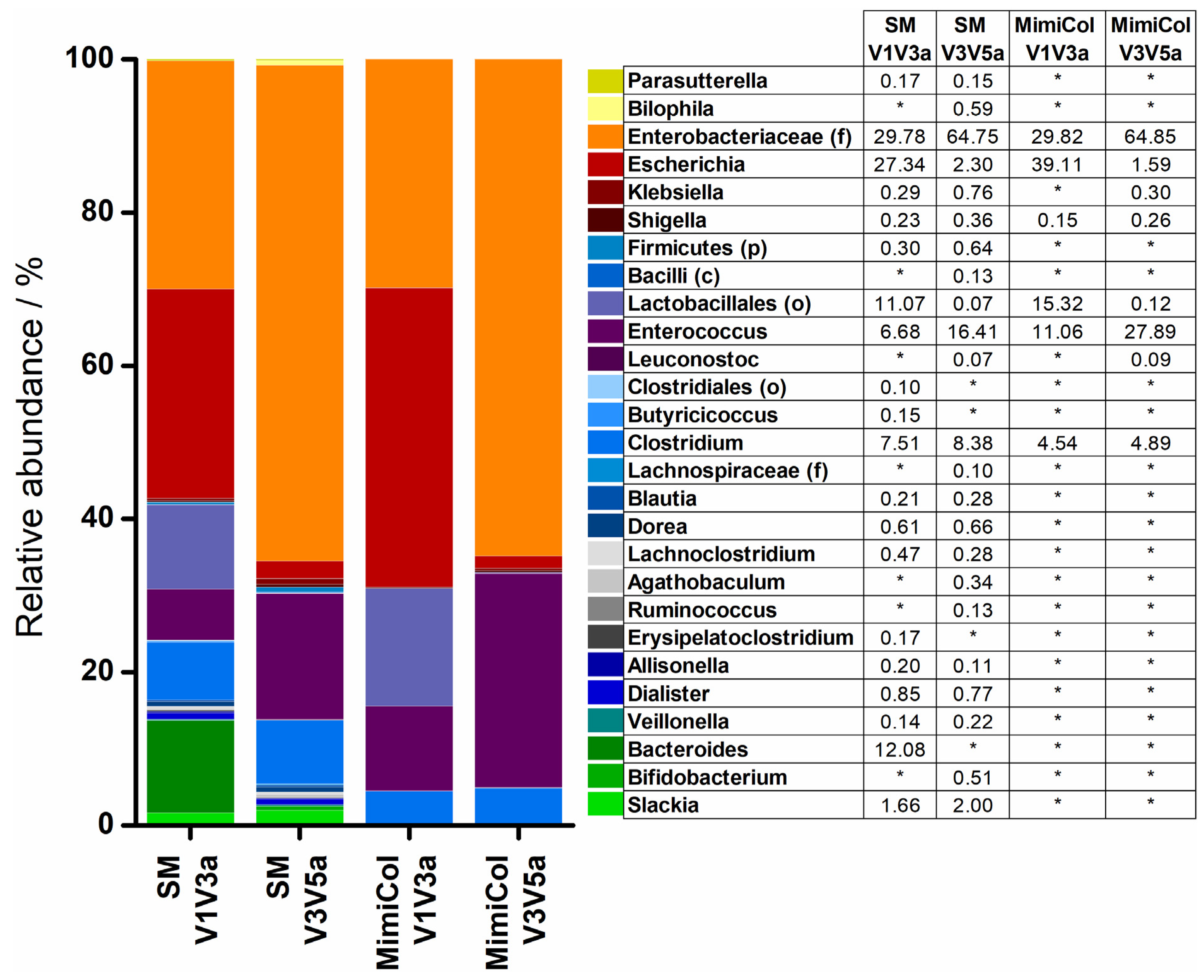
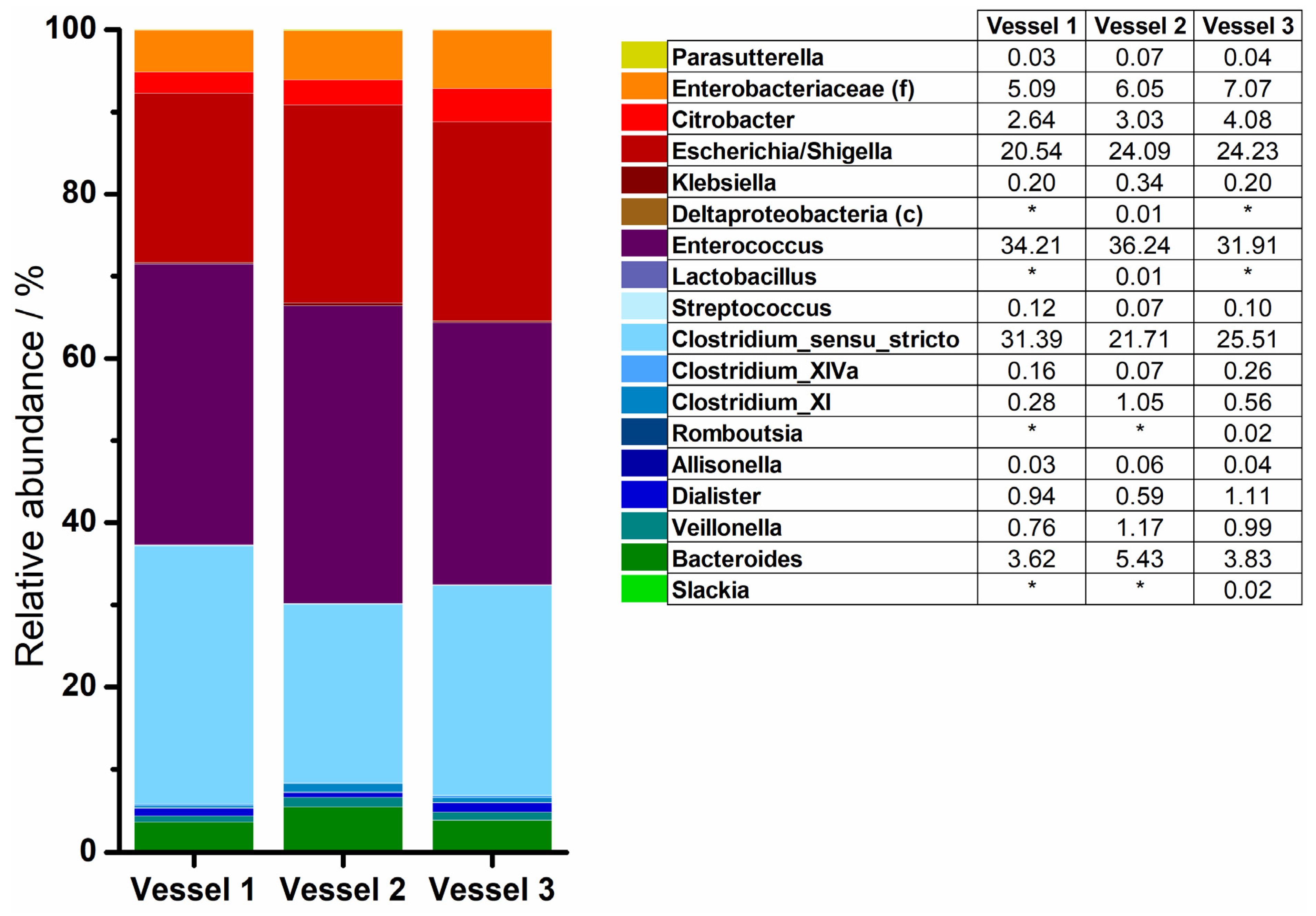
3.3. 16S rRNA Sequencing
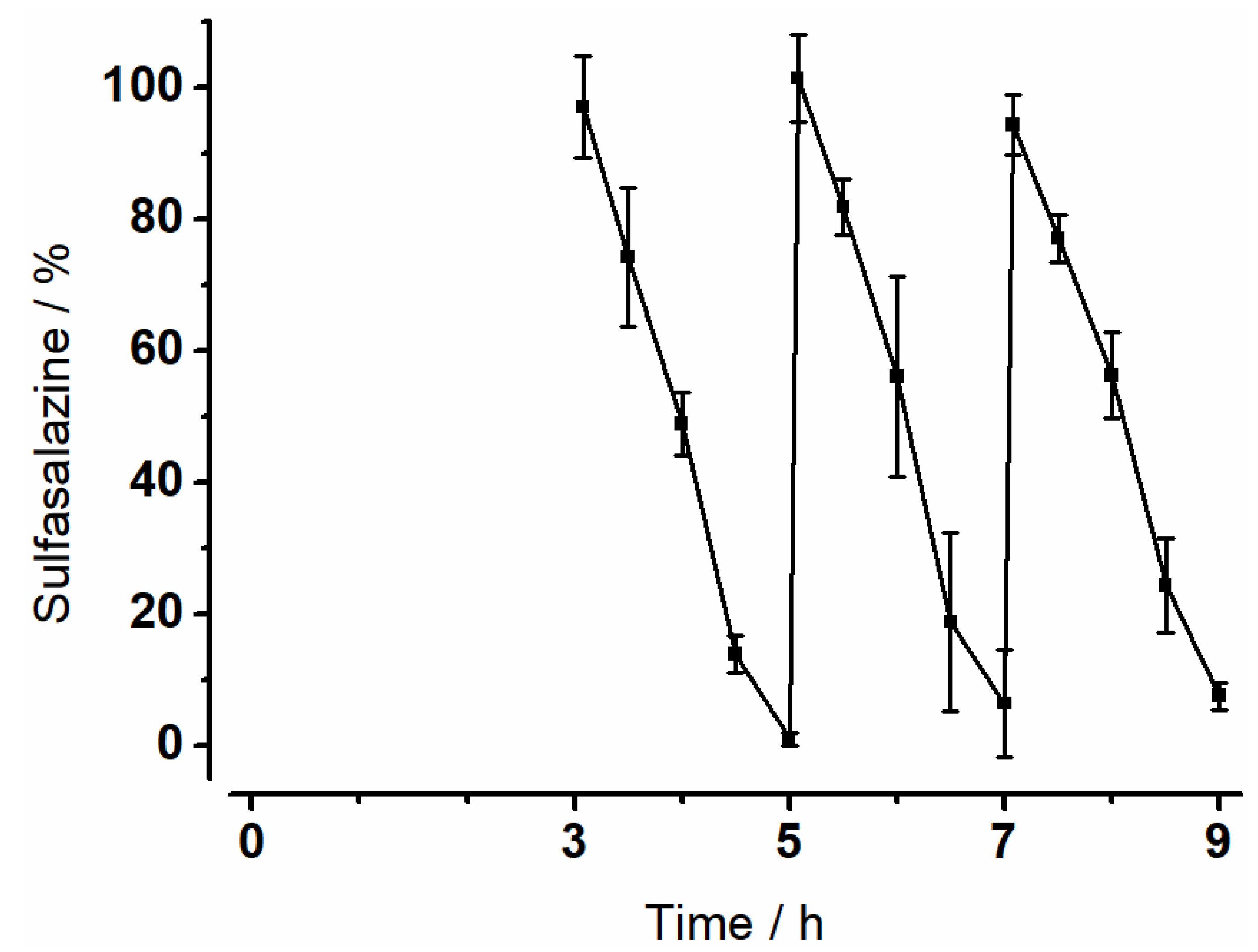
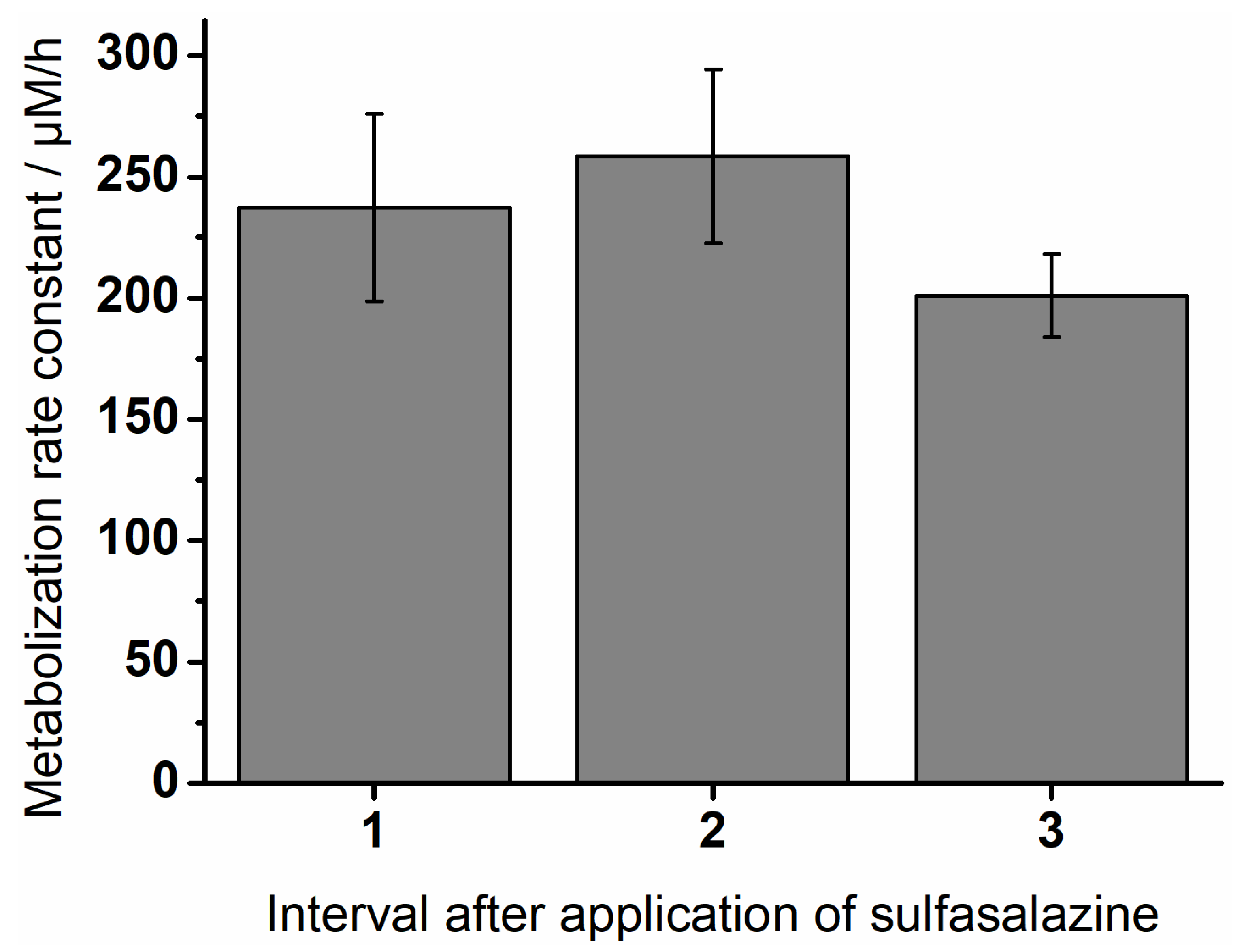
3.4. Degradation of Sulfasalazine
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Nissen, L.; Casciano, F.; Gianotti, A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: An updated review. FEMS Microbiol. Lett. 2020, 367, fnaa097. [Google Scholar] [CrossRef] [PubMed]
- Kotla, N.G.; Rana, S.; Sivaraman, G.; Sunnapu, O.; Vemula, P.K.; Pandit, A.; Rochev, Y. Bioresponsive drug delivery systems in intestinal inflammation: State-of-the-art and future perspectives. Adv. Drug Deliv. Rev. 2018, 146, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Goldin, B.R. Intestinal microflora: Metabolism of drugs and carcinogens. Ann. Med. 1990, 22, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, V.V.; Jayaprakash, R.; Mathew, S.T. Colon specific drug delivery systems: A review on various pharmaceutical approaches. J. Appl. Pharm. Sci. 2012, 2, 163–169. [Google Scholar]
- Scheline, R.R. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol. Rev. 1973, 25, 451–532. [Google Scholar]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- Minekus, M.; Phillipe, M.; Havenaar, R.; Huis in’t Veld, J.H.J. A Multicompartmental Dynamic Computer- controlled Model Simulating the Stomach and Small Intestine. Altern. Lab. Anim. 1995, 23, 197–209. [Google Scholar] [CrossRef]
