Abstract
Cationic liposomes (CLs) are effective carriers of a variety of therapeutics. Their applications as vectors of nucleic acids (NAs), from long DNA and mRNA to short interfering RNA (siRNA), have been pursued for decades to realize the promise of gene therapy, with approvals of the siRNA therapeutic patisiran and two mRNA vaccines against COVID-19 as recent milestones. The long-term goal of developing optimized CL-based NA carriers for a broad range of medical applications requires a comprehensive understanding of the structure of these vectors and their interactions with cell membranes and components that lead to the release and activity of the NAs within the cell. Structure–activity relationships of lipids for CL-based NA and drug delivery must take into account that these lipids act not individually but as components of an assembly of many molecules. This review summarizes our current understanding of how the choice of the constituting lipids governs the structure of their CL–NA self-assemblies, which constitute distinct liquid crystalline phases, and the relation of these structures to their efficacy for delivery. In addition, we review progress toward CL–NA nanoparticles for targeted NA delivery in vivo and close with an outlook on CL-based carriers of hydrophobic drugs, which may eventually lead to combination therapies with NAs and drugs for cancer and other diseases.
1. Introduction
Amphiphilic molecules, i.e., molecules with a polar, hydrophilic headgroup and a hydrophobic tail or tails, spontaneously self-assemble in water, primarily due to hydrophobic interactions [1]. The resulting assemblies expose the headgroups to the aqueous environment while avoiding exposure of the tails, which instead form their own hydrophobic environment. This understanding, which is now commonplace, originated only after Bangham and Horne discovered liposomes (also referred to as unilamellar or multilamellar vesicles; Figure 1) while studying suspensions of phospholipids by electron microscopy [2]. Liposomes, consisting of closed assemblies of bilayers of lipids, resembled cell membranes in the electron micrographs, thereby confirming that the makeup of these biological structures is lipid-based. Bangham and Horne’s work further demonstrated that lipid bilayer membranes can accommodate hydrophobic molecules while forming a barrier for polar, hydrophilic molecules. Thus, these studies also confirmed the hypothesis that the lipids of plasma membranes provide the permeability barrier that is essential to biological membrane function.
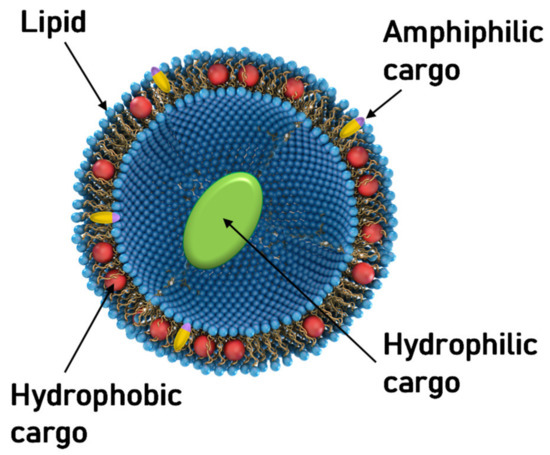
Figure 1.
Schematic depiction of a unilamellar liposome consisting of a self-assembly of amphiphilic lipid molecules. The liposome can transport cargo at three distinct sites: within its hydrophobic bilayer (red spheres), within its hydrophilic aqueous interior (green ellipsoid), and at the amphipathic interface (yellow/blue rods).
The structure of liposomes makes it evident that they can accommodate cargo at three different sites (Figure 1): their interior (hydrophilic), within the bilayer (hydrophobic), and at the interface (amphiphilic). Figure 1 schematically represents cargo at these sites as a green ellipsoid, red spheres, and yellow/blue rods, respectively. Soon, the potential of liposomes to deliver protein and small-molecule therapeutics was recognized and studied [3,4,5,6,7].
Only later, the use of cationic liposomes (CLs) to deliver nucleic acids (NAs) was pioneered by Phillip Felgner and coworkers [8,9] They demonstrated that positively charged CLs could simultaneously form complexes with negatively charged NAs and bind the anionic sulfate groups of cell-surface proteoglycans on mammalian cells [10], triggering not only uptake by cells but also the expression of the delivered DNA [8] and RNA [9]. Soon after the publication of the seminal papers by Felgner et al., the dominant structure proposed for CL–DNA complexes was a “bead-on-string” model of DNA strands decorated with distinctly attached CLs [11]. As described in detail below, CL–NA complexes instead spontaneously form a variety of novel self-assembled (liquid crystalline) structures [12,13,14,15] with distinct structure-activity relations [14,15,16,17,18].
In the wake of the landmark paper by Felgner et al., countless cationic lipids have been synthesized and studied [19,20,21,22,23,24,25,26,27,28,29,30,31]. In addition, numerous aspects of the formation and action of complexes of CLs with DNA and other nucleic acids (NAs) have been investigated [32,33,34,35,36,37,38,39,40,41,42,43]. These worldwide research efforts with CL vectors have the aim of improving efficacy both in vitro (at the cell level) and in vivo.
CL-based vectors are now widely used for in vitro delivery of NAs, e.g., in functional genomics. However, the main motivation in the field has been the use of CL-based NA vectors in gene therapeutics [44,45,46], a concept explored in numerous clinical trials [45,47,48]. Major discoveries in the biochemical arena, such as gene silencing via RNA interference (RNAi) [49,50] induced by double-stranded small interfering RNA (siRNA) [18,51,52,53,54,55,56,57,58] and microRNA [59], gene editing via CRISPR/Cas-9 [60,61,62], and expression of base-modified mRNA [63,64,65,66,67,68,69,70], have greatly expanded the therapeutic potential of NAs. The first culmination of these efforts was the FDA approval of a lipid-based siRNA vector (patisiran/ONPATTRO®, Alnylam Pharmaceuticals) in 2018 for treatment of the polyneuropathy caused by hereditary transthyretin amyloidosis [71,72]. Patisiran delivers an siRNA targeting a conserved sequence within the transthyretin mRNA. Its action decreases hepatic production of mutant transthyretin to prevent the buildup of amyloid protein in peripheral nerves. Recently, the scale of this first breakthrough was dwarfed by the success and use (in potentially billions of patients) of two lipid-based mRNA vaccines [73] against SARS-CoV2. Building on decades of work on both the lipid vector [72,74,75] and the mRNA payload [70], the vaccines developed by Pfizer/BioNTech [76,77,78] and Moderna [79,80] became the first vaccines approved against COVID-19 in December 2020. The lipid compositions of the vaccine nanoparticles are similar to that of patisiran [81,82], albeit employing ionizable cationic lipids with branched rather than unsaturated tails and different PEG-lipids (Figure 2 and Figure 3).
