Abstract
Crustins are an antimicrobial peptide (AMP) family that plays an important role in innate immunity in crustaceans. It is important to discover new AMPs from natural sources to expand the current database. Here, we identified and characterized a new crustin family member, named AaCrus1, from Amphibalanus amphitrite. AaCrus1 shares high identity (48.10%) with PvCrus, a Type I crustin of Penaeus vannamei that possesses a whey acidic protein (WAP) domain. AaCrus1 contains 237 amino acids and eight cysteine residues forming conserved ‘four-disulfide core’ structure. Our recombinant AaCrus1 (rAaCrus 1) could inhibit the growth of two Gram-positive bacteria (Staphylococcus aureus, Bacillus sp. T2) and four Gram-negative bacteria (Vibrio parahaemolyticus, Vibrio harveyi, Vibrio anguillarum, Vibrio alginolyticus) with a minimum inhibitory concentration of 3.5–28 μM. It can further induce agglutination of both Gram-positive and Gram-negative bacteria. rAaCrus1 can bind to bacteria and damage bacterial cell membranes. Furthermore, rAaCrus1 disrupted biofilm development of S. aureus and V. parahaemolyticus. Our discovery and characterization of this new crustin can be further optimized as a good alternative to antibiotics.
1. Introduction
The advent of antibiotics effectively treats diseases caused by bacterial infections. Furthermore, antibiotics are used in aquaculture to treat pathogenic infections in aquaculture animals. Vibrio sp. infection is one of the main diseases hindering the development of aquaculture, and some Vibrio sp. can cause serious foodborne illness in humans and animals [1]. However, the overdosed antibiotics and global interconnectedness has accelerated the emergence of antibiotic-resistant strains and ecological pollution. The World Health Organization has urgently called upon scientists to find novel effective alternatives to antibiotics. Drug-resistant Vibrio parahaemolyticus strains have been detected in China, South Korea and other places [1]. Antimicrobial peptides (AMPs), also known as host defense peptides, are a class of short peptides with biological activity that are widely present in nature [2,3]. As an important part of the biological innate immune system, AMPs have broad-spectrum antibacterial properties, which have a mild inhibitory or killing effect on Gram-negative bacteria, Gram-positive bacteria, fungi, viruses and parasites [4]. AMPs are less likely to increase the resistance of bacteria and are excellent alternatives to antibiotics [2,5,6].
Crustaceans are ancient animals with a large number and variety which are widely distributed in the ocean, freshwater ponds and lakes. Crustaceans mainly use humoral immunity, cellular immunity and other innate immune methods to combat pathogenic microorganisms. As part of their immune system, AMPs from crustaceans display remarkable diversity in their structural and genetic composition, which can serve as future templates for novel antimicrobial agents [7,8]. Since the first crustacean AMP was discovered in Carcinus maenas in 1996 [9], there are around 70 crustacean AMPs now recorded in the ADP database (http://aps.unmc.edu/AP/, accessed on 1 December 2021). Due to its species diversity, more new crustacean antibacterial peptides are being discovered annually [10,11].
Crustins, discovered in crustaceans, are generally cationic AMPs with 6–22 kDa and contain twelve conserved cysteine residues, eight of which comprise a typical whey acid protein (WAP) domain [12]. The WAP domain consist of a four-disulfide bond core arrangement at the C-terminus and is potentially associated with multiple functions, such as antimicrobial activity and proteinase inhibition [13,14]. The WAP domain at the C-terminus is very conservative, but the length and amino acid sequence at the N-terminus are highly variable [13,14]. As shown in Figure 1, according to the sequence difference in N-terminal, crustins were divided initially into four members [10,11]. The N-terminal has a cysteine-rich region that is to be found in type I crustin [15]. Type II crustins have a glycine-rich region between the cysteine-rich region and the signal peptide region [12]. Type III crustins have no cysteine-rich region and glycine-rich region, only a proline-rich region and WAP domain [16]. Type IV crustins have two WAP domains [17]. Though the four types of crustins all show good antimicrobial activity, different types of crustins with various structures cause differences in antibacterial effects. Some studies found that crustins may inhibit bacterial growth through protease inhibitory or cell membrane damage [11,14].
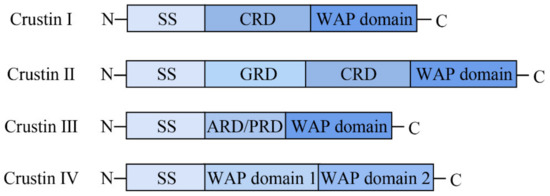
Figure 1.
Schematic representation (not to scale) of the structural organization of the four crustin Types (I to IV) found in crustaceans.
It is generally believed that the WAP domain of the four disulfide bonds in crustins is the key area for the antibacterial function [12,14]. The WAP domain of a crustin (ReCrus1) found in Rimicaris exoculata has an independent and different division of labor from other regions. The other regions are responsible for the binding of crustin to bacteria, and the WAP domain is responsible for killing bacteria [14]. Since AMPs with glycine and lysine rich are highly selective for Gram-negative bacteria [18], the glycine-rich region may be one of the reasons for the antibacterial effect of type II crustin. Some crustins have agglutinating activity by cross-linking with bacterial surface components to form a crystal lattice [19,20,21,22]. However, there are very few studies on crustins from non-decapod crustaceans [12,19].
A. amphitrite (Crustacea: Cirripedia) is a small striped barnacle commonly found in harbors and on ship hulls around the world. The microbial community and the development of barnacles influence each other, and the microbial community can affect the development of barnacles, promote or prevent barnacle larvae’s settlement and colonization [20,21]. Barnacles secrete lectin, serotonin and other substances to transform the microbial community around the microorganisms, and at the same time, need to use innate immunity to resist the invasion of pathogenic microorganisms [20]. Therefore, there may be AMPs with excellent antibacterial effects in barnacles. In order to find novel crustins in A. amphitrite, we constructed a crustin protein sequence database and compared the protein sequence of A. amphitrite in the NCBI database, thus identifying a new crustin member of the crustins family. With recombinant expression of the new crustin in Escherichia coli, we obtained a pure crustin protein without purification tag, named recombinant AaCrus1 (rAaCrus1). To further explore the AaCrus1 application in aquaculture industry, we evaluated its antibacterial activity and investigated the possible antibacterial mechanism against multiple Vibrio pathogens in aquaculture, including Vibrio parahaemolyticus, Vibrio harveyi, Vibrio anguillarum and Vibrio alginolyticus.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
The Gram-positive bacteria (Staphylococcus aureus, Bacillus sp. T2) and the Gram-negative bacteria (Vibrio parahaemolyticus, Vibrio harveyi, Vibrio anguillarum, Vibrio alginolyticus) were a gift from Professor Chaogang Wang [21,22]. They were originally glycerol bacteria and were stored at –80 °C. Before the experiment, the strains (glycerol bacteria) were cultured with Luria-Bertan (LB) medium shaken at 200 rpm, 37 °C overnight (usually about 10 h).