- Minekus, M.; Smeets-Peeters, M.; Bernalier, A.; Marol-Bonnin, S.; Havenaar, R.; Marteau, P.; Alric, M.; Fonty, G.; Huis in’t Veld, J.H.J. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl. Microbiol. Biotechnol. 1999, 53, 108–114. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S.; Gibson, G.R. Validation of a Three-Stage Compound Continuous Culture System for Investigating the Effect of Retention Time on the Ecology and Metabolism of Bacteria in the Human Colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, M.; Axenstrand, M.; Håkansson, Å.; Marefati, A.; Pedersen, B.L. In vitro methods to study colon release: State of the art and an outlook on new strategies for better in-vitro biorelevant release media. Pharmaceutics 2019, 11, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.A.; Varum, F.; Al-Gousous, J.; Hofmann, M.; Page, S.; Langguth, P. In Vitro Methodologies for Evaluating Colon-Targeted Pharmaceutical Products and Industry Perspectives for Their Applications. Pharmaceutics 2022, 14, 291. [Google Scholar] [CrossRef] [PubMed]
- Beeck, R.; Glöckl, G.; Krause, J.; Schick, P.; Weitschies, W. Mimicking the dynamic Colonic microbiota in vitro to gain a better understanding on the in vivo metabolism of xenobiotics: Degradation of sulfasalazine. Int. J. Pharm. 2021, 603, 120704. [Google Scholar] [CrossRef]
- Koziolek, M.; Grimm, M.; Becker, D.; Iordanov, V.; Zou, H.; Shimizu, J.; Wanke, C.; Garbacz, G.; Weitschies, W. Investigation of pH and Temperature Profiles in the GI Tract of Fasted Human Subjects Using the Intellicap System. J. Pharm. Sci. 2015, 104, 2855–2863. [Google Scholar] [CrossRef]