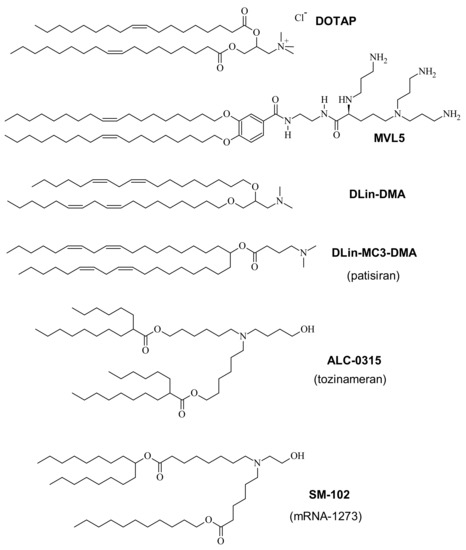
Figure 2.
Structures of the cationic lipids DOTAP (1+; 1,2-dioleoyl-trimethylammonium propane chloride) [83], MVL5 (5+) [84], DLin-DMA [85], and the cationic lipids used in FDA-approved CL–NA nanoparticle formulations. Tozinameran and mRNA-1273 are names for the COVID-19 vaccines developed by Pfizer/BioNTech and Moderna, respectively. The positive charge on DOTAP is present independent of pH, while the amine groups of the other lipids acquire their charge by protonation below a certain pH. To highlight this fact, those lipids are sometimes called “ionizable” lipids. MVL5 is a commercially available multivalent ionizable lipid.
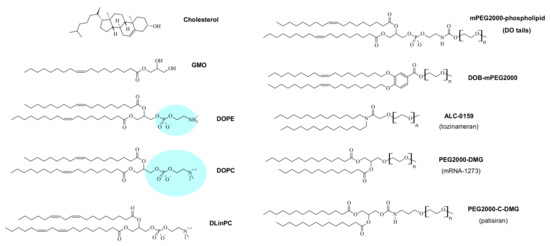
Figure 3.
Structures of selected neutral (“helper”) lipids and PEG-lipids used in CL-based NA vectors. The molecular weight of the PEG chain of all depicted lipids is 2000 g/mol (n = 45). The phospholipids DOPC, DOPE, and DLinPC are zwitterionic, while GMO and cholesterol are uncharged. The blue highlighting represents the large difference in hydration for DOPE and DOPC. DLinPC: 1,2-dilinoleoyl-sn-glycero-phosphocholine; DO: dioleoyl; DOPC: 1,2-dioleoyl-sn-glycero-phosphocholine; DOPE: 1,2-dioleoyl-sn-glycero-phosphoethanolamine; GMO: glycerol monooleate.
With these successes, lipid-based synthetic NA vectors have stepped out of the shadow of viral vectors. Engineered viruses had been preferred in early clinical trials of gene therapy [47,48]. However, their use has resulted in severe side effects from immune reactions and insertional mutagenesis [86,87,88,89]. Furthermore, the finite capsid size of viral vectors limits the length of their NA cargo to a maximum of ≈20–30 kbp. Thus, engineered viruses are incapable of delivering larger human genes, let alone genes together with noncoding regulatory sequences (often exceeding 100 kbp) or human artificial chromosomes (>1 Mbp) [90,91,92]. In contrast, synthetic vectors such as CL–NA complexes are easier to produce, less immunogenic, and safer [93], and their formation by self-assembly imposes no fundamental limit on the size of the incorporated NA.
Without discounting the many breakthroughs that were required for their success, the recently approved CL–NA therapeutics represent low-hanging fruit in terms of delivery challenges. For example, there are no FDA-approved lipid-based vectors that incorporate elements for targeted delivery, which remains a major challenge. The development of efficient carriers of NAs remains the bottleneck for broadening their applications [53,94] and fully realizing the potential of gene therapy, and to do so will require major additional efforts in both basic and applied science. While a lot of progress has been made studying the mechanism of delivery and transfection by CL-based NA carriers [29,32,33,35,38,39,40,41,42,95,96,97], much more work remains before a comprehensive understanding of their interactions with membranes and other cell components such as the cytoskeleton can allow a rational design of effective vectors.
Establishing structure–activity relationships for cationic lipids for NA delivery is more challenging than for small molecule drugs. Instead of a single molecule interacting with, e.g., the binding pocket of a protein, a large number of lipids joins together in a dynamic assembly before interacting with the NA to form the active principle that engages with the target tissue or cell. In addition, the local environment may affect the CL–NA assembly and the interactions with its target. Thus, changes to the structure of the lipid may affect the efficacy of the NA vector in ways that are amplified by the process of self-assembly. With this review, we aim to provide a guide for parsing through the overwhelming number of lipids and formulations by relating the lipid structure to properties of lipid self-assembly. Some of these properties are only peripherally known to chemists designing these lipids because they stem from the biophysics and physical chemistry of lipids as well as from colloid science.
The importance of charge in preparing effective CL-based NA vectors was appreciated early on. In addition to the fact that complex formation with CLs protects NAs from degradation, an early rationale for utilizing cationic rather than neutral or negative liposomes to deliver DNA was that CL–DNA complexes designed to have an overall positive charge (i.e., with a lipid/DNA charge ratio > 1) would electrostatically adsorb to anionic mammalian cells, thus leading to cellular binding and more efficient uptake [8]. Here, the lipid/DNA charge ratio refers to the number of charges on the lipid divided by the number of charges on the DNA (under the conditions that the lipid and DNA are combined).
2. Lipid Shape and Membrane Curvature Elastic Energy Determine Their Self-Assembled Structures
It has been appreciated in surfactant and lipid science for over four decades that the shape of an amphiphile plays a major role in determining the structure of its self-assemblies [1,98,99,100,101]. Figure 4 illustrates the concept of lipid shape and the resulting assemblies with molecular models of a few lipids for illustration. The area of the lipid’s headgroup relative to that of the tails determines the lipid’s shape, and arrangement of these shapes with the constraint of exposing tails only to themselves and headgroups to the exterior aqueous environment naturally leads to the self-assembled structure. These assemblies, in turn, constitute the lipid building blocks of resulting CL–NA complexes.
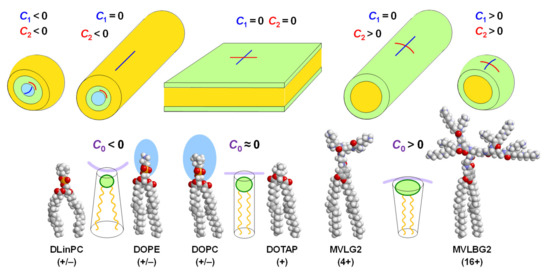
Figure 4.
Relation of lipid shape and the resulting self-assemblies. Inverse-cone-shaped lipids give rise to inverse micellar structures (left); cylindrical lipids (center), where the headgroup area approximately matches the projected area of the tails, prefer (nearly) flat membranes (lamellar phases and large vesicles, depending on water excess); cone-shaped lipid (right) prefer micellar structures such as cylinders of varying length and spheres. Blue and red lines indicate the membrane curvatures C1 and C2, respectively, of the assemblies. Purple lines behind the schematic depictions of lipids with different shapes indicate the spontaneous curvature C0 of their assemblies. See Figure 2, Figure 3 and Figure 4 for the chemical structures of the displayed lipids.