2.2. Prediction and Identification of Crustin Sequences
A. amphitrite genome information (NCBI access number: GCF_019059575.1) and 90 crustin sequences of Penaeus vannamei (49), Penaeus monodon (23) and Portunus trituberculatus (18) were obtained from NCBI. A local database was built using crustin sequences from other species by makeblastdb. We subsequently employed BLASTP (e-value: 1 × 10−5) to identify putative crustin sequences from A. amphitrite using the local database. Sequence homology searches were performed by the Protein BLAST algorithm in the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 December 2021). Signal peptide was identified by SignalP 5.0 [23]. Multiple alignments of amino acid sequences were built with ClustalW (https://www.genome.jp/tools-bin/clustalw, accessed on 1 December 2021), and the output pattern was visualized by DNAMAN 8 (Lynnon Biosoft, San Ramon, CA, USA). The neighbor-joining phylogenetic tree was performed by MEGA-X with 1000 bootstraps as assess reliability (Mega Limited, Auckland, New Zealand). ITOL6.3.2 was used to modify the evolutionary tree [24]. The full-length atomic model of rAaCrus1 was constructed with iterative template-based fragment assembly simulations using I-TASSER [25]. Protein physicochemical properties were predicted by ProtParam (https://web.expasy.org/protparam/, accessed on 1 December 2021).
2.3. Expression and Purification of Recombinant AaCrus1 (rAaCrus1)
The mature peptide of AaCrus1 was fused to the C-terminus of the His-SUMO tag, and prokaryotic expression was performed in E. coli BL21 (DE3) (TransGen Biotech, Beijing, China). The nucleotides in AaCrus1 (NCBI access number: VIIS01000966.1) were optimized according to the codon preference of E. coli, and BamHI and XhoI restriction sites were added at both ends of the sequence. The nucleotide sequence of the mature peptide of AaCrus1 is 726 bp. The gene was synthesized by the chemical synthesis method by General Biosystems (Anhui) Company (Chuzhou, China), and the synthesized DNA was ligated with pSmartI-SUMO vector through BamHI and XhoI restriction sites, and pSmartI-SUMO was provided by General Biosystems (Anhui) Company (Chuzhou, Anhui, China). The resulting recombinant plasmid was named pSmartI-SUMO-AaCrus1 (Supplementary Figure S1). pSmart-SUMO-AaCrus1 plasmid size is 6274 bp. AaCrus1EF: 5′-TTA AGA TTC TTG TAC GAC GG-3′ and AaCrus1ER: 5′-TGC TAG TTA TTG CTC AGC GG-3′ were primers. The PCR was performed at 95 °C for 3 min, with 35 cycles of 95 °C, 30 s, 51 °C, 30 s, 72 °C, 1 min and 72 °C, 5 min. General Biosystems (Anhui) Company delivers pSmartI-SUMO-AaCrus1 in the form of lyophilized powder. pSmartI-SUMO-AaCrus1 was transformed into E. coli BL21 (DE3) by heat shock method, and positive single clones were screened with solid LB medium contain kanamycin (50 µg/mL). Positive single clones confirmed by PCR and sequencing were used for recombinant protein production [26]. The positive signal clones were inoculated in LB medium containing kanamycin, and the bacterial solution was shaken at 200 r/min at 37 °C for overnight incubation. The next day, the bacterial liquid cultured overnight was added to fresh LB medium at a volume ratio of 1:100, the bacterial liquid was shaken at 37 °C at 200 r/min, and the bacterial liquid was cultured to OD600 = 0.6. Recombinant protein expression was induced by the addition of isopropyl-β-d-thiogalactoside (IPTG) to an ultimate concentration of 0.8 mM at 37 °C for 6 h. The cells were broken by ultrasonic means for 40 min (power: 195 W, ultrasonic for 2 s, pause for 4 s) and centrifuged at 4 °C for 30 min at 10,000 r/min to separate the supernatant and precipitate. The crude protein in the uninduced cells and the induced cells was extracted for 12% SDS-PAGE, and the crude protein in the induced cells was subjected to SDS-PAGE and Western Blot. The His-SUMO-AaCrus1 protein from the supernatant was isolated and purified by Ni-column affinity chromatography. The His-Sumo-AaCrus1 protein was dialyzed against 1× PBS for 24 h at 4 °C. His-SUMO-AaCrus1 protein was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For Western Blot, proteins were transferred onto a polyvinylidene fluoride membrane (PVDF), which was blocked with 5% skim milk in 1× TBST. The blot was incubated with His-Tag Mouse Monoclonal Antibody (1:1000, Beyotime, Shanghai, China) and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000, Beyotime, Shanghai, China), respectively. Detection was performed with the BeyoECL Plus (Beyotime, Shanghai, China) according to the manufacturer’s instructions. In order to completely remove the SUMO tag, 1 U SUMO (purchased from General Biosystems (Anhui) Company (Chuzhou, China)) enzyme was used at 4 °C for 6 h. The SUMO tag, which contained His tag, was intercepted by Ni-column affinity chromatography, and the unattached liquid was rAaCrus1 protein solution without tag. The purified rAaCrus1 protein was analyzed by SDS-PAGE. The protein concentration was determined using the BSA Protein Assay Kit (Beyotime, Shanghai, China) according to the manufacturer’s instruction. Finally, the protein solution was separated and lyophilized to obtain protein powder stored at −80 °C.
2.4. Identify rAaCrus1 by Liquid Chromatograph Mass Spectrometer (LC-MS)
The rAaCrus1 protein band obtained by SDS-PAGE gel electrophoresis was cut and placed in a 1.5 mL Eppendorf centrifuge tube, 50% volume fraction of acetonitrile was added, and the color was decolorized overnight at 37 °C with 200 r/min shaking. Using pure acetonitrile to make the colloid white and hard, the supernatant was discarded, trypsin digestion added, and then the covering solution (ammonium bicarbonate) was added in a 37 °C water bath for 16 h. After digestion by trypsin Eppendorf centrifuge tube, the solution was transferred to a new 1.5 mL, the extract added (water and anhydrous acetonitrile mixed in a volume ratio of 1:4, add 0.5% of the total volume of formic acid), sonicated for 10 min, centrifuged, and then the extract was vacuum dried for 4 h to obtain protein powder. The protein powder was dissolved with the sample solution (water and anhydrous acetonitrile in a volume ratio of 1:49, then add 0.5% formic acid) to dissolve the protein powder, vortexed and shaken 13,000× g at room temperature for 15 min, and finally, the LC-MS (TRIPLETOF 5600+, AB SCIEX, Singapore) was used to identify rAaCrus1.
2.5. Antimicrobial Activity of the rAaCrus1
In order to find as much novel potential rAaCrus1 as possible, we thus performed a modified CLSI microtiter plate assay to test the minimum inhibition concentration (MIC) with 104 CFU mL−1. The bacteria were cultured with the method described above until OD600 reached 0.4, and then diluted into 104 CFU mL−1 with Mueller–Hinton Broth medium. rAaCrus1 were dissolved with 1× PBS, and 20 μL peptide was mixed with 80 μL diluted strains in a 96 well plate. A serious final concentration of the peptide (56 μM, 28 μM, 14 μM, 7 μM, 3.5 μM, 1.75 μM, 0.875 μM) was used for MIC, and 1× PBS was the negative control. The plate was incubated at 37 °C for 18 h and OD600 was checked at 0 h and 18 h. The lowest concentration at which no growth of the indicator was observed was considered as the MIC of the peptide. The assay was performed with three times biological replicates and three times technical replicates.
