Development of a Combined Lipid-Based Nanoparticle Formulation for Enhanced siRNA Delivery to Vascular Endothelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Characterization of LNP
2.3. siRNA Integrity in Serum
2.4. Cell Culture and In Vitro Transfection
2.5. Cell Association with LNP and siRNA Cargo Release in HUVEC
2.6. Gene Expression Analysis by qPCR
2.7. Protein Expression Analysis by Western Blot
2.8. Cell Viability
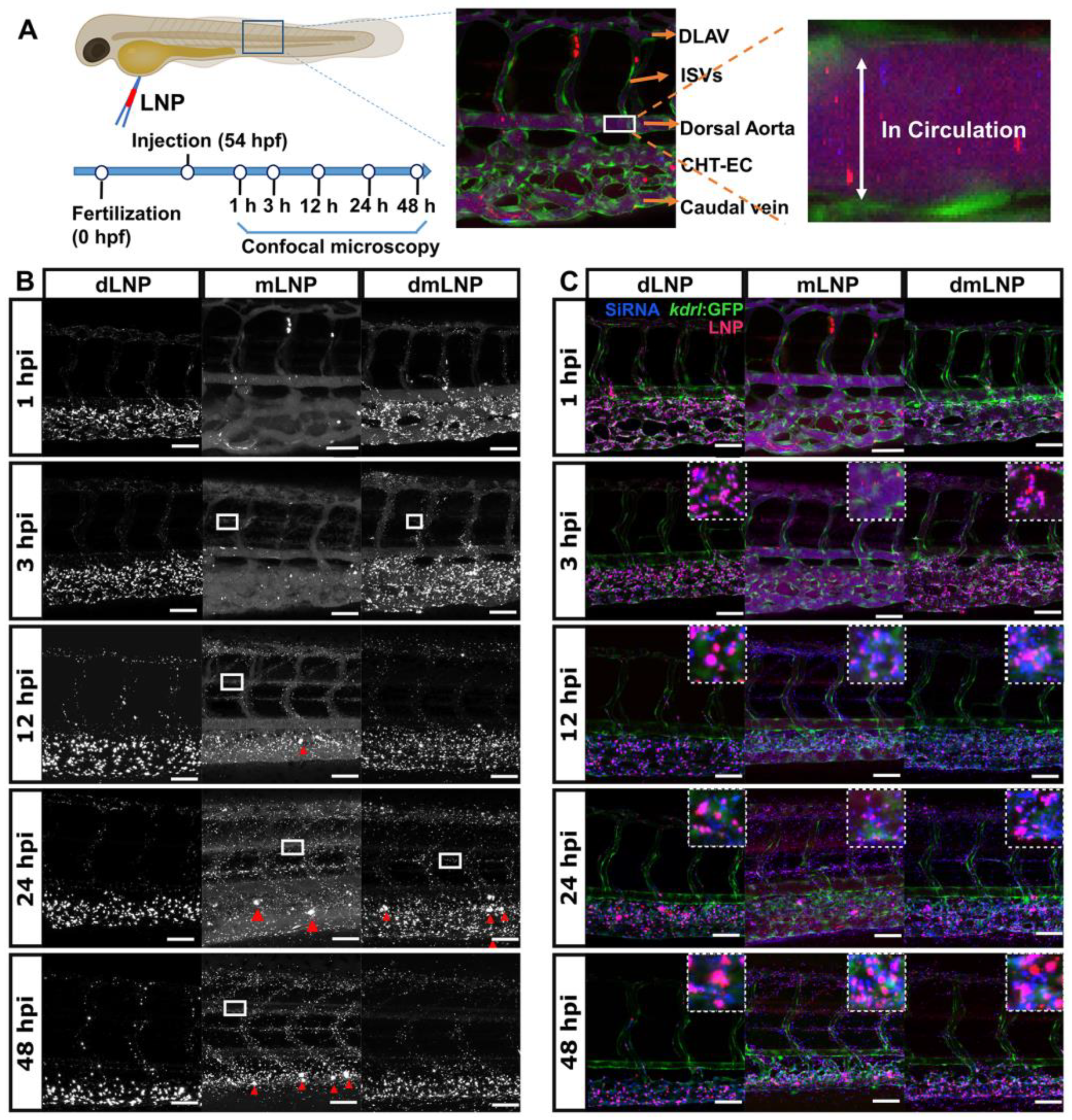
2.9. Zebrafish Husbandry and Injections
2.10. Zebrafish Imaging and Quantification of GFP Silencing in Zebrafish Embryos
2.11. Statistical Analysis
3. Results
3.1. Preparation and Characterization of LNP with Different Cationic Lipid Compositions
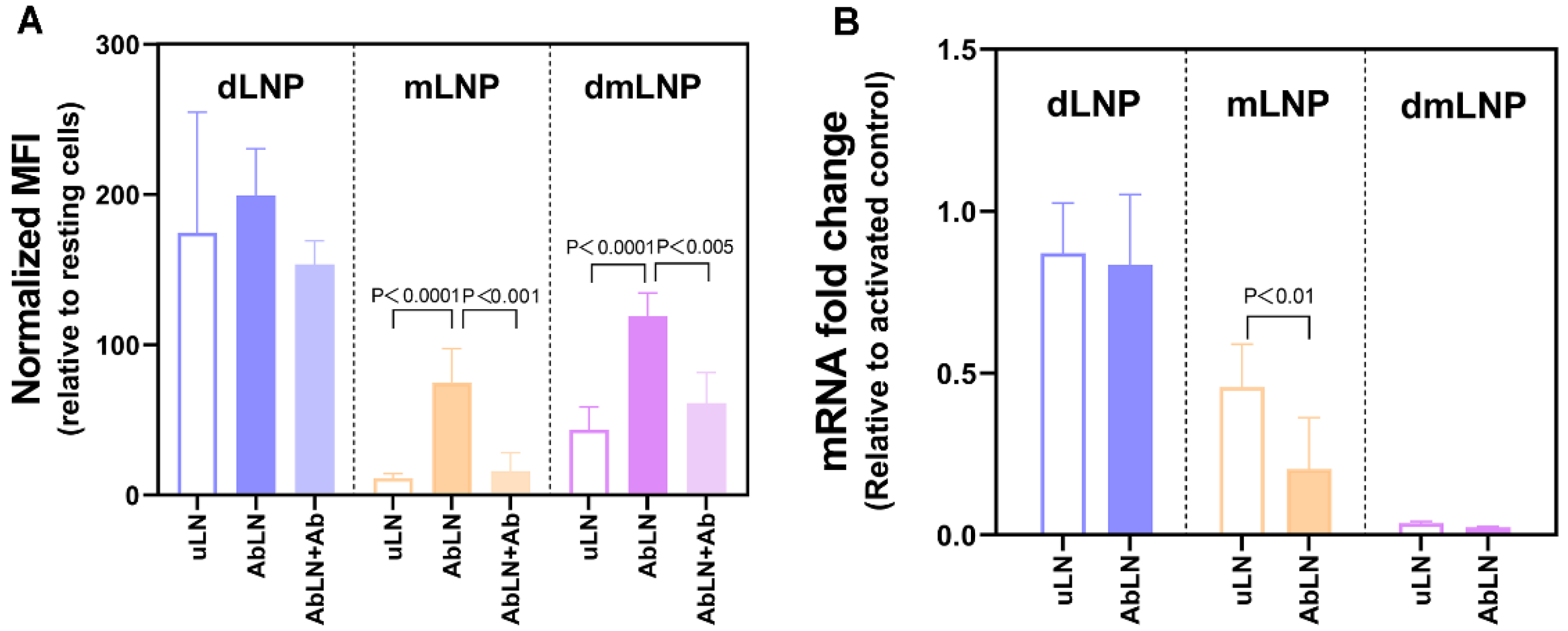
3.2. Influence of the Cationic Lipid Composition on LNP-Cell Association and Gene Silencing in HUVEC
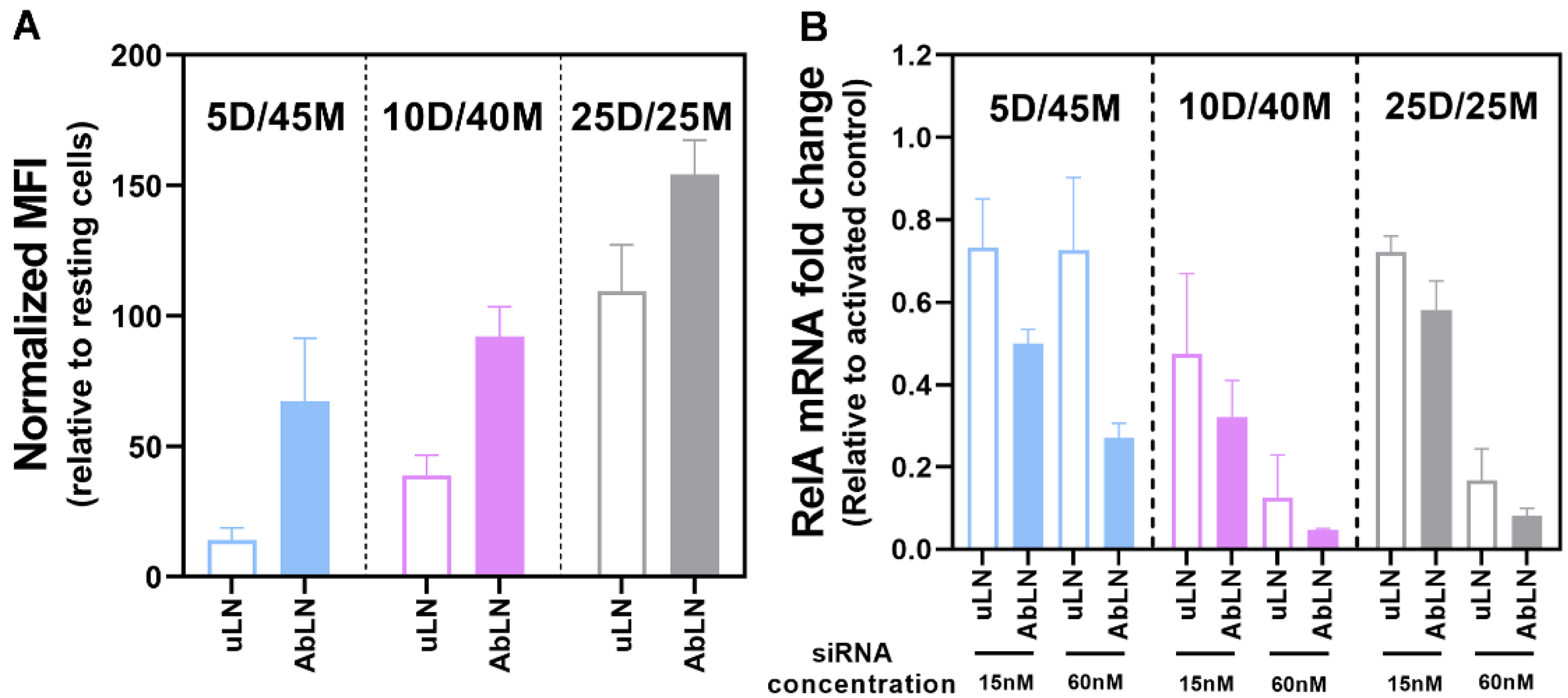
3.3. Influence of the DOTAP/MC3 Ratios in the dmLNP Formulations on Cell Association and Gene Silencing in EC
3.4. Intracellular Release of Encapsulated siRNA from LNP in EC
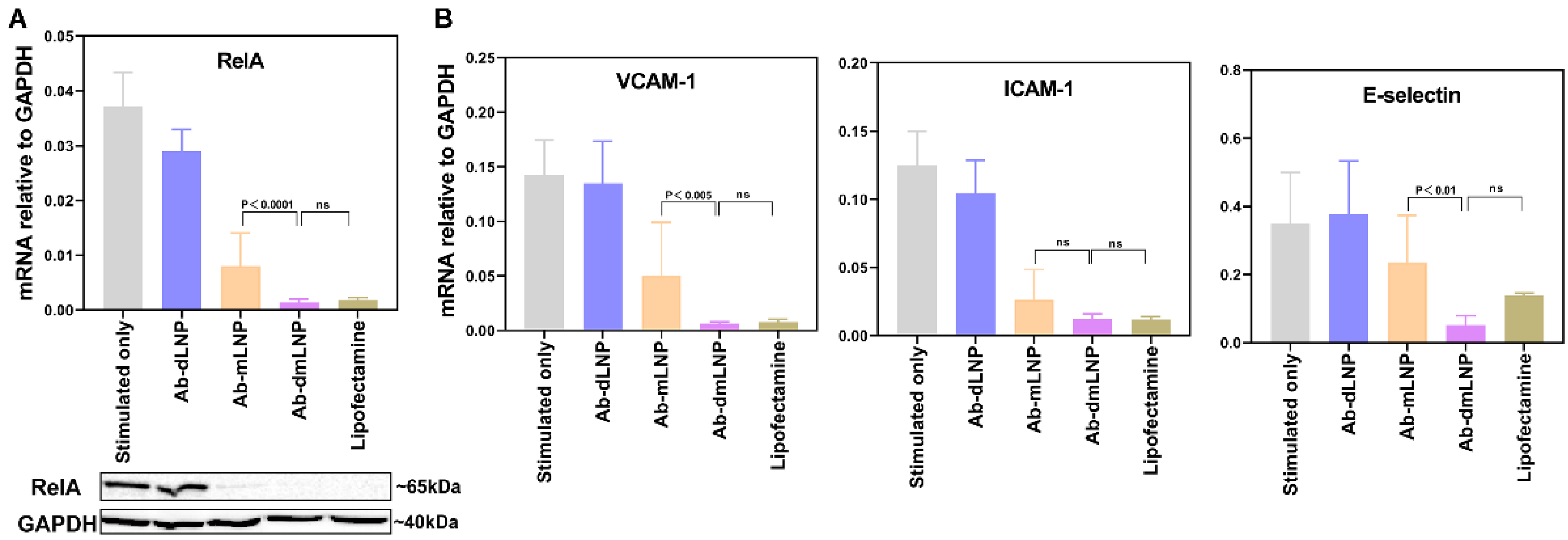
3.5. Anti-Inflammatory Potential of LNP/siRNARelA in Inflammatory-Activated EC In Vitro
3.6. In Vivo LNP/siRNA Behavior in Zebrafish Embryos
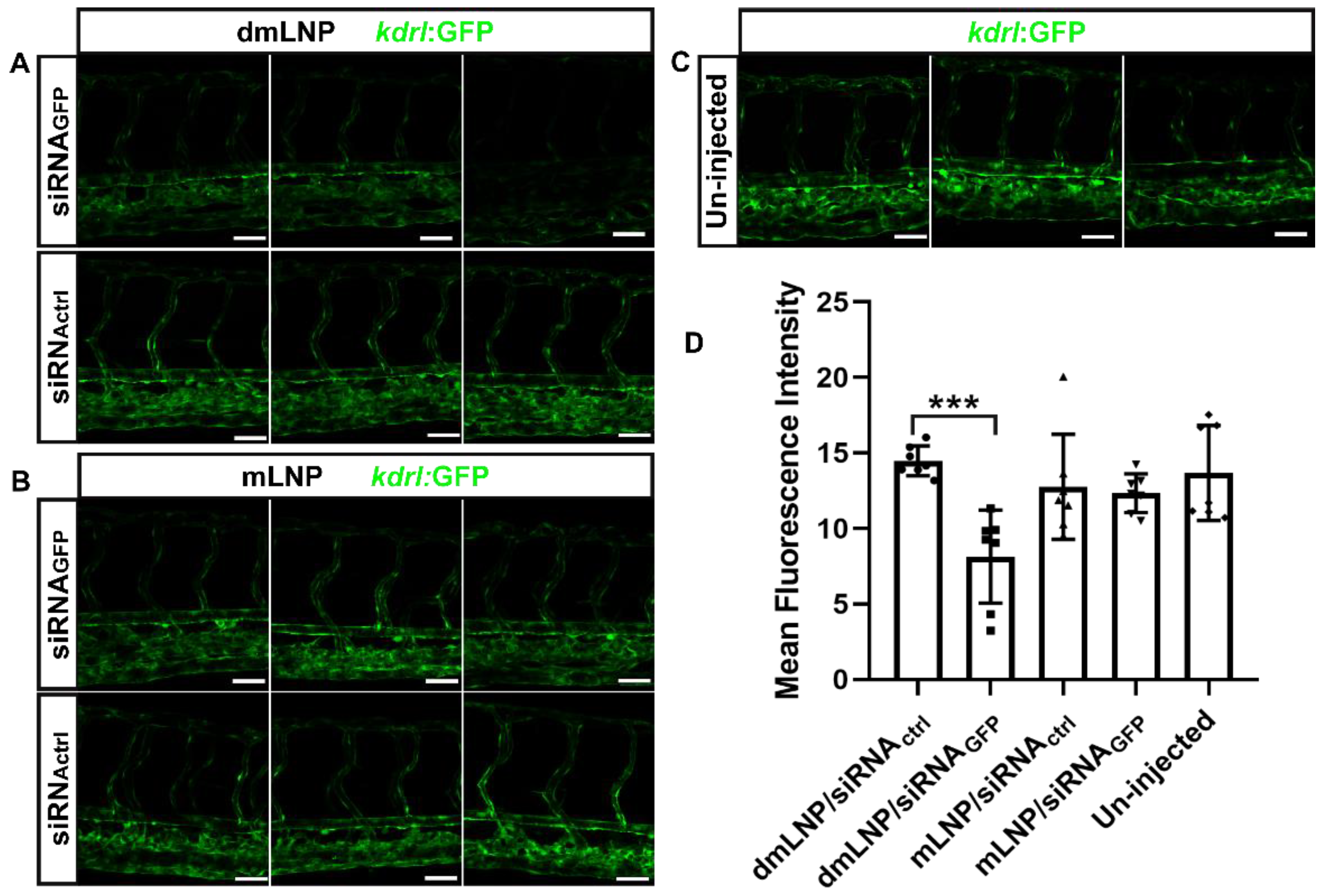
3.7. Gene Silencing Potency of MC3 Based LNP Loaded with siRNA against GFP in Zebrafish
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cahill, P.A.; Redmond, E.M. Vascular endothelium—Gatekeeper of vessel health. Atherosclerosis 2016, 248, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Krüger-Genge, A.; Blocki, A.; Franke, R.-P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef] [PubMed]