A quantitative (but nonetheless semiempirical) parameter for describing lipid shape and the resulting assemblies is the “packing parameter”, v/a0lc. This dimensionless parameter is the ratio of the average area of the tails, expressed as the volume of the tails (v) over their maximum effective length (lc) and the optimal headgroup area (a0) [1]. For the lipids in Figure 4, the packing parameter increases from left to right, with v/a0lc = 1 for lipids with a cylindrical shape. As an example of the usefulness of the packing parameter, consider the cationic lipids used in the COVID-19 vaccines (Figure 2) in comparison with hypothetical lipids of the same structure (i.e., the same a0) but without the branches in the tails. Adding branching will increase v while leaving lc unaffected and thus decrease the packing parameter, meaning an increased propensity for the formation of the inverse structures depicted on the left in Figure 4.
The physical formalism to describe lipid membranes and their elastic properties uses the membrane curvatures, defined as the inverse of the membrane radii (Ci = 1/Ri) in two orthogonal directions (Figure 4). For example, C1 = C2 = 0 for a flat membrane; C1 = C2 = 1/Rsph for a spherical membrane with radius Rsph; and C1 = 0, C2 = Rcyl for a cylindrical membrane with radius Rcyl. Negative curvature is also possible, e.g., for the surface of an inverse micelle (Figure 4, top left). The shape of a lipid determines the spontaneous (in other words, preferred) curvature of the membrane, termed C0. Introducing further the membrane modulus κ (a measure of the bending stiffness of the membrane) and the Gaussian modulus κG (a measure of the propensity for (κG > 0) or resistance to (κG < 0) forming saddle shapes, see Figure 6), the elastic free energy of a membrane per unit area, E/A, is
with C = C1 + C2 being the mean curvature of the membrane and C1C2 being termed the Gaussian curvature of the membrane [102,103,104]. The elastic moduli κ and κG can be related to the interactions between neighboring lipids in the membrane using harmonic spring models [105,106].
E/A = 0.5 κ (C − C0)2 + κG C1C2,
The first term in Equation (1) describes the free energy cost of bending a membrane away from its spontaneous curvature, which increases as the membrane stiffness (κ) increases. As mentioned above, the shape of a lipid determines C0 [1,107]. Intuitively it may be easier to consider the spontaneous radius of curvature, R0, in some cases. Lipids with a headgroup area that is approximately equal to the projected area of their tails have a cylindrical shape. This corresponds to a spontaneous curvature C0 = 0 for the bilayer (Figure 4, center), and these lipids tend to assemble into lamellar bilayer structures such as lamellar lyotropic phases (at low water content) or large uni- and multilamellar vesicles (in excess water). Examples of such lipids are the neutral lipid DOPC (as well as other related phosphocholines with saturated tails) and the monovalent cationic lipid DOTAP. The molecular models in Figure 4 show that the steric size of the headgroup of DOTAP is much smaller than that of DOPC (or DOPE, see below), but the charge of the headgroup and the electrostatic repulsion between lipids that it induces as well as its hydration must be taken into account when considering the shape of the lipid in an aqueous environment.
The shape of a lipid with a headgroup area that is smaller than the tail area is described as an inverse cone. For lipids of this shape, C0 < 0, and they tend to form inverse micelles (Figure 4, left) that assemble further into structures such as the inverse hexagonal (HII) phase [107,108] for inverse cylindrical micelles or cubic phases for inverse spherical micelles. A well-known example of such lipids is DOPE, which differs from DOPC only in bearing an amino- instead of a trimethylammonium-group in its headgroup. This results in a larger hydration of the DOPC versus the DOPE headgroup and thus a difference in lipid shape (indicated by the blue color in Figure 3 and Figure 4), again highlighting that the structure of lipids must be considered in the context of their surroundings.
Instead of a decrease in headgroup size, an increase in the average cross-sectional area of the lipid tails can also change lipid shape from cylinder to inverse cone. An example of this is the lipid DLinPC, which bears two cis double bonds in its linoleoyl tails, compared to the single double bond in each tail of DOPC. As illustrated in Figure 4, the additional bend in the tail cannot be compensated for by gauche conformations in the rest of the chain, and the tail area increases. Consistent with this, CLs containing DLinPC showed a propensity to form the inverse hexagonal phase [109]. Similarly, increasing chain unsaturation facilitated the formation of the inverse hexagonal phase for a series of cationic lipids with C18 tails [85]. Of these lipids, the one with linoleyl tails (DLin-DMA; Figure 2), has a structure very similar to the cationic lipid used in the patisiran formulation (Figure 2, DLin-MC3-DMA; note the tails with two cis double bonds). It is intriguing to note in this context that the branching of the tails of the cationic lipids used in the mRNA vaccines against COVID-19 (Figure 2) also suggests that they have an increased cross-sectional area.
Cone-shaped lipids have a headgroup with an area that is larger than that of the tail. In this case, C0 > 0, and the lipids usually assemble into cylindrical or spheroidal micellar structures (Figure 4, right). This shape is common for amphiphiles with a single tail, but their high toxicity prevents their use in therapeutics. Lipids with two tails can be cone-shaped if they have a headgroup with a high charge and/or large steric size. An extreme example is the custom-synthesized lipid MVLBG2 [14,30] with a headgroup that bears 16 positive charges at full protonation (Figure 5). Lipids with a more moderate charge, such as DOGS [110] and MVL5 [111], have also shown a propensity for micellar structures. The headgroup of PEG-lipids is large because of the steric size of the polymer chain (depending on PEG length), and lipid mixtures containing them also form micellar structures, depending on the fraction of PEG-lipid (see below) [112,113,114].
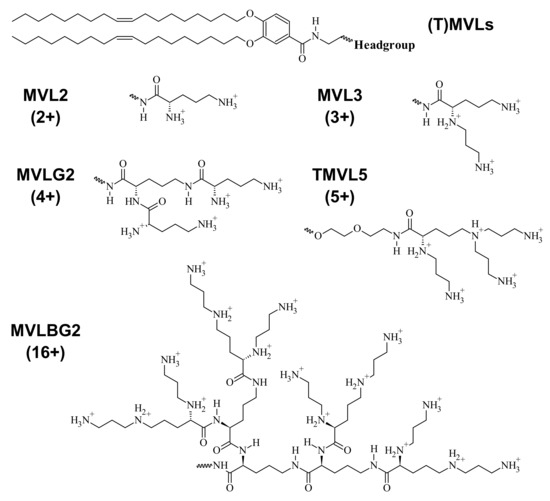
Figure 5.
Chemical structure and maximum charge of custom synthesized multivalent lipids ((T)MVLs) mentioned in the text [14,17,30,84].