2.6. Molecular Dynamics (MD) Simulations
Given the importance of peptide–membrane interaction under computational simulations [27,28]. A series of MD simulations were performed to study the interactions between AaCrus1 and membrane. AaCrus1 was described with the GROMOS 54a7 force field [29]. The membrane was modelled with 128 dipalmitoylphosphatidylcholine (DPPC) molecules in a united-atom force field by Berger et al. [30]. The solvent was represented by SPC water [31]. Two systems were simulated: AaCrus1 along and AaCrus1 membrane. A solvent box of 7.56 × 7.56 × 7.56 nm3 and 6.42 × 6.44 × 12.00 nm3 was built for the former and latter systems, respectively. In both systems, ions were added to maintain the neutral charge.
All MD simulations were carried out with GROMACS 2019.3 [32]. The cutoff for the short-range part of the Lennard–Jones and Coulomb interaction was set to 1.5 nm. The particle mesh Ewald algorithm [33,34] was used to compute the long-range part of the Coulomb interaction (with a grid space of 0.12 nm and a spline of order 4). The temperature was controlled at 323 K (above the phase transition temperature of DPPC) using the Nosé–Hoover algorithm [35,36] with the time constant of 0.5 ps. The pressure was maintained at 1 bar with the Parrinello–Rahman algorithm [37] in a semi-isotropic way where the time constant and compressibility was set to 5 ps and 5 × 10−5 (kJ·mol−1·nm−3)−1, respectively. All bonds were fixed using the LINCS algorithm [38], thus a time step of 2 fs could be applied. For each system, energy was first minimized, then was equilibrated under NVT condition for 200 ns, followed by an NPT simulation for 400 ns. The last 200 ns of trajectories were used for analysis.
2.7. Microorganism-Binding Assay
Western Blot experiment was used to detect the binding activity of rAaCrus1 to bacteria. Briefly, the microorganisms (1 × 108 CFU) in a 1.5 mL centrifuge tube were incubated in 200 μL of His-SUMO-AaCrus1(20 μM) by gentle rotation for 30 min at room temperature. The cells were collected and washed three times with 1× Tris Buffered Saline (TBS) and then resuspended. After the centrifugation at 10,000 rpm for 5 min, the bacteria and supernatant were directly loaded on SDS-PAGE for analysis, respectively. rAaCrus1 was the positive control. Finally, proteins were transferred onto a polyvinylidene fluoride membrane (PVDF), which was blocked with 5% skim milk in 1× TBST. The blot was incubated with His-Tag Mouse Monoclonal Antibody (1:1000, Beyotime, Shanghai, China) and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000, Beyotime, Shanghai, China), respectively. Detection was completed with the BeyoECL Plus (Beyotime, Shanghai, China), according to the manufacturer’s instructions.
2.8. Agglutinating Assay
To understand the function mechanism of rAaCrus1, six microorganisms described previously were selected for the agglutination assay. Briefly, six microorganisms were cultured at mid-logarithmic phase, centrifuged at 5000× g for 5 min, washed with 1× TBS four times and resuspended in 1× TBS to 108 CFU·mL−1. The microorganism suspension (30 μL) was incubated with a series of dilutions of rAaCrus1 in the range of 0.875–56 μM in 1× TBS (30 μL) with the absence or presence of 5 mM CaCl2 at 37 °C for 1 h. Compared to the negative control (400 μg/mL BSA), the minimal agglutinating concentration (MAC) in this research was identified as the lowest protein concentration harvesting apparent agglutination function. Agglutination was then observed under a Leica DMi1 Microscope (Leica, Wetzlar, Germany). All treatments were performed with three times biological replicates and three times technical replicates.
2.9. Electron Microscopy
The electron microscope was carried out with reference to previous studies [14]. S. aureus, V. anguillarum and V. alginolyticus were cultured in Luria–Bertan medium to mid-log phase. Then, the bacteria were collected and resuspended to 106 CFU/mL in 1× PBS. The bacteria were cultured at 2 × MIC rAaCrus1 concentration for 2 h on round coverslip in 24-well plates. After treatment, the cells were fixed with 5% glutaraldehyde in PBS (pH 7.4) for 2 h at 4 °C, followed by washing three times with 1× PBS. The bacteria treated with 1× PBS was the control. The bacteria were dehydrated in a series of concentrations of ethanol (30, 50, 70, 80, 90 and 100%) for 10 min at 4 °C and incubated with isoamyl acetate for 10 min [14]. The cells were sputter-coated with platinum (MC1000, Hitachi, Tokyo, Japan) after critically point-dried (Hitachi-HCP, Hitachi, Tokyo, Japan), and examined with scan electron microscopy (SEM) (APREO S, Thermo, Eindhoven, The Netherlands).
2.10. PI Staining Assay
S. aureus, V. anguillarum and V. alginolyticus were cultured as described above at their 2 × MIC concentration for 2 h. Then, the samples were stained with a PI staining kit (Sangon, Shanghai, China) according to the manufacturer’s instructions. The cells were observed with a fluorescence microscope (BX51, Olympus, Tokyo, Japan).
2.11. Biofilm Inhibition
S. aureus and V. parahaemolyticus were cultured according to the method described above. After the test strain was cultured at OD600 of 0.4, it was transferred to a 96-well plate treated with TC, and 200 μL of bacterial liquid was added to each well and cultured for 12 h. The bacterial solution was aspirated and the plate was washed with sterile PBS (pH 7.5). Then, rAaCrus1 solution (10–50 μM) was added to the wells and incubated at 37 °C for 24 h. After discarding the rAaCrus1 solution, the 96-well plates were washed with sterile 1× PBS (pH 7.5), then 100 μL 0.2% crystal violet staining solution (w/v) was added to each well and washed with sterile H2O after staining for 15 min. Once dry, 100 μL of absolute ethanol was added to each well, and the absorbance at a wavelength of 570 nm was measured. For microscopic analysis, the strains to be tested were cultured to an OD600 of 0.4, then transferred to a 24-well plate, where round coverslips were placed, and 200 μL of bacterial liquid was added to each well and cultured for 12 h. The round coverslips were then removed and washed with sterile PBS (pH 7.5). Then, round coverslips with biofilm were placed in a new 24-well plate, and rAaCrus1 solution (10–50 μM) was added to the 24-well plate so that it covered the round coverslips. Round coverslips were collected after incubating the 24-well plate at 37 °C for 24 h. After crystal violet staining, stained glass slides were placed on glass slides with the biofilm facing up and observed under a light microscope at magnification of 40× [39].
2.12. Statistical Analysis
GraphPad Prism 7.0 (GraphPad, San Diego, CA, USA) was selected for statistical analysis. Statistical significance was determined with one-way analysis of variance (ANOVA). All data are presented as mean ± SD. p value < 0.05 was considered statistically significant.
3. Results
3.1. Sequence and Structure Characterization of AaCrus1
The amino acid sequence of AaCrus1 (NCBI access number: KAF0303230,1) contains 257 residues with a calculated molecular weight of 26.63 kDa and a predicted pI of 8.47 by ProtParam. The CDS sequence encoding AaCrus1 is 774 bp. The nucleotide sequence of the mature peptide of AaCrus1 is 726 bp.