- Tuǧcu-Demiröz, F.; Acartürk, F.; Takka, S.; Konuş-Boyunaǧa, Ö. In-vitro and in-vivo evaluation of mesalazine-guar gum matrix tablets for colonic drug delivery. J. Drug Target. 2004, 12, 105–112. [Google Scholar] [CrossRef]
- Yang, L. Biorelevant dissolution testing of colon-specific delivery systems activated by colonic microflora. J. Control. Release 2008, 125, 77–86. [Google Scholar] [CrossRef]
- Haksworth, G.; Drasar, B.S.; Hill, M.J. Intestinal Bacteria and the Hydrolysis of Glycosidic Bonds. J. Med. Microbiol. 1971, 4, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Neau, S.H. In vitro degradation of chitosan by bacterial enzymes from rat cecal and colonic contents. Biomaterials 2002, 23, 2761–2766. [Google Scholar] [CrossRef]
- Karrout, Y.; Neut, C.; Wils, D.; Siepmann, F.; Deremaux, L.; Flament, M.P.; Dubreuil, L.; Desreumaux, P.; Siepmann, J. Novel polymeric film coatings for colon targeting: Drug release from coated pellets. Eur. J. Pharm. Sci. 2009, 37, 427–433. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tse, H.; Yuen, K.-Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934. [Google Scholar] [CrossRef] [Green Version]
- Dieterich, W.; Schink, M.; Zopf, Y. Microbiota in the Gastrointestinal Tract. Med. Sci. 2018, 6, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Xie, L.; Li, Y.; Wei, C. More than 9,000,000 unique genes in human gut bacterial community: Estimating gene numbers inside a human body. PLoS ONE 2009, 4, e6074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badley, A.D.; Camilleri, M.; O’Connor, M.K. Noninvasive measurement of human ascending colon volume. Nucl. Med. Commun. 