As mentioned above, the environment of a lipid must be taken into account when considering its shape, and changing the conditions can lead to a change in shape and thus the structure of the lipid assembly. One example of this are changes in pH, which may increase or reduce the charge of a lipid’s headgroup, depending on the lipid structure. Another is the salt concentration in the aqueous environment; higher salt concentrations screen electrostatic interactions, effectively reducing the headgroup size.
The spontaneous curvature of a mixture of lipids is equal to the sum of the spontaneous curvatures of the individual lipids, C0,i, weighted with their molar fraction, xi, provided that complete mixing of lipids in the membrane takes place (see below). Thus, for a mixture of two lipids, C0 = x1C0,1 + x2C0,2, with xi = ni/(n1 + n2) and n1, n2 the number of the two lipids in the membrane. An illustration of this is provided by mixtures of cone-shaped MVLBG2 with cylindrical DOPC. These mixtures form sheets (vesicles), cylindrical micelles that shorten with increasing content of MVLBG2, and spherical micelles at low, intermediate, and very high contents of MVLBG2, respectively [115].
An example on the other side of the curvature spectrum are mixtures of DOPE and DOPC, which form lamellar or inverse micellar structures depending on their composition [101].
The second term in Equation (1) contains the Gaussian modulus, κG, and the Gaussian curvature, C1C2. Depending on the sign of their Gaussian modulus, membranes will prefer to form shapes of either positive or negative Gaussian curvature to minimize their elastic free energy. Examples of shapes with a positive Gaussian curvature (C1C2 > 0) are the outer (C1 > 0, C2 > 0) and inner (C1 < 0, C2 < 0) monolayer of a spherical vesicle, while flat bilayers (C1 = C2 = 0) and cylindrical micelles (C1 > 0, C2 = 0) have a Gaussian curvature of zero. Membranes with a positive Gaussian modulus κG > 0 will favor saddle-shaped surfaces with C1C2 < 0. These include the surfaces of bicontinuous cubic phases (Figure 6, left) and membrane pores (Figure 6, right; note the resemblance of the shape to a saddle) [103,107,116]. Thus, a consideration of membrane elasticity suggests that cubic phase-forming lipids (i.e., with κG > 0) favor the formation of pores (resulting from the fusion of two membranes that face each other in close proximity) [116,117,118]. Membrane fusion leading to pore formation has been mechanistically associated with the formation of bicontinuous cubic phases [116,117,118]. Because escape from the endosome after being internalized by the cell, which requires pore formation, is a major barrier to successful NA delivery, this is a highly relevant insight to the development of efficient CL-based NA vectors.
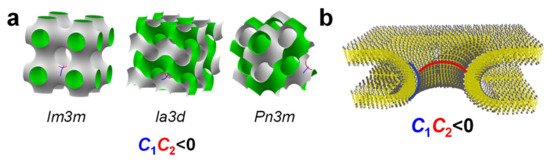
Figure 6.
(a) Structures of a cubic phase with Im3m symmetry and two inverse bicontinuous cubic phases, the gyroid (QIIG, space group Ia3d) and the diamond (QIID, space group Pn3m) cubic phases. These structures consist of a single continuous lipid bilayer interface (with saddle-shaped negative Gaussian curvature C1C2 < 0) dividing space into two disconnected water channels. The drawn surfaces, with one side colored gray and the other colored green, represent the shape of the water–bilayer interface [119,120]. (b) Schematic depiction of a membrane pore, illustrating its saddle-shaped negative Gaussian curvature. Adapted with permission from [121]. Copyright 2011 American Chemical Society.
The Gaussian modulus (κG) of the bilayer membrane may be driven positive in lipids with extreme inverse cone shapes. Simple elasticity theories, which treat the curvature elasticity of the flat lipid bilayer as the sum of the elasticities of two oppositely oriented monolayers, have shown that one may relate the Gaussian modulus of a bilayer to that of the monolayer via the equation [104,117]
κG,bilayer = 2κG,monolayer − 4δC0,monolayerκmonolayer.
Here, the bending rigidity of the bilayer κbilayer = 2κmonolayer, and δ is the monolayer thickness. The equation implies that lipids with inverse cone shape (C0,monolayer < 0) contribute a positive term to the Gaussian modulus of the bilayer. Thus, one may drive a transition from the inverse hexagonal to the bicontinuous (inverse) cubic phase (κG,bilayer > 0) with lipids with sufficiently negative C0,monolayer. Support of this analysis is found in phase diagrams of lipid/water mixtures where the bicontinuous (inverse) cubic phase with κG,bilayer > 0 is often located near an HII phase with C0,monolayer < 0 [15,120,121]. Thus, designing vectors containing lipids with extreme inverse cone shapes (with a propensity for formation of bicontinuous cubic phases) may be considered a rational approach to overcoming the endosomal escape barriers of vectors because of the acquired fusogenic properties.
3. The Lamellar LαC Phase of Cationic Liposome–DNA Complexes
While their opposite charge makes it appear intuitive that cationic liposomes and anionic NAs form complexes, the fact is that both already are associated with neutralizing counterions in solution. The primary driving force for their spontaneous self-assembly is the entropy gained by the release of positive counterions that are tightly bound to DNA (referred to as Manning condensation [122]) and negative counterions near the cationic liposome surface within the Guoy–Chapman layer [1]. As the cationic membranes neutralize the phosphate groups on the DNA, those small counterions gain in entropy because they are now free to diffuse and no longer bound to the DNA or cationic membranes. This driving force is also important in the assembly of other oppositely charged macro-ions, e.g., cationic polymers and DNA [123] or CLs and anionic proteins.
Initially, complexes of CLs and DNA were thought to consist of intact liposomes associated with DNA in a “bead-on-string” or “spaghetti and meatballs” structure [11]. Many subsequent studies have shown that such highly disordered assemblies are at best a short-lived intermediate in the process of CL–DNA complex formation. Ultimately, a complete topological transition from liposomes into collapsed condensates in the form of distinct liquid crystalline self-assemblies occurs [12,13,14,15,124]. The internal structure of the CL–DNA complexes as initially revealed by synchrotron small-angle X-ray scattering (SAXS) studies [12] is consistent with images from cryogenic electron microscopy studies [124,125,126].
As mentioned above, membrane shape is often determined by the spontaneous curvature of the membrane, which is related to the molecular shape of the constituent lipids. The shape of the membrane component, in turn, often determines the structure of the resulting CL–NA complex. Bilayer sheets, reflecting a spontaneous curvature of C0=0, as building blocks give rise to the most common phase of CL–DNA complexes. This phase was also the first one to be discovered [12,124]: the lamellar, LαC, phase (Figure 7a), consisting of a multilamellar arrangement of DNA monolayers intercalated (sandwiched) between cationic membranes.
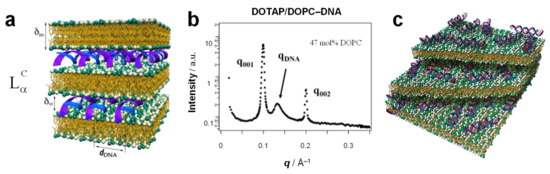
Figure 7.