AaCrus1 possesses a signal peptide in the N-terminus (residues 1 to 20) and a WAP domain in the C-terminus, in which a ‘four-disulfide core’ structure can be formed by Cys212–Cys241, Cys219–Cys245, Cys228–Cys240 and Cys234–Cys251. Protein BLAST showed that AaCrus1 shares the highest identity (48.10%) with PvCrus, a Type I crustin of Penaeus vannamei (GenBank accession No. QOL09947.1). Sequence alignment between AaCrus1 and representative type I crustins showed that they both have eight cysteines that form a tetradisulfide core structure on the WAP domain (Figure 2A). In addition, four cysteines before WAP domain (corresponding to Cys180, Cys184, Cys198 and Cys199 in AaCrus1) were also conserved among the Type I crustins (Figure 2A). Compared with other type I crustins, there is a longer sequence between the signal peptide region and the cysteine-rich region. Phylogenetic analysis showed that AaCrus1 was grouped into the clade of type I crustins (Figure 2B). The predicted protein structure of AaCrus1 contains random coils, α-helix and β-pleated sheet (Figure 2C).
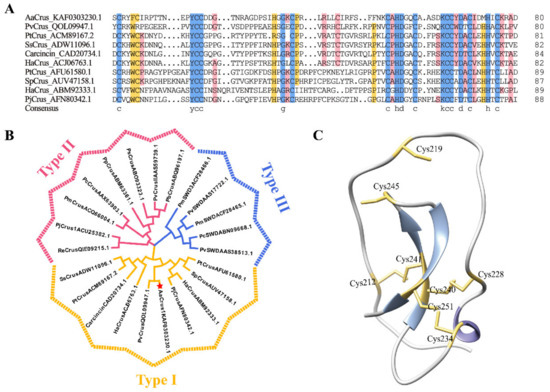
Figure 2.
Sequence alignments and phylogenetic and structural analysis of AaCrus1. (A) Alignment of AaCrus1 WAP domain and cysteine-rich domain with Type I crustins (shown in Supplementary Table S2). The consensus residues are shaded red; the residues that are ≥75% identical among the aligned sequences are shaded blue. The consensus residues are shaded red; the residues that are ≥75% identical among the aligned sequences are shaded blue. (B) Phylogenetic analysis of AaCrus1 homologues. The phylogenetic tree was constructed with MEGA-X using the neighbor-joining method. Numbers beside the internal branches indicate bootstrap values based on 1000 replications. The GenBank accession numbers of the crustins are indicated after crustins’ names. The crustin information used to construct the evolutionary tree is in Supplementary Table S3. (C) The predicted structure of Crus1 was built using I-TASSER. The cysteines in the WAP domain are shown in yellow, β-pleated sheet is shown in blue and α-helix is shown in purple.
3.2. Recombinant Expression and Purification
As SDS-PAGE analysis (Figure 3A) showed no significant difference in the protein expressed by the bacteria before and after the induction of IPTG. Western Blot analysis (Figure 3B) was further performed. Western Blot analysis showed that there are obvious bands between 35 kDa and 55 kDa, which are in line with the size of His-SUMO-AaCrus1 consisting of His-SUMO (~15 kDa) and AaCrus1 (26.63 kDa). The 200 µM imidazole eluent can elute His-SUMO-AaCrus1 from the Ni-column (Figure 3C, lane 6). After the treatment of SUMO enzyme, rAaCrus1 without His-SUMO tag was obtained and characterized in SDS-PAGE at 25–35 kDa (Figure 3D).
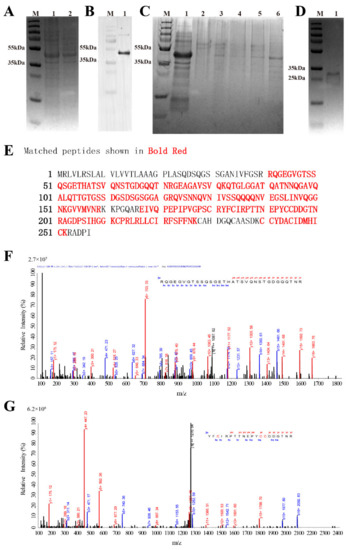
Figure 3.
The acquisition process and MS spectrum analysis of AaCrus1. (A) SDS-PAGE analysis of recombinant AaCrus1(rAaCrus1) expressed with a SUMO tag in E. coli. Lane M, protein marker; lane 1, total protein obtained from E. coli with IPTG induction; lane 2, total protein obtained from E. coli without induction. (B) Western Blot analysis of rAaCrus1 expressed with a SUMO tag in E. coli. Lane M, protein marker; lane 1, total protein obtained from E. coli with IPTG induction. (C) rAaCrus1 purified with nickel column chromatography. Lane M, protein marker; lane 1, protein not caught by the nickel column; lane 2, equilibration buffer; lane 3, eluent with 50 mM Imidazole; lane 4, eluent with 100 mM Imidazole, lane 5, eluent with 150 mM Imidazole; lane 6, eluent with 200 mM Imidazole. (D) SDS-PAGE analysis of rAaCrus1 without SUMO tag. Lane M, protein marker; lane 1, rAaCrus1 without SUMO tag. (E) Alignment of mass spectrometry results with rAaCrus1 sequence. (F) MS spectrum of “RQGEGVGTSSQSGETHATSVQNSTGDGQQTNR”. (G) MS spectrum of “YFCIRPTTNEPYCCDDGTNR”.
3.3. LC-MS Identification Results
Using Mascot software to compare the amino acid sequence of rAaCrus1, as shown in Figure 3E–G, a total of three peptides were detected, and the amino acid coverage reached 75%. It showed that with the prokaryotic expression obtained the fusion protein, the tag was successfully removed by the SUMO enzyme, and rAaCrus1 without foreign amino acids was obtained.
3.4. Antimicrobial Activity of rAaCrus1
The antibacterial activity of rAaCrus1 was tested against two Gram-positive and four Vibrio species, all of which were Gram-negative, by measuring the minimum inhibitory concentration (MIC) of each bacterium. As shown in Table 1, rAaCrus1 exhibited apparent inhibitory activities against Gram-positive bacteria and against Gram-negative bacteria. For Gram-negative bacteria, rAaCrus1 at a concentration of 7 μM can effectively inhibit the growth of V. alginolyticus. The most potent activity was detected against V. parahemolyticus and V. harveyi, with MIC of 28 μM. Compared with Gram-negative bacteria, AaCrus1 has a lower MIC value for the two Gram-positive bacteria used in this study; both are 3.5 μM.

Table 1.
The minimal inhibitory concentration (MIC) of AaCrus1 and ampicillin against Gram-positive and Gram-negative bacteria.