1993, 14, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Koziolek, M.; Schneider, F.; Grimm, M.; Mode, C.; Seekamp, A.; Roustom, T.; Siegmund, W.; Weitschies, W. Intragastric pH and pressure profiles after intake of the high-caloric, high-fat meal as used for food effect studies. J. Control. Release 2015, 220, 71–78. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 1991, 70, 443–459. [Google Scholar] [CrossRef]
- Clarridge, J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Thingholm, L.B.; Skiecevičiene, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in Vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef]
- Frost, F.; Kacprowski, T.; Rühlemann, M.; Pietzner, M.; Bang, C.; Franke, A.; Nauck, M.; Völker, U.; Völzke, H.; Dörr, M.; et al. Long-term instability of the intestinal microbiome is associated with metabolic liver disease, low microbiota diversity, diabetes mellitus and impaired exocrine pancreatic function. Gut 2021, 70, 522–530. [Google Scholar] [CrossRef]
- Sousa, T.; Paterson, R.; Moore, V.; Carlsson, A.; Abrahamsson, B.; Basit, A.W. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 2008, 363, 1–25. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A readily prepared medium for the cultivation of the lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Moore, W.E.C.; Cato, E.P.; Holdeman, L.V. Some current concepts in intestinal bacteriology. Am. J. Clin. Nutr. 1978, 31, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P.; Pochart, P.; Doré, J.; Béra-Maillet, C.; Bernalier, A.; Corthier, G. Comparative Study of Bacterial Groups within the Human Cecal and Fecal Microbiota. Appl. Environ. Microbiol. 