- Aird, W.C. Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2012, 2, a006429. [Google Scholar] [CrossRef] [PubMed]
- Leus, N.G.J.; Talman, E.G.; Ramana, P.; Kowalski, P.S.; Woudenberg-Vrenken, T.E.; Ruiters, M.H.J.; Molema, G.; Kamps, J.A.A.M. Effective siRNA delivery to inflamed primary vascular endothelial cells by anti-E-selectin and anti-VCAM-1 PEGylated SAINT-based lipoplexes. Int. J. Pharm. 2014, 459, 40–50. [Google Scholar] [CrossRef]
- Gholizadeh, S.; Visweswaran, G.R.R.; Storm, G.; Hennink, W.E.; Kamps, J.A.A.M.; Kok, R.J. E-selectin targeted immunoliposomes for rapamycin delivery to activated endothelial cells. Int. J. Pharm. 2018, 548, 759–770. [Google Scholar] [CrossRef]
- Xu, S.; Ilyas, I.; Little, P.J.; Li, H.; Kamato, D.; Zheng, X.; Luo, S.; Li, Z.; Liu, P.; Han, J.; et al. Endothelial Dysfunction in Atherosclerotic Cardiovascular Diseases and Beyond: From Mechanism to Pharmacotherapies. Pharmacol. Rev. 2021, 73, 924–967. [Google Scholar] [CrossRef]
- Rajendran, P.; Rengarajan, T.; Thangavel, J.; Nishigaki, Y.; Sakthisekaran, D.; Sethi, G.; Nishigaki, I. The Vascular Endothelium and Human Diseases. Int. J. Biol. Sci. 2013, 9, 1057–1069. [Google Scholar] [CrossRef]
- Jamwal, S.; Sharma, S. Vascular endothelium dysfunction: A conservative target in metabolic disorders. Agents Actions 2018, 67, 391–405. [Google Scholar] [CrossRef]
- Bordy, R.; Totoson, P.; Prati, C.; Marie, C.; Wendling, D.; Demougeot, C. Microvascular endothelial dysfunction in rheumatoid arthritis. Nat. Rev. Rheumatol. 2018, 14, 404–420. [Google Scholar] [CrossRef]