3.5. MD Simulations
The root–mean–square distances (RMSDs) and radii of gyration (Rg) of crustin in aqueous solution and membrane are plotted in Figure 4A,B. RMSDs reveal that both systems are already in equilibrium. Due to the membrane constraint, AaCrus1 is less flexible than in aqueous solution. The mean Rg of AaCrus1 in aqueous solution and membrane are 1.05 nm and 1.22 nm, respectively, indicating that in membrane, AaCrus1 is more extended. In Figure 4C,D, the root–mean–square fluctuations (RMSFs) for both systems are given. In aqueous solution, while the beta sheet region is highly rigid, loop regions exhibit different flexibility. Loop Cys212-Leu218 is almost as rigid as the beta sheet region. The disulfide bond between Cys212 and Cys241 (2.04 Å) plays an important role in stabilizing this region. Loop Ser206-Cys212 is the most flexible part in AaCrus1, and it exhibits the largest conformation change upon binding to the membrane. Loop Ser206-Phe222, deeply inserted into the membrane, anchors AaCrus1 to the membrane (see Figure 4E,F); thus, it is probably an essential part in determining the antimicrobial function of AaCrus1.
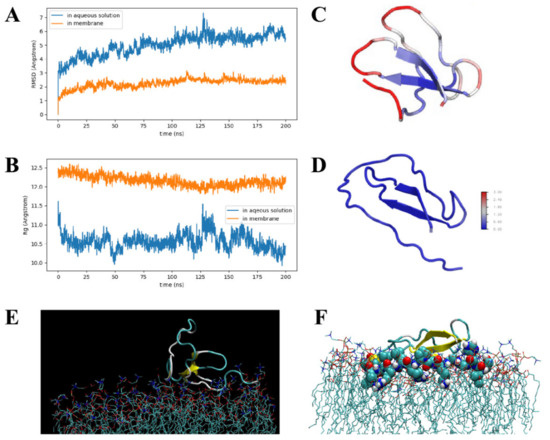
Figure 4.
Molecular dynamics simulation. (A)The root–mean–square distances (RMSDs) of AaCrus1 in aqueous solution and membrane. (B) The radii of gyration (Rg) of AaCrus1 in aqueous solution and membrane. (C) The root–mean–square fluctuations (RMSFs) of AaCrus1 in aqueous solution. (D) The root–mean–square fluctuations (RMSFs) of AaCrus1 in membrane. (E) AaCrus1 and membrane binding simulation (initial stage). (F) AaCrus1 and membrane binding simulation (medium stage).
3.6. Microorganism-Binding Activity of rAaCrus1
The results showed that rAaCrus1 could strongly bind to two Gram-positive bacteria (S. aureus, Bacillus sp. T2) and four Gram-negative bacteria (V. parahaemolyticus, V. harveyi, V. anguillarum, V. alginolyticus). These results indicated that rAaCrus1 might be binding to bacterial cells (Figure 5), which contribute to their antibacterial activity.
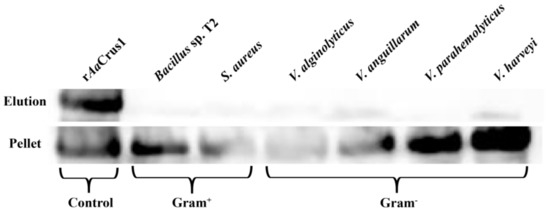
Figure 5.
Bacterial agglutination and microorganism-binding activity of rAaCrus1. rAaCrus1 was detected by Western Blot assay after treatment with bacteria (Gram+ and Gram−). rAaCrus1 was taken as a positive control. Up panel, elution fractions; bottom panel, final pellet fractions.
3.7. Agglutination Activity
After the agglutination test, rAaCrus1 had a good agglutination effect on Gram-positive bacteria and Gram-negative bacteria. According to Table 2 and Supplementary Figure S2, we found that Ca2+ can increase the agglutination effect of AaCrus1. The agglutination effect of rAaCrus1 on Gram-positive bacteria is better than that on Gram-negative bacteria.

Table 2.
Agglutinating activity of AaCrus1. Minimal agglutinating concentration is defined as the lowest protein concentration obtained with obvious agglutination, compared with the negative control.
3.8. Effects of rAaCrus1 on the Bacterial Morphology
After being treated with rAaCrus1 for two hours, the surfaces of S. aureus, V. anguillarum and V. alginolyticus were uneven, the cells had severely shrunk and obvious cracks appeared on the surface of the cells to release the intracellular components (Figure 6 and Supplementary Figure S3).
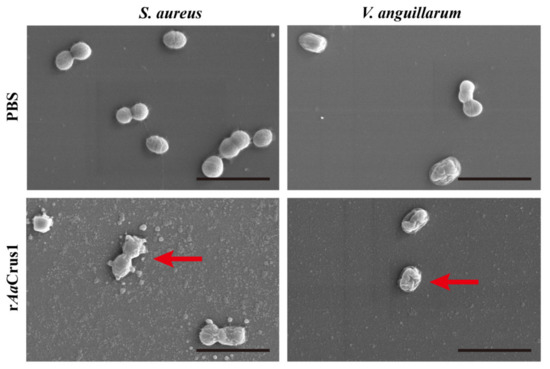
Figure 6.
Morphological and cell membrane integrity changes of the bacterial cells treated with rAaCrus1. Morphological changes of the bacterial cells treated with rAaCrus1. About 106 CFU·mL−1 bacteria were incubated with 2 × MIC of AaCrus1 for 2 h and observed under scan electron microscopy. PBS were used as control. The scales are 3 μm. At the red arrow, the suspected bacterial content can be seen flowing out, and the cell surface shrinks.
3.9. Cell Membrane Permeability
Propidium iodide (PI) cannot pass through living cell membranes, but it can pass through damaged cell membranes to stain the nucleus, as we described in previous reports [39,40]. PI staining showed that after rAaCrus1 treated S. aureus, V. anguillarum and V. alginolyticus, a large amount of PI was able to penetrate bacterial cells (Supplementary Figure S3), indicating that rAaCrus1 could cause bacterial cell membrane destabilization.
3.10. Biofilm Inhibition
According to Figure 7, rAaCrus1 disrupted development of S. aureus (Gram-positive bacteria) and V. parahaemolyticus (Gram-positive bacteria) biofilms. The higher the concentration of rAaCrus1, the better the disrupting effect on the biofilm development.
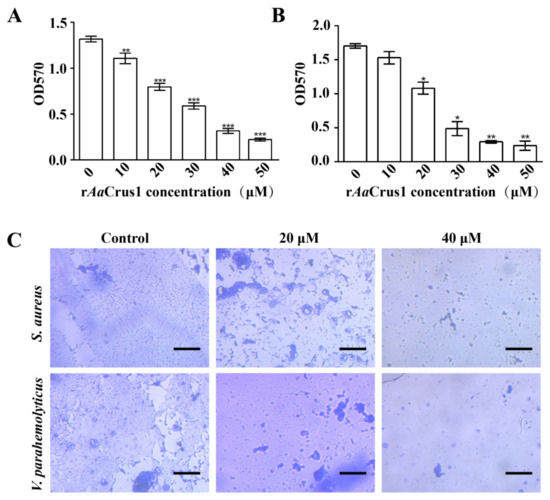
Figure 7.