(a) Schematic of the LαC phase of CL–DNA complexes, which consists of lipid bilayers of thickness δm alternating with DNA monolayers of thickness δw. The interlayer spacing, which gives rise to the series of peaks labeled q00i in SAXS, is d = δw + δm = 2π/q001. From [13]. Reprinted with permission from AAAS. (b) Example of the typical SAXS pattern resulting from CL–DNA complexes in the lamellar (LαC) phase. The Bragg reflections at q001 and q002 result from the multilamellar structure (see part b). The broad DNA–DNA correlation peak at qDNA reflects the ordered arrangement of the DNA rods (see part b) with an average interaxial spacing dDNA = 2π/qDNA. Complexes were formed from a DOTAP/DOPC (53:47, mol:mol) lipid mixture and λ-phage DNA. Reprinted with permission from [12]. (c) Schematic of the lamellar phase of CL–siRNA complexes. Note the lack of orientation order for the short siRNA rods. Reprinted with permission from [18]. Copyright 2007 American Chemical Society.
The characteristic signature of the lamellar phase in SAXS is a series of evenly spaced, sharp peaks at q = q00L, with q00L= L × q001. (Figure 7b) The lamellar spacing d, which comprises the thickness of the membrane (δm) and the water gap (δw), can be obtained from the position of the first peak as d = δm + δw = 2π/q001. Typical dimensions of a lamellar phase (measured for DOTAP/DOPC–DNA complexes [12]) are d = 64 Å, with δm = 39 Å [12] and δw = d − δm = 25 Å, which is approximately equal to the thickness of one monolayer of B-DNA (diameter ≈ 20 Å) including a hydration shell. (The membrane thickness was independently determined using SAXS of the lamellar Lα phase formed by the same lipids without DNA at an increasing volume fraction of water in the one-phase regime [127,128,129,130].)
In many cases, SAXS of the lamellar phase also reveals a broader peak at qDNA (Figure 7b) that arises from DNA–DNA correlations and gives the spacing between the DNA rods as dDNA = 2π/qDNA. Unlike the interlamellar spacing, the DNA spacing has been observed to vary widely depending on the composition of the membranes, in particular on the membrane charge density (due to the requirement of local charge neutrality in the complex [12]). The lower limit of dDNA at high membrane charge density is ≈ 25 Å, corresponding to close packing of DNA chains with one hydration shell, but values as high as dDNA ≈ 55 Å have been observed at low membrane charge density for DOTAP/DOPC–DNA complexes [12,131,132]. The DNA is electrostatically adsorbed on the cationic membrane, forming a two-dimensional (2D) smectic phase (i.e., a finite-size one-dimensional array of chains), as shown by quantitative line-shape analysis of the DNA correlation peak [133,134]. This makes the LαC phase a hybrid liquid crystalline phase because the lipids form a 3D smectic phase while the DNA forms a 2D smectic phase.
The LαC structure has been observed in CL–DNA complexes containing a broad variety of DNA: linear and circular (plasmid DNA) [16]. monodisperse (λ-phage DNA) [12] and polydisperse (e.g., calf thymus DNA) [135], long and very short [136]. Stable lamellar complexes form in water as well as cell culture medium [16] and disassemble only at very high salt concentrations (around 1 M monovalent salt, dependent on the membrane charge density) [131]. CLs formed from a wide variety of cationic lipids form the lamellar phase [21,137,138,139]. Even the strongly cone-shaped lipid MVLBG2 gives rise to LαC complexes when mixed with a large amount of DOPC (90 mol%) [14].
NAs other than DNA also form lamellar phases. In the case of siRNA (≈19–27 bp RNA double strands with two nucleotide 3′-overhangs), no RNA–RNA correlation peak is observed, suggesting that the siRNAs are not ordered (aligned) within the aqueous layer (Figure 7c) [18]. When siRNA was replaced with similar short DNA confined between membranes of the LαC phase, unexpectedly large end-to-end interactions between 11 bp DNA rods set in when their overhangs were reduced from 10 or 5 to 2 thymidines. This, in turn, led to the formation of a novel 2D columnar nematic liquid crystalline phase with finite-length columns consisting of stacks of on average four short DNA molecules [136].
Single-stranded mRNA, like DNA and short double-stranded siRNA, forms lamellar CL–NA complexes when complexed with DOPC/DOTAP membranes [140]. However, the interactions in CL–mRNA complexes are more complex compared with those in CL–DNA complexes in several aspects. Firstly, the electrostatic interactions of mRNA with cationic lipid membranes are biased by hydrophobic interaction of the exposed nucleic acid bases, and secondly, mRNA is more flexible and prone to form secondary structures than DNA [141], The exact structural features of mRNA in complexes with cationic lipid moieties remain undetermined. Nevertheless, the properties of lipids and CL–NA phases described in this review are expected to apply also to the understanding of the principles of self-assembly in CL–mRNA nanoparticles (NPs).
Other phases that are closely related to the LαC phase but less relevant to delivery applications have also been reported. Three-dimensional columnar phases are characterized by the order of the DNA chains not only within a single layer but also between layers. Such phases have been observed for complexes of CL membranes in the “gel” phase with chain-ordered lipids with long DNA [142,143,144]. The additional ordering of DNA occurs due to charge-based DNA–DNA repulsion. Cationic lipids have reduced mobility in the gel phase, which limits their ability to screen electrostatic forces between DNA rods in different layers. Alternatively, the 3D ordering of DNA may be due to static membrane undulations that are coherent between layers. Recently, 3D long-range order was also observed for short, blunt DNA (which stacks into columns) confined between membranes containing lipids with chain-melted tails. This unexpected ordering is mediated by coherent membrane undulations across layers [145]. Theories of the lamellar phase of CL–DNA complexes also predict the existence of a novel “sliding columnar” phase, where the DNA rods have long-range orientation order from layer to layer but are not positionally correlated between layers [146,147,148].
4. The Inverse Hexagonal (HIIC) Phase
The inverse micellar building blocks resulting from lipids with preferred curvature C0 < 0, such as DOPE (Figure 4), favor the formation of the inverted hexagonal phase of CL–DNA complexes (Figure 8a). In this phase, termed HIIC, DNA chains occupy the aqueous interior of inverse cylindrical micelles, and those micelles are arranged on a hexagonal lattice, forming a 2D columnar liquid crystalline phase.
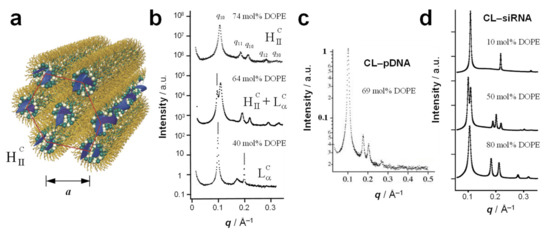
Figure 8.