- Pasquier, J.; Ghiabi, P.; Chouchane, L.; Razzouk, K.; Rafii, S.; Rafii, A. Angiocrine endothelium: From physiology to cancer. J. Transl. Med. 2020, 18, 52. [Google Scholar] [CrossRef]
- Luxen, M.; van Meurs, M.; Molema, G. Unlocking the Untapped Potential of Endothelial Kinase and Phosphatase Involvement in Sepsis for Drug Treatment Design. Front. Immunol. 2022, 13, 867625. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, P.S.; Leus, N.G.J.; Scherphof, G.L.; Ruiters, M.H.J.; Kamps, J.A.A.M.; Molema, G. Targeted siRNA delivery to diseased microvascular endothelial cells-Cellular and molecular concepts. IUBMB Life 2011, 63, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mangala, L.S.; Rodriguez-Aguayo, C.; Kong, X.; Lopez-Berestein, G.; Sood, A.K. RNA interference-based therapy and its delivery systems. Cancer Metastasis Rev. 2018, 37, 107–124. [Google Scholar] [CrossRef]
- Yang, J.; Li, Q.; Yang, X.; Feng, Y.; Ren, X.; Shi, C.; Zhang, W. Multitargeting Gene Delivery Systems for Enhancing the Transfection of Endothelial Cells. Macromol. Rapid Commun. 2016, 37, 1926–1931. [Google Scholar] [CrossRef]
- Green, J.J.; Shi, J.; Chiu, E.; Leshchiner, E.S.; Langer, R.; Anderson, D.G. Biodegradable Polymeric Vectors for Gene Delivery to Human Endothelial Cells. Bioconjug. Chem. 2006, 17, 1162–1169. [Google Scholar] [CrossRef]
- Adrian, J.E.; Morselt, H.W.M.; Süss, R.; Barnert, S.; Kok, J.W.; Ásgeirsdóttir, S.A.; Ruiters, M.H.J.; Molema, G.; Kamps, J.A.A.M. Targeted SAINT-O-Somes for improved intracellular delivery of siRNA and cytotoxic drugs into endothelial cells. J. Control. Release 2010, 144, 341–349. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Lintermans, L.L.; Morselt, H.W.M.; Leus, N.G.J.; Ruiters, M.H.J.; Molema, G.; Kamps, J.A.A.M. Anti-VCAM-1 and Anti-E-selectin SAINT-O-Somes for Selective Delivery of siRNA into Inflammation-Activated Primary Endothelial Cells. Mol. Pharm. 2013, 10, 3033–3044. [Google Scholar] [CrossRef]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. 2012, 124, 8657–8661. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; van der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef]
- Ponti, F.; Campolungo, M.; Melchiori, C.; Bono, N.; Candiani, G. Cationic lipids for gene delivery: Many players, one goal. Chem. Phys. Lipids 2021, 235, 105032. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Kos, P.; Tieu, V.; Zhou, K.; Siegwart, D.J. Development of Cationic Quaternary Ammonium Sulfonamide Amino Lipids for Nucleic Acid Delivery. ACS Appl. Mater. Interfaces 2018, 10, 2302–2311. [Google Scholar] [CrossRef] [PubMed]
- Böttger, R.; Pauli, G.; Chao, P.-H.; AL Fayez, N.; Hohenwarter, L.; Li, S.-D. Lipid-based nanoparticle technologies for liver targeting. Adv. Drug Deliv. Rev. 2020, 154–155, 79–101. [Google Scholar] [CrossRef]
- Qi, X.; Zhao, W.; Zhuang, S. Comparative study of the in vitro and in vivo characteristics of cationic and neutral liposomes. Int. J. Nanomed. 2011, 6, 3087–3098. [Google Scholar] [CrossRef]
- Kamps, J.A.A.M.; Swart, P.J.; Morselt, H.W.M.; Pauwels, R.; De Béthune, M.-P.; De Clercq, E.; Meijer, D.K.; Scherphof, G.L. Preparation and characterization of conjugates of (modified) human serum albumin and liposomes: Drug carriers with an intrinsic anti-HIV activity. Biochim. Biophys. Acta (BBA)-Biomembr. 1996, 1278, 183–190. [Google Scholar] [CrossRef]