Biofilm inhibitory effect of AaCrus1. Biofilm inhibitory concentration of rAaCrus1 (0–50 μg/mL) on S. aureus (A) and V. parahaemolyticus (B). *, Compared with control (0 μM), p value < 0.01; **, Compared with control (0 μM), p value < 0.001; ***, Compared with control (0 μM), p value < 0.0001. (C) Control and treated with the different concentrations of rAaCrus1 on S. aureus and V. parahaemolyticus biofilm development. The scales are 50 μm.
4. Discussion
Crustins were first purified from the blood cells of the shore crab C. maenas and have a good antibacterial effect on Gram-positive bacteria. To date, a variety of crustins have been found in crustaceans such as amphipods, decapods and isopods. Generally, type I crustins are mainly active against Gram-positive bacteria [41,42]. Type II and type III crustins are active against both Gram-positive and Gram-negative bacteria [9,43]. Type IV crustins have protease inhibitory activity and antibacterial activity [44]. Most crustins with good antibacterial effect usually work at a concentration below 50 μM [41,42]. As non-decapod crustaceans, the habitat of A. amphitrite is more complex than decapod crustaceans such as crabs and prawns, suggesting that A. amphitrite encounter more various pathogenic microorganisms. It has served as an important source to discover new crustins.
After the construction and comparison of the protein database of crustins of A. amphitrite, we discovered an atypical crustin consisting of 257 amino acids, named AaCrus1. Its molecular weight is 26.67 kDa, higher than the previously reported crustins. Despite having the same WAP domain as other crustins, AaCrus1 additionally has a cysteine-rich region before the WAP domain. The predicted structure model shows that AaCrus1, similar to other crustins [42], has a helix, a strand and some coils. There are 160 amino acids between the signal peptides and the cysteine-rich region of AaCrus1, which exceeds the number of amino acids in this region of type I crustins listed in Figure 2A and Supplementary Table S2. There are 28 glycines between the signal peptide 1 and the cysteine-rich region of AaCrus, however, BLAST and phylogenetic analysis showed that AaCrus1 is closely related to PvCrus (GenBank accession: QOL09947.1) and belongs to type I crustins. It is less than 50% similar to the decapod crustacean crustins (Supplementary Table S1). The molecular weight and sequences of AaCrus1 are different from the reported crustins, possibly because A. amphitrite are not decapod crustaceans.
Given the discovery of AaCrus1, a new crustin, we further applied a recombinant strategy to obtain a full sequence of the peptide. Compared with artificially synthesized AMPs [45], the prokaryotic recombinant expression system is low in cost and can form correctly folded proteins/peptides. Fusion expression AMPs with a SUMO tag can eliminate the toxicity of AMPs to prokaryotic hosts, prevent degradation by proteases and increase solubility [46,47]. The final AMPs were obtained by removing the SUMO tag by enzymatic digestion and purification. SDS-PAGE and LC-MS characterisation of the purified AMPs further confirmed the recombinant products, rAaCrus1. The missing region from Cys228 to Lys239 under LC-MS analysis may be caused by the highly constructed four-disulfide bond for difficulty of enzymatic digestion. MD simulations shown that disulfide bonds between Cys212 and Cys241 play an important role in stabilizing this region. These results indicate that the structure of AaCrus1 assembled by the prokaryotic expression system is consistent with the prediction.
Based on the results towards multiple Vibrio pathogens commonly existing in the aquaculture industry, our antibacterial activity result showed that rAaCrus1 is similar to other crustins and has broad-spectrum antibacterial activity against Gram-positive and Gram-negative bacteria. Bacterial agglutination test, microbial binding test, SEM and PI staining results indicate that rAaCrus1 may achieve antibacterial status by agglutinating bacteria and damaging cell membranes.
Agglutinating bacteria is one of the ways in which AMPs work. Positively charged AMPs can neutralize the charge of certain surface components (lipopolysaccharide, lipoteichoic acid or peptidoglycan) of bacteria, reducing the electrostatic repulsion between bacteria, leading to agglutination [22,48]. The positively charged rAaCrus1 (with lysine and arginine) can neutralize the anions on the surface of the bacteria and reduce the electrostatic repulsion on the surface of the bacteria. The binding of rAaCrus1 to bacteria was detected by Western Blot technology. It was found that rAaCrus1 can bind strongly to the tested bacteria; rAaCrus1 may bind to one or more bacterial surface components and interact with the bacterial surface. This may be the reason why rAaCrus1 can agglutinate two kinds of Gram-positive bacteria and four kinds of Gram-negative bacteria.
Some AMPs can exert antibacterial effects across the bacterial plasma membrane with the help of lipoteichoic acid and wall teichoic acid on the bacterial surface [49,50]. The peptidoglycan on the bacterial membrane can attract cationic AMPs to the bacterial cell membrane and promote insertion into the membrane, which will eventually lead to membrane leakage [51,52]. Our MD simulations showed that the coil part of the AaCrus1 structure first binds tightly to the phosphate surface of the upper layer of the bacterial cell membrane, and then Ser206-Phe222 in the WAP domain binds to the cell membrane and deeply inserts into it and anchors it. This may be one of the reasons for the strong binding of rAaCrus1 and bacteria in the microbial binding experiment. Electron microscopy further showed that the bacteria treated with rAaCrus1 showed cell surface atrophy and the contents flowed out, indicating that the bacterial cell membrane was damaged. The results of PI staining showed that rAaCrus1 changed the permeability of the bacterial cell plasma membrane. Therefore, we believe that the antibacterial mode of action of AaCrus1 may be that its WAP domain and glycine-rich region both play a role in binding to bacteria. The WAP domain and the cationic amino acid residues in it are deeply embedded in the cell membrane and cause the bacterial membrane to penetrate. Finally, AaCrus1 crosses to the bacterial cell plasma membrane, and the WAP domain continues to function to kill the bacteria.
Bacteria in the biofilm are a thousand times more resistant to antibiotics than scattered bacteria [53]. Some AMPs can inhibit the development of biofilms by degrading or preventing the production of important biofilm matrix components [54]. It is also possible to prevent the initial attachment to the surface by inhibiting swimming or swarm movement or interfering with flagella assembly to inhibit the development of biofilms. Through crystal violet staining, we found that AaCrus1 can inhibit the development of biofilms. Like other crustins, AaCrus1 may have protease inhibitory activity and affect the production of biofilm matrix components.
5. Conclusions
In this study, we discovered and characterized a new crustin, AaCrus1 (recombinant version rAaCrus1). By using makeblastdb for build a local database of crustins, retrieving crustins from barnacle genomes using blastp and the local crustins database, we found new non-decapod crustacean crustin. The recombinant expression further assisted us to obtain rAaCrus1 with His-SUMO tag, followed by the His-SUMO tag removal. All the recombinant analogues were characterized by SDS-PAGE, Western Blot and LC-MS. The bioassay showed its potent and broad-spectrum antibacterial activity and bacterial agglutination activity. Additional mechanistic study and computational modelling further suggested its possible antibacterial mechanism. AaCrus1 can help organisms resist microbial attack by agglutinating cells and can also bind to bacterial surfaces and disrupt bacterial cell membranes, ultimately leading to bacterial death. Our results suggest that this new AaCrus1 can serve as a template for the further development and design of promising antimicrobial agents and their applications in aquaculture.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pharmaceutics14020413/s1, Table S1: Blast result of AaCrus1 with other crustins in NCBI. Table S2: Information on the crustins used in the sequence alignment of Figure 2A. Table S3: Information on the crustin-I used to construct the evolutionary tree. Table S4: Information on the crustin-II used to construct the evolutionary tree. Table S5: Information on the crustin-III used to construct the evolutionary tree. Table S6: MIC of rAaCrus1 with tag and rAaCrus1. Figure S1: pSmartI-SUMO-AaCrus1 map. Figure S2: Agglutination of bacteria (Gram+ and Gram−) induced by rAaCrus1. Figure S3: The effect of Aarus1 on bacterial cell membrane integrity. Supplementary data: The nucleotide sequence of codon-optimized AaCrus1.