2001, 67, 4939–4942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic metabolism in the genus Lactobacillus: Impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef] [Green Version]
- Stalons, D.R.; Thornsberry, C.; Dowell, V.R. Effect of Culture Medium and Carbon Dioxide Concentration on Growth of Anaerobic Bacteria Commonly Encountered in Clinical Specimens. Appl. Microbiol. 1974, 27, 1098–1104. [Google Scholar] [CrossRef]
- Peppercorn, M.A. Sulfasalazine. Pharmacology, clinical use toxicity, and related new drug development. Ann. Intern. Med. 1984, 101, 377–386. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Hanauer, S.B. Systematic review: The pharmacokinetic profiles of oral mesalazine formulations and mesalazine pro-drugs used in the management of ulcerative colitis. Aliment. Pharmacol. Ther. 2003, 17, 29–42. [Google Scholar] [CrossRef]
- Volin, M.V.; Harlow, L.A.; Woods, J.M.; Campbell, P.L.; Asif Amin, M.; Tokuhira, M.; Koch, A.E. Treatment with sulfasalazine or sulfapyridine, but not 5-Aminosalicylic acid, inhibits basic fibroblast growth factor-induced endothelial cell chemotaxis. Arthritis Rheum. 1999, 42, 1927–1935. [Google Scholar] [CrossRef]