- Böttcher, C.; Van Gent, C.; Pries, C. A rapid and sensitive sub-micro phosphorus determination. Anal. Chim. Acta 1961, 24, 203–204. [Google Scholar] [CrossRef]
- Peterson, G.L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977, 83, 346–356. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Zwiers, P.J.; Morselt, H.W.M.; Kuldo, J.M.; Leus, N.G.; Ruiters, M.H.J.; Molema, G.; Kamps, J.A.A.M. Anti-VCAM-1 SAINT-O-Somes enable endothelial-specific delivery of siRNA and downregulation of inflammatory genes in activated endothelium in vivo. J. Control. Release 2014, 176, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Kowalski, P.S.; Morselt, H.W.M.; Schepel, I.; Jongman, R.M.; Aslan, A.; Ruiters, M.H.J.; Zijlstra, J.; Molema, G.; Van Meurs, M.; et al. Endothelium-targeted delivery of dexamethasone by anti-VCAM-1 SAINT-O-Somes in mouse endotoxemia. PLoS ONE 2018, 13, e0196976. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.-W.; Beis, D.; Mitchell, T.; Chen, J.-N.; Stainier, D.Y.R. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005, 132, 5199–5209. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [PubMed]
- Arias-Alpizar, G.; Bussmann, J.; Campbell, F. Zebrafish Embryos as a Predictive Animal Model to Study Nanoparticle Behavior in vivo. Bio-Protocol 2021, 11, e4173. [Google Scholar] [CrossRef]
- Campbell, F.; Bos, F.L.; Sieber, S.; Arias-Alpizar, G.; Koch, B.E.; Huwyler, J.; Kros, A.; Bussmann, J. Directing Nanoparticle Biodistribution through Evasion and Exploitation of Stab2-Dependent Nanoparticle Uptake. ACS Nano 2018, 12, 2138–2150. [Google Scholar] [CrossRef]