Author Contributions
Conceptualization, C.S. and W.L.; methodology, C.S., X.X. and W.Z.; software, J.Z.; validation, W.Z. and X.X.; data curation, C.S. and W.L.; writing—original draft preparation, W.Z. and C.S.; writing—review and editing, C.S. and W.L.; visualization, T.Y. and Q.Z.; supervision, Z.H. and Y.X.; project administration, Z.H. and C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Key R&D Program of China (2018YFA0902500), the National Natural Science Foundation of China (41706137, 41876188), Shenzhen Basic Research Projects (JCYJ20180507182405562), Guangxi Innovation Drive Development Special Fund (Gui Ke AA18242047), CAS Key Laboratory of Science and Technology on Operational Oceanography (No. OOST2021-07), the Natural Science Foundation of SZU (000002110258) and the Graduate Education Innovation Project (Guangdong province-2020JGXM094, SZUGS2019JG13). Zhangli Hu received funding for the Construction of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) and Shenzhen Special Fund for Sustainable Development.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data available are reported in the article.
Acknowledgments
We acknowledge the technical support of Biosciences Central Research Facility, Shenzhen University with LC-MS. We thank Instrument Analysis Center of Shenzhen University for the assistance with SEM analysis.
Conflicts of Interest
The authors declare that there are no conflict of interest.
References
- Loo, K.-Y.; Letchumanan, V.; Law, J.W.-F.; Pusparajah, P.; Goh, B.-H.; Ab Mutalib, N.-S.; He, Y.-W.; Lee, L.-H. Incidence of Antibiotic Resistance in Vibrio spp. Rev. Aquac. 2020, 12, 2590–2608. [Google Scholar] [CrossRef]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides with Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically Modified and Conjugated Antimicrobial Peptides against Superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, C.; Marquette, A.; Bechinger, B. The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. In Antimicrobial Peptides; Matsuzaki, K., Ed.; Springer Nature: Singapore, 2019; pp. 33–64. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.-R.; Jhong, J.-H.; Wang, Z.; Chen, S.; Wan, Y.; Horng, J.-T.; Lee, T.-Y. Characterization and Identification of Natural Antimicrobial Peptides on Different Organisms. Int. J. Mol. Sci. 2020, 21, 986. [Google Scholar] [CrossRef] [Green Version]
- Leite, M.L.; da Cunha, N.B.; Costa, F.F. Antimicrobial Peptides, Nanotechnology, and Natural Metabolites as Novel Approaches for Cancer Treatment. Pharmacol. Ther. 2018, 183, 160–176. [Google Scholar] [CrossRef]
- Smith, V.J.; Chisholm, J.R.S. Antimicrobial proteins in crustaceans. In Phylogenetic Perspectives on the Vertebrate Immune System; Beck, G., Sugumaran, M., Cooper, E.L., Eds.; Springer: Boston, MA, USA, 2001; pp. 95–112. [Google Scholar] [CrossRef]
- Zanjani, T.N.; Miranda-Saksena, M.; Cunningham, L.A.; Dehghani, F. Antimicrobial Peptides of Marine Crustaceans: The Potential and Challenges of Developing Therapeutic Agents. Curr. Med. Chem. 2018, 25, 2245–2259. [Google Scholar] [CrossRef]
- Matos, G.M.; Rosa, R.D. On the Silver Jubilee of Crustacean Antimicrobial Peptides. Rev. Aquac. 2021, 1–19. [Google Scholar] [CrossRef]
- Tincu, J.A.; Taylor, S.W. Antimicrobial Peptides from Marine Invertebrates. Antimicrob. Agents Chemother. 2004, 48, 3645–3654. [Google Scholar] [CrossRef] [Green Version]
- Destoumieux-Garzón, D.; Rosa, R.D.; Schmitt, P.; Barreto, C.; Vidal-Dupiol, J.; Mitta, G.; Gueguen, Y.; Bachère, E. Antimicrobial Peptides in Marine Invertebrate Health and Disease. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150300. [Google Scholar] [CrossRef] [Green Version]
- Smith, V.J.; Fernandes, J.M.O.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP Domain-Containing Antibacterial Proteins from Crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Albores, F.; Martínez-Porchas, M. Crustins Are Distinctive Members of the WAP-Containing Protein Superfamily: An Improved Classification Approach. Dev. Comp. Immunol. 2017, 76, 9–17. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Sun, Y.; Sun, L. A Crustin from Hydrothermal Vent Shrimp: Antimicrobial Activity and Mechanism. Marine Drugs 2021, 19, 176. [Google Scholar] [CrossRef]
- Li, M.; Ma, C.; Zhu, P.; Yang, Y.; Lei, A.; Chen, X.; Liang, W.; Chen, M.; Xiong, J.; Li, C. A New Crustin Is Involved in the Innate Immune Response of Shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 94, 398–406. [Google Scholar] [CrossRef]
- Yang, L.; Luo, M.; Guo, Z.; Zuo, H.; Weng, S.; He, J.; Xu, X. A Shrimp Gene Encoding a Single WAP Domain (SWD)-Containing Protein Regulated by JAK-STAT and NF-KB Pathways. Dev. Comp. Immunol. 2020, 104, 103537. [Google Scholar] [CrossRef]
- Tassanakajon, A.; Somboonwiwat, K.; Amparyup, P. Sequence Diversity and Evolution of Antimicrobial Peptides in Invertebrates. Dev. Comp. Immunol. 2015, 48, 324–341. [Google Scholar] [CrossRef]
- Ilić, N.; Novković, M.; Guida, F.; Xhindoli, D.; Benincasa, M.; Tossi, A.; Juretić, D. Selective Antimicrobial Activity and Mode of Action of Adepantins, Glycine-Rich Peptide Antibiotics Based on Anuran Antimicrobial Peptide Sequences. Biochim. Biophys. Acta Biomembr. 2013, 1828, 1004–1012. [Google Scholar] [CrossRef] [Green Version]
- Sruthy, K.S.; Nair, A.; Puthumana, J.; Antony, S.P.; Singh, I.S.B.; Philip, R. Molecular Cloning, Recombinant Expression and Functional Characterization of an Antimicrobial Peptide, Crustin from the Indian White Shrimp, Fenneropenaeus indicus. Fish Shellfish Immunol. 2017, 71, 83–94. [Google Scholar]
- Khandeparker, L.; Anil, A.C.; Desai, D.V. Immuno-Modulation of Settlement Cues in the Barnacle, Amphibalanus amphitrite: Significance of Circulating Haemocytes. Hydrobiologia 2019, 830, 229–241. [Google Scholar] [CrossRef]