- De Gunzburg, J.; Ducher, A.; Modess, C.; Wegner, D.; Oswald, S.; Dressman, J.; Augustin, V.; Feger, C.; Andremont, A.; Weitschies, W.; et al. Targeted adsorption of molecules in the colon with the novel adsorbent-based Medicinal Product, DAV132: A proof of concept study in healthy subjects. J. Clin. Pharmacol. 2015, 55, 10–16. [Google Scholar] [CrossRef]
- Kennedy, M.; Chinwah, P.; Wade, D.N. A Pharmacological Method of Measuring Mouth Caecal Transit Time in Man. Br. J. Clin. Pharmacol. 1979, 8, 372–373. [Google Scholar] [CrossRef] [Green Version]
- Broesder, A.; Bircan, S.Y.; de Waard, A.B.; Eissens, A.C.; Frijlink, H.W.; Hinrichs, W.L.J. Formulation and In Vitro Evaluation of Pellets Containing Sulfasalazine and Caffeine to Verify Ileo-Colonic Drug Delivery. Pharmaceutics 2021, 13, 1985. [Google Scholar] [CrossRef] [PubMed]
- Kellow, J.E.; Borody, T.J.; Phillips, S.F.; Haddad, A.C.; Brown, M.L. Sulfapyridine appearance in plasma after salicylazosulfapyridine. Another simple measure of intestinal transit. Gastroenterology 1986, 91, 396–400. [Google Scholar] [CrossRef]
- Rafii, F.; Cerniglia, C.E. Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ. Health Perspect. 1995, 103, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Rafii, F.; Franklin, W.; Cerniglia, C.E. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl. Environ. Microbiol. 1990, 56, 2146–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| In Vivo [17] | MimiCol3 | |
|---|---|---|
| Volume | 170 ± 40 mL ascending colon [24] | 150 mL |
| Motility | Pendulum movements, segmentations and fast mass transfer | Shaking at 100 rpm |
| pH | 5–8 (mean pH 6.5 ± 0.3) [25] | 6.2 ± 0.25 pH profile starting at pH 7.4 |