- Eberlein, J.; Herdt, L.; Malchow, J.; Rittershaus, A.; Baumeister, S.; Helker, C.S. Molecular and Cellular Mechanisms of Vascular Development in Zebrafish. Life 2021, 11, 1088. [Google Scholar] [CrossRef]
- Lou, G.; Anderluzzi, G.; Schmidt, S.T.; Woods, S.; Gallorini, S.; Brazzoli, M.; Giusti, F.; Ferlenghi, I.; Johnson, R.N.; Roberts, C.W.; et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: The impact of cationic lipid selection. J. Control. Release 2020, 325, 370–379. [Google Scholar] [CrossRef]
- Patel, S.; Ryals, R.C.; Weller, K.K.; Pennesi, M.E.; Sahay, G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release 2019, 303, 91–100. [Google Scholar] [CrossRef]
- Wang, J.; Lu, Z.; Wientjes, M.G.; Au, J.L.-S. Delivery of siRNA Therapeutics: Barriers and Carriers. AAPS J. 2010, 12, 492–503. [Google Scholar] [CrossRef]
- Basha, G.; Novobrantseva, T.I.; Rosin, N.; Tam, Y.Y.C.; Hafez, I.M.; Wong, M.K.; Sugo, T.; Ruda, V.M.; Qin, J.; Klebanov, B.; et al. Influence of Cationic Lipid Composition on Gene Silencing Properties of Lipid Nanoparticle Formulations of siRNA in Antigen-Presenting Cells. Mol. Ther. 2011, 19, 2186–2200. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.J.; Tam, Y.Y.C.; Hafez, I.; Sandhu, A.; Chen, S.; Ciufolini, M.A.; Nabi, I.R.; Cullis, P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Sousa, D.A.; Gaspar, R.; Ferreira, C.J.O.; Baltazar, F.; Rodrigues, L.R.; Silva, B.F.B. In Vitro CRISPR/Cas9 Transfection and Gene-Editing Mediated by Multivalent Cationic Liposome–DNA Complexes. Pharmaceutics 2022, 14, 1087. [Google Scholar] [CrossRef] [PubMed]
- Yung, B.C.; Li, J.; Zhang, M.; Cheng, X.; Li, H.; Yung, E.M.; Kang, C.; Cosby, L.E.; Liu, Y.; Teng, L.; et al. Lipid Nanoparticles Composed of Quaternary Amine–Tertiary Amine Cationic Lipid Combination (QTsome) for Therapeutic Delivery of AntimiR-21 for Lung Cancer. Mol. Pharm. 2016, 13, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Dalby, B.; Cates, S.; Harris, A.; Ohki, E.C.; Tilkins, M.L.; Price, P.J.; Ciccarone, V.C. Advanced transfection with Lipofectamine 2000 reagent: Primary neurons, siRNA, and high-throughput applications. Methods 2004, 33, 95–103. [Google Scholar] [CrossRef]
- Sieber, S.; Grossen, P.; Detampel, P.; Siegfried, S.; Witzigmann, D.; Huwyler, J. Zebrafish as an early stage screening tool to study the systemic circulation of nanoparticulate drug delivery systems in vivo. J. Control. Release 2017, 264, 180–191. [Google Scholar] [CrossRef]
- Fenaroli, F.; Westmoreland, D.; Benjaminsen, J.; Kolstad, T.; Skjeldal, F.M.; Meijer, A.H.; van der Vaart, M.; Ulanova, L.; Roos, N.; Nyström, B.; et al. Nanoparticles as Drug Delivery System against Tuberculosis in Zebrafish Embryos: Direct Visualization and Treatment. ACS Nano 2014, 8, 7014–7026. [Google Scholar] [CrossRef]
- Pattipeiluhu, R.; Arias-Alpizar, G.; Basha, G.; Chan, K.Y.T.; Bussmann, J.; Sharp, T.H.; Moradi, M.; Sommerdijk, N.; Harris, E.N.; Cullis, P.R.; et al. Anionic Lipid Nanoparticles Preferentially Deliver mRNA to the Hepatic Reticuloendothelial System. Adv. Mater. 2022, 34, 2201095. [Google Scholar] [CrossRef]