(a) Schematic of the inverted hexagonal (HIIC) phase of CL–DNA complexes. In this phase, inverse cylindrical micelles containing DNA (i.e., DNA coated with a lipid monolayer) are arranged on a hexagonal lattice. The average spacing between the inverse micelles, a, can be obtained from the SAXS profile as a = 4π/√3q10. From [13]. Reprinted with permission from AAAS. (b) Example of the characteristic SAXS pattern of CL–DNA complexes in the HIIC phase (top profile). Also shown are characteristic SAXS patterns of CL–DNA complexes transitioning from the LαC phase (bottom profile) to coexisting LαC and HIIC phases (middle profile) and eventually the HIIC phase (top profile) as the content of DOPE in the membranes of the DOTAP/DOPE–DNA complexes increases. From [13]. Reprinted with permission from AAAS. (c) SAXS profile of complexes of DOTAP/DOPE (69 mol% DOPE) with plasmid DNA, revealing that the complexes are in the HIIC phase. Reprinted from [16], Copyright 2003, with permission from the Biophysical Society. (d) SAXS of DOTAP/DOPE–siRNA complexes reveals the formation of the lamellar phase at low content of DOPE (top), the inverse hexagonal phase at high content of DOPE (bottom), and coexistence of the two phases in a narrow regime of intermediate DOPE content. Reprinted with permission from [18]. Copyright 2007 American Chemical Society.
The typical SAXS peaks arising from the 2D hexagonal lattice of HIIC CL–DNA complexes are labeled q10, q11, q20, q12, q30 (Figure 8b, 74 mol% DOPE). From these, the unit cell spacing may be calculated as a = 4π/√3q10. Given that evenly spaced peaks similar to those at q10, q20, q30, etc. are also observed for the lamellar phase, the existence of a peak at q11, i.e., q10 × √3 can be considered a key indicator for distinguishing the two phases. A typical unit cell spacing for the HIIC phase is a ≈ 67.4 Å (DOTAP/DOPE–DNA complexes with 74 mol% DOPE) [13]. Assuming a lipid monolayer thickness of ≈ 19.5 Å (half of δm above), this yields a diameter of the interior of the inverse micelles of ≈ 28 Å, sufficient for a DNA molecule with approximately two hydration shells.
The HIIC phase appears in the DNA complexes of DOTAP/DOPE CLs beyond a threshold content of DOPE (because for DOTAP alone, C0 = 0) [13]. As shown in Figure 8b, the SAXS profile exhibits only the features of the LαC phase a DOPE content of 40 mol%, but at 74 mol% DOPE, a pure HIIC phase is observed (with the coexistence of the HIIC and LαC phases at 64 mol% DOPE). These complexes were prepared from long, linear λ-phage DNA.
Complexes of DOTAP/DOPE CLs with reporter gene-carrying plasmid DNA (Figure 8c) [16] as well as with siRNA (Figure 8d) [18] also formed inverse hexagonal phases at sufficiently high membrane content of DOPE. This further supports the hypothesis that the membrane structure, guided by the lipid shape, is the main determinant of the structure of CL–NA assemblies. Similar to DOTAP/DOPE–DNA complexes (Figure 8b), varying the preferred curvature of the lipid mixture forming CL–siRNA by means of changing the ratio of DOTAP (C0 = 0) and DOPE (C0 < 0), makes it possible to steer the phase of the resulting CL–siRNA complexes toward lamellar (high percentage of DOTAP) or inverse hexagonal (high percentage of DOPE), with the coexistence of the two phases at an intermediate composition (Figure 8d) [18]. The SAXS profile of such a mixture of coexisting phases is a superimposition of the profiles of the phases, with the integrated intensity proportional to the amount of phase present in the sample.
5. Hexagonally Ordered Cylindrical Micelles Embedded in a DNA Honeycomb Lattice: The HIC Phase
When the lipid assemblies are cylindrical micelles (for lipids with positive preferred curvature, C0 > 0), CL–DNA complexes may form the hexagonal (HIC) phase (Figure 9a). In this liquid crystalline structure, the cylindrical lipid micelles form a 2D hexagonal lattice. They are surrounded by the DNA, which forms a three-dimensional continuous substructure with honeycomb symmetry [14]. This is an interesting contrast to the isolated DNA rods (1D) and sheets (2D) in the HIIC and LαC phases, respectively (Figure 7a and Figure 8a).
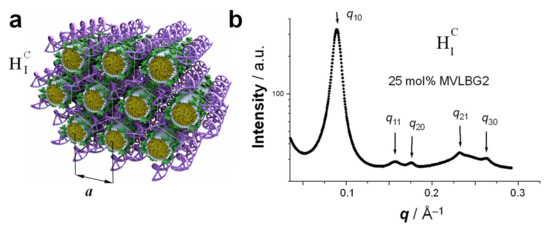
Figure 9.
(a) Schematic of the HIC phase of CL-DNA complexes. In this phase, cylindrical micelles formed by membranes containing strongly cone-shaped lipids such as MVLBG2 are arranged on a hexagonal lattice and surrounded by the oppositely charged DNA chains which form a honeycomb structure. (b) Synchrotron SAXS pattern of MVLBG2/DOPC–DNA complexes at 25 mol% of the highly charged lipid MVLBG2 (Figure 5). As described in the text, the SAXS peaks index to a 2D hexagonal lattice. Reprinted with permission from [14]. Copyright 2006 American Chemical Society.
The HIC phase was first observed in a narrow composition range of CL–DNA complexes containing ≈25 mol% MVLBG2 (with the remainder DOPC) in their membranes. MVLBG2 is a highly charged (16+) multivalent cationic lipid with a dendritic headgroup [14,30]. A distorted HIC phase was observed at higher contents of MVLBG2 and other highly charged (8+) dendritic lipids [115]. However, the headgroups of most other, even multivalent (up to 5+) cationic lipids, appear to be too small to force the formation of the cylindrical micelles that make up these phases.
The characteristic pattern of SAXS peaks of the HIC phase (Figure 9b) resembles that of the HIIC phase (Figure 8b) because of their identical symmetry. However, the lattice spacing observed for the HIC phase of MVLBG2/DOPC–DNA complexes is fairly large at a = 81.5 Å (compared to 67.4 Å for the HIIC phase of DOTAP/DOPE–DNA complexes, see above). The diameter of the hydrophobic core of the rod-shaped micelles is around 40 Å, as estimated from the thickness of a bilayer formed by lipids with identical (oleoyl) tails [14]. The DNA, headgroups, water, and counterions occupy the remaining space. For MVLBG2/DOPC–DNA complexes with higher contents (30–50 mol%) of MVLBG2, SAXS reveals a distorted hexagonal HIC phase [115].
6. Cubic Lipid Phases with Embedded Nucleic Acid
In some cases, the incorporation of NA disrupts the preferred phase of the lipid component because it cannot otherwise be accommodated in the assembly. For example, the lipid GMO [149] (Figure 3) readily forms bicontinuous cubic phases such as the gyroid QIIG with space group Ia3d (Figure 6a); this holds true even when GMO is mixed with (relatively small amounts of) cationic lipids such as DOTAP or MVL5 [15,121,150]. Both of these cubic phases with added cationic lipid are able to incorporate functional siRNA to generate a novel bicontinuous double gyroid cubic lipid phase, with the siRNA residing its two water channels (Figure 10a) [15,121]. The name of the phase indicates the fact that both the water and the lipid subphases are continuous in three dimensions, and that there are two independent water channels of gyroid symmetry (Ia3d space group). Figure 10b shows SAXS profiles for two compositions, illustrating the large number of peaks observed for this phase. For this phase, the lattice spacing aQ = 2π√6/q211. A gyroid cubic structure has also been proposed for siRNA complexes of a mixture of GMO with divalent cationic gemini lipids [151]. More recently, cubic CL–siRNA with Im3m symmetry have been discovered when a small amount of PEG-GMO lipid was added to DOTAP/GMO mixtures at very high GMO content (95 mol%) [152,153].
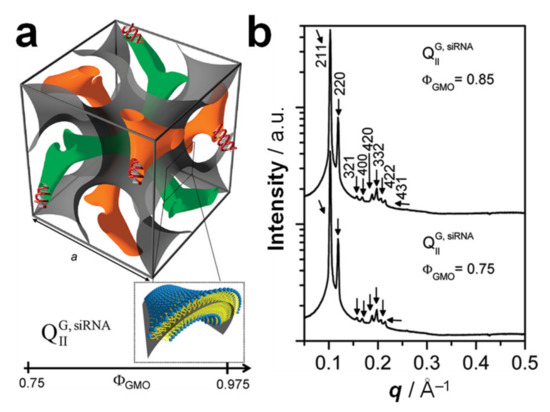
Figure 10.
(a) Schematic depiction of the double-gyroid cubic phase of CL–siRNA complexes labeled (QIIG, siRNA). The two intertwined but independent water channels are shown in green and orange. For clarity, the lipid membrane separating the two water channels is represented by a gray surface corresponding to its center (see inset). Note the negative Gaussian curvature of the bilayer, C1C2 < 0. (b) Synchrotron SAXS data obtained for DOTAP/GMO–siRNA at a DOTAP/GMO molar ratio of 15/85 (top) and 25/75 (bottom). The large number of peaks reveals the body-centered gyroid cubic structure (space group Ia3d). Reprinted with permission from [15]. Copyright 2010 American Chemical Society.
When CLs of the compositions that form double-gyroid cubic CL–siRNA complexes are mixed with DNA, complexes in the HIIC phase are formed. This likely occurs because the energetic cost of bending the DNA to conform to the highly curved channels of the double-gyroid phase is greater than the cost of rearranging the lipid phase into the related HIIC phase. In fact, studies with a series of short DNA duplexes of varied length and end structure (“sticky”, repulsive, and no overhangs) revealed that stacking of these short pieces of DNA can induce the HIIC phase. Thus, counteracting the stacking with repulsive (nonpairing) overhangs or increased temperature shifted the equilibrium towards the gyroid cubic phase [154].
7. Transfection Efficiency and the Structure of CL–DNA Complexes
7.1. The Early Rise of DOPE and Its Relation to Complex Structure
Early studies revealed that CL–DNA complexes displayed increased transfection efficiency (TE) if the cationic lipid was combined with a neutral lipid. (TE is a measure of the extent of expression of an exogenous gene that is transferred into the cell). The term “helper lipid” was coined for these lipids, and DOPE, probably initially chosen because of its reputation as a “fusogenic” lipid, proved much more successful than DOPC [155]. Considering the propensity of DOPE for inverse hexagonal structures, this suggested a correlation between the liquid crystalline structure of CL–DNA complexes and their TE. A series of studies designed to elucidate this correlation indeed found important structure-dependent differences. As a function of increasing the content of cationic lipid, the TE of the DNA complexes of a series of cationic lipids mixed with DOPC increased to a maximum level before decreasing again (Figure 11a). At its maximum, the TE of these lamellar complexes was the same as that of DOTAP/DOPE complexes in the HIIC phase. However, in terms of the ratio of cationic lipid to helper lipid, the position of the maximum in TE shifts (Figure 11a), depending on the charge of the cationic lipid and the size of its headgroup. This means that it is easy to miss the most effective composition when only testing a few ratios of cationic to helper lipids, as was done in early investigations. In contrast, the TE of CL–DNA complexes in the HIIC phase is large over a wide range of compositions (Figure 11b). This means that the early investigations were very likely to find a cationic lipid/DOPE mixture with high TE, even if only a few compositions were tested.
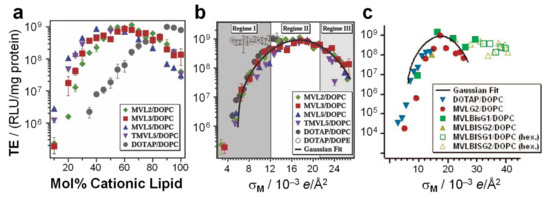
Figure 11.
The membrane charge density (σM, the average charge per unit area of the membrane) is a universal parameter for the transfection efficiency (TE) of lamellar CL–DNA complexes, but not for nonlamellar (HIIC or HIC) complexes. (a) TE plotted as a function of molar fraction cationic lipid for DNA complexes of MVL2, MVL3, MVL5, TMVL5, and DOTAP mixed with DOPC (see Figure 2, Figure 3 and Figure 5). (b) The same data as in (a), but plotted as a function of σM collapses onto a universal, bell-shaped curve as a function of σM (the solid line is a Gaussian fit to the data). TE data for DOTAP/DOPE complexes (open circles, HIIC phase) deviates from the universal curve, indicative of a distinctly different transfection mechanism for the inverted hexagonal phase. Three regimes of transfection efficiency are highlighted as described in the text. The membrane charge density can be written as σM = [1 − Φnl/(Φnl + rΦcl)]σcl. Here, r = Acl/Anl is the ratio of the headgroup areas of the cationic and the neutral lipid; σcl = eZ/Acl is the charge density of the cationic lipid with valence Z; Φnl and Φcl are the mole fractions of the neutral and cationic lipids, respectively. The membrane charge density was calculated using Anl = 72 Å2, rDOTAP = 1, rMVL2 = 1.05 ± 0.05, rMVL3 = 1.30 ± 0.05, rMVL5 = 2.3 ± 0.1, rTMVL5 = 2.5 ± 0.1, ZDOTAP = 1, ZMVL2 = 2.0 ± 0.1, ZMVL3 = 2.5 ± 0.1, ZMVL5 = ZTMVL5 = 4.5 ± 0.1 [17]. (c) TE for DNA complexes of MVLG2 (4+), MVLBisG1 (8+), MVLBisG2 (16+), and DOTAP mixed with DOPC (see Figure 2, Figure 3 and Figure 5) plotted as a function of membrane charge density. Filled symbols are for lamellar complexes, while empty symbols are for complexes in the HIC or distorted HIC phases. Again, the data for nonlamellar complexes deviates from the universal curve for lamellar complexes, indicating different transfection mechanisms. Parts (a,b) adapted from [17] with permission from John Wiley & Sons, Ltd. Part (c) adapted with permission from [115]. Copyright 2009 American Chemical Society.