| Microbiota | 1011 CFU/mL [26] | Standard microbiota derived from a fecal sample of a healthy human volunteer (2.82 × 1010 CFU) |
| Temperature | 37 °C | 37 °C |
| Anaerobic atmosphere | Anaerobic, microaerophilic | Head space gassing with N2 |
| Bacterial Sub-Population | Agar Medium |
|---|---|
| Enterobacteria | MacConkey agar |
| Lactobacilli | deMan, Rogosa and Sharpe (MRS) agar |
| Bifidobacteria | Bifidus Selective Medium (BSM) agar |
| Clostridia | Modified reinforced clostridial medium |
| Bacteroides | Modified Schaedler agar |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beeck, R.; Dols, A.; Schneider, F.; Seradj, D.-S.; Krause, J.; Schick, P.; Weitschies, W. An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3. Pharmaceutics 2022, 14, 1049. https://doi.org/10.3390/pharmaceutics14051049
Beeck R, Dols A, Schneider F, Seradj D-S, Krause J, Schick P, Weitschies W. An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3. Pharmaceutics. 2022; 14(5):1049. https://doi.org/10.3390/pharmaceutics14051049
Chicago/Turabian StyleBeeck, Regine, Annemarie Dols, Felix Schneider, Dariah-Sohreh Seradj, Julius Krause, Philipp Schick, and Werner Weitschies. 2022. "An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3" Pharmaceutics 14, no. 5: 1049. https://doi.org/10.3390/pharmaceutics14051049
APA StyleBeeck, R., Dols, A., Schneider, F., Seradj, D.-S., Krause, J., Schick, P., & Weitschies, W. (2022). An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3. Pharmaceutics, 14(5), 1049. https://doi.org/10.3390/pharmaceutics14051049