| Category | Sample | Size (nm) | Polydispersity Index (PDI) | siRNA Encapsulation Efficiency (EE, %) | Ab Conjugated/LNP (µg/µmol TL) |
|---|---|---|---|---|---|
| uLN | dLNP | 88 ± 16 | 0.11 ± 0.05 | 98 ± 1 | - |
| mLNP | 59 ± 7 | 0.21 ± 0.09 | 91 ± 8 | - | |
| dmLNP | 63 ± 9 | 0.28 ± 0.11 | 97 ± 1 | - | |
| AbLN | Ab-dLNP | 139 ± 25 | 0.22 ± 0.05 | 98 ± 1 | 100 ± 19 |
| Ab-mLNP | 118 ± 10 | 0.34 ± 0.06 | 91 ± 2 | 85 ± 15 | |
| Ab-dmLNP | 110 ± 6 | 0.34 ± 0.09 | 90 ± 7 | 92 ± 23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Bi, D.; Plantinga, J.A.; Molema, G.; Bussmann, J.; Kamps, J.A.A.M. Development of a Combined Lipid-Based Nanoparticle Formulation for Enhanced siRNA Delivery to Vascular Endothelial Cells. Pharmaceutics 2022, 14, 2086. https://doi.org/10.3390/pharmaceutics14102086
He Y, Bi D, Plantinga JA, Molema G, Bussmann J, Kamps JAAM. Development of a Combined Lipid-Based Nanoparticle Formulation for Enhanced siRNA Delivery to Vascular Endothelial Cells. Pharmaceutics. 2022; 14(10):2086. https://doi.org/10.3390/pharmaceutics14102086
Chicago/Turabian StyleHe, Yutong, Dongdong Bi, Josée A. Plantinga, Grietje Molema, Jeroen Bussmann, and Jan A. A. M. Kamps. 2022. "Development of a Combined Lipid-Based Nanoparticle Formulation for Enhanced siRNA Delivery to Vascular Endothelial Cells" Pharmaceutics 14, no. 10: 2086. https://doi.org/10.3390/pharmaceutics14102086
APA StyleHe, Y., Bi, D., Plantinga, J. A., Molema, G., Bussmann, J., & Kamps, J. A. A. M. (2022). Development of a Combined Lipid-Based Nanoparticle Formulation for Enhanced siRNA Delivery to Vascular Endothelial Cells. Pharmaceutics, 14(10), 2086. https://doi.org/10.3390/pharmaceutics14102086