7.2. Membrane Charge Density as a Universal Parameter for Transfection by Lamellar CL–DNA Complexes
Importantly, the work described above also revealed that the membrane charge density, σM, is a universal parameter predictive of the TE of lamellar CL–DNA complexes [16,17]. The membrane charge density is defined as the average charge per unit area of the membrane. It can be considered a lipid-independent way to describe how highly charged a membrane is. In contrast, variations in headgroup charge and size mean that membranes at the same mol fraction of cationic lipid can carry very different numbers of charges per area. Figure 11a,b illustrates how the membrane charge density unifies the dependence of TE on composition. Figure 11a plots the TE against the molar fraction of cationic lipid, for DNA complexes of monovalent DOTAP and a series of custom-synthesized multivalent lipids ((T)MVLs, Figure 5) with systematically varied headgroup charge (between 2+ and 5+) mixed with DOPC [17,84] The amount of DNA and the charge of cationic lipid to DNA were kept constant for all data points (at an optimal charge ratio of 2.8). While the shape of the curves is similar, the maxima appear at different molar ratios of cationic lipid/DOPC. In contrast, when the TE of the complexes is plotted against the membrane charge density σM (Figure 11b), the data merge onto a single (universal) curve. This identifies σM as a universal parameter for transfection by lamellar CL–DNA complexes (i.e., a predictor of TE). The solid curve in Figure 11b is the fit of a Gaussian curve to the TE data which yields the optimal charge density σM* = 17.0 ± 0.1 × 10–3 e/Å2 (see the caption of Figure 11 for the fitting equation) [17].
The shape of the universal curve suggests three distinct regimes of TE for lamellar CL–DNA complexes (highlighted by different backgrounds in Figure 11b), which can be related to two barriers to transfection in a simple model to explain the data. At low membrane charge density, an increase in σM (regime I, dark gray) enhances fusion between the cationic membranes of lamellar complexes (trapped in endosomes) and anionic endosomal membranes. This facilitates the release of complexes into the cell cytoplasm and thereby transfection. In contrast, at very high membrane charge density (regime III, light gray), an increase in σM progressively inhibits cationic membranes from efficiently releasing the DNA following an endosomal escape. These opposing trends give rise to the optimal regime II around σM*, where σM is large enough to promote fusion with and escape from the endosome but not too large to prevent the subsequent release of DNA from the complex [17].
In contrast to the strong dependence of the TE of lamellar CL–DNA complexes on σM, nonlamellar complexes show high TE independent of σM. This is true both for HIIC complexes (Figure 11b, open circles, DOTAP/DOPE–DNA complexes) [16,17] and complexes in the HIC and distorted HIC phases [14,115] (Figure 11c) and indicates that their mechanism of action must be distinct from that of lamellar complexes, including different interactions of the complexes with cellular membranes and organelles [16,17]. For example, the high TE of HIIC complexes in regime I is likely related to the efficient fusion between the membranes of complexes containing DOPE and cellular membranes (e.g., those of the endosome) [16].
Note that because of the high content of DOPE required to induce the phase, only the regime of low σM is accessible with HIIC complexes. In contrast, the high charge of the lipids that form the HIC and distorted HIC phases mean that these phases only appear at high σM. (At low contents of the same lipids, C0 is dominated by that of DOPC and lamellar phases are formed.) The contiguous substructure of DNA in these assemblies may be the reason that their efficiency remains high even at extremely high membrane charge density.
7.3. Highly Efficient Gene Silencing with Cubic CL–siRNA Complexes
As mentioned above, cubic phase forming lipids favor saddle-splay-shaped surfaces with negative Gaussian curvature (C1C2 < 0) which are also found in membrane pores. Pore formation is a possible mechanism for escaping the endosome after endocytosis of complexes, a rate-limiting step in transfection [156,157,158]. In fact, for a series of cationic phospholipids with varied tails, the difference in their propensity to form cubic phases with lipids found in cellular membranes has been invoked as a possible explanation of the differences in transfection efficiency [21,138,139]. Another approach was based on the hypothesis that cubic-phase CL–NA complexes would exhibit enhanced membrane fusion with cellular membrane barriers such as endosomal membranes because the constituting lipids promote the formation of pores with the desired negative Gaussian curvature [15,116,121]. The enhanced fusion, in turn, would result in efficient cytoplasmic siRNA delivery and thus gene silencing.
The cubic gyroid phase of CL–siRNA complexes indeed turned out to be capable of efficiently delivering siRNA, resulting in sequence-specific gene silencing without toxicity even at very low membrane charge density (Figure 12) [15,121]. This is in contrast to lamellar CL–siRNA complexes, which require relatively high membrane densities to enable endosomal escape, and supports the idea that a different mechanism of fusion is operating. The low cytotoxicity is notable because DOPE, which forms the nonlamellar HIIsiRNA phase at appropriate membrane compositions, exhibits significant toxicity at the high amounts of lipid required for siRNA delivery [18].
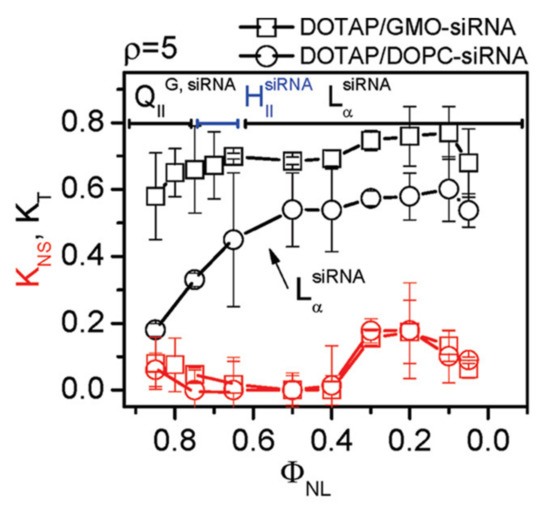
Figure 12.
Sequence-specific gene silencing of CL–siRNA complexes incorporating the cubic-phase forming lipid GMO (Figure 2) is strongly improved compared to DOTAP/DOPC–siRNA complexes. Optimal silencing corresponds to KT (total (specific and nonspecific) gene knockdown; black lines and symbols) approaching 1 while KNS (nonspecific gene knockdown, red lines, and symbols) is minimal. DOTAP/GMO–siRNA complexes (squares) are in the gyroid cubic phase (QIIG,siRNA) at a high mole fraction of neutral lipid (ΦNL) where KT is high and KNS is low