- Rajitha, K.; Nancharaiah, Y.V.; Venugopalan, V.P. Role of Bacterial Biofilms and Their EPS on Settlement of Barnacle (Amphibalanus Reticulatus) Larvae. Int. Biodeterior. Biodegrad. 2020, 150, 104958. [Google Scholar] [CrossRef]
- Zhou, L.; Li, G.; Jiao, Y.; Huang, D.; Li, A.; Chen, H.; Liu, Y.; Li, S.; Li, H.; Wang, C. Molecular and Antimicrobial Characterization of a Group G Anti-Lipopolysaccharide Factor (ALF) from Penaeus monodon. Fish Shellfish Immunol. 2019, 94, 149–156. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Chen, J.; Li, J.; Shang, C.; Wang, C. Structural Characteristics, Prokaryotic Expression and Activity Analysis of Antimicrobial Peptide ALFPm10 from Penaeus monodon. Int. J. Peptide Res. Ther. 2022, 28, 1–10. [Google Scholar] [CrossRef]
- Li, W.; Lin, F.; Hung, A.; Barlow, A.; Sani, M.-A.; Paolini, R.; Singleton, W.; Holden, J.; Hossain, M.A.; Separovic, F.; et al. Enhancing Proline-Rich Antimicrobial Peptide Action by Homodimerization: Influence of Bifunctional Linker. Chem. Sci. 2022. [Google Scholar] [CrossRef]
- Lin, B.; Hung, A.; Li, R.; Barlow, A.; Singleton, W.; Matthyssen, T.; Sani, M.-A.; Hossain, M.A.; Wade, J.D.; O’Brien-Simpson, N.M.; et al. Systematic Comparison of Activity and Mechanism of Antimicrobial Peptides against Nosocomial Pathogens. Eur. J. Med. Chem. 2022, 231, 114135. [Google Scholar] [CrossRef]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and Testing of the GROMOS Force-Field Versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Berger, O.; Edholm, O.; Jähnig, F. Molecular Dynamics Simulations of a Fluid Bilayer of Dipalmitoylphosphatidylcholine at Full Hydration, Constant Pressure, and Constant Temperature. Biophys. J. 1997, 72, 2002–2013. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction models for water in relation to protein hydration. In Intermolecular Forces, Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry, Jerusalem, Israel, 13–16 April 1981; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; pp. 331–342. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N Log (N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Nosé, S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Li, W.; O’Brien-Simpson, N.M.; Yao, S.; Tailhades, J.; Reynolds, E.C.; Dawson, R.M.; Otvos, L., Jr.; Hossain, M.A.; Separovic, F.; Wade, J.D. C-Terminal Modification and Multimerization Increase the Efficacy of a Proline-Rich Antimicrobial Peptide. Chem. Eur. J. 2017, 23, 390–396. [Google Scholar] [CrossRef]
- Li, W.; O’Brien-Simpson, N.M.; Tailhades, J.; Pantarat, N.; Dawson, R.M.; Otvos, L.; Reynolds, E.C.; Separovic, F.; Hossain, M.A.; Wade, J.D. Multimerization of a Proline-Rich Antimicrobial Peptide, Chex-Arg20, Alters Its Mechanism of Interaction with the Escherichia coli Membrane. Chem. Biol. 2015, 22, 1250–1258. [Google Scholar] [CrossRef]
- Sperstad, S.V.; Haug, T.; Paulsen, V.; Rode, T.M.; Strandskog, G.; Solem, S.T.; Styrvold, O.B.; Stensvåg, K. Characterization of Crustins from the Hemocytes of the Spider Crab, Hyas araneus, and the Red King Crab, Paralithodes camtschaticus. Dev. Comp. Immunol. 2009, 33, 583–591. [Google Scholar] [CrossRef]
- Smith, V.J.; Dyrynda, E.A. Antimicrobial Proteins: From Old Proteins, New Tricks. Mol. Immunol. 2015, 68, 383–398. [Google Scholar] [CrossRef] [Green Version]
- Bandeira, P.T.; Vernal, J.; Matos, G.M.; Farias, N.D.; Terenzi, H.; Pinto, A.R.; Barracco, M.A.; Rosa, R.D. A Type IIa Crustin from the Pink Shrimp Farfantepenaeus paulensis (CrusFpau) Is Constitutively Synthesized and Stored by Specific Granule-Containing Hemocyte Subpopulations. Fish Shellfish Immunol. 2020, 97, 294–299. [Google Scholar] [CrossRef]
- Suthianthong, P.; Pulsook, N.; Supungul, P.; Tassanakajon, A.; Rimphanitchayakit, V. A Double WAP Domain-Containing Protein PmDWD from the Black Tiger Shrimp Penaeus monodon Is Involved in the Controlling of Proteinase Activities in Lymphoid Organ. Fish Shellfish Immunol. 2011, 30, 783–790. [Google Scholar] [CrossRef]
- Li, W.; O’Brien-Simpson, N.M.; Hossain, M.A.; Wade, J.D. The 9-Fluorenylmethoxycarbonyl (Fmoc) Group in Chemical Peptide Synthesis—Its Past, Present, and Future. Aust. J. Chem. 2020, 73, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Panavas, T.; Sanders, C.; Butt, T.R. SUMO Fusion Technology for Enhanced Protein Production in Prokaryotic and Eukaryotic Expression Systems. In SUMO Protocols; Ulrich, H.D., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 303–317. [Google Scholar]
- Shimokawa-Falcão, L.H.A.L.; Caporrino, M.C.; Barbaro, K.C.; Della-Casa, M.S.; Magalhães, G.S. Toxin Fused with SUMO Tag: A New Expression Vector Strategy to Obtain Recombinant Venom Toxins with Easy Tag Removal inside the Bacteria. Toxins 2017, 9, 82. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Li, S.; Li, F.; Xiang, J. Structure and Bioactivity of a Modified Peptide Derived from the LPS-Binding Domain of an Anti-Lipopolysaccharide Factor (ALF) of Shrimp. Marine Drugs 2016, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Koprivnjak, T.; Weidenmaier, C.; Peschel, A.; Weiss, J.P. Wall Teichoic Acid Deficiency in Staphylococcus aureus Confers Selective Resistance to Mammalian Group IIA Phospholipase A2 and Human β-Defensin 3. Infect. Immun. 2008, 76, 2169–2176. [Google Scholar] [CrossRef] [Green Version]
- Martin, N.I.; Breukink, E. The Expanding Role of Lipid II as a Target for Lantibiotics. Future Microbiol. 2007, 2, 513–525. [Google Scholar] [CrossRef]
- De Kruijff, B.; van Dam, V.; Breukink, E. Lipid II: A Central Component in Bacterial Cell Wall Synthesis and a Target for Antibiotics. Prostaglandins Leukot. Essent. Fat. Acids 2008, 79, 117–121. [Google Scholar] [CrossRef]
- Fernández, L.; Breidenstein, E.B.M.; Hancock, R.E.W. Creeping Baselines and Adaptive Resistance to Antibiotics. Drug Resist. Updates 2011, 14, 1–21. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm Activity of Host Defence Peptides: Complexity Provides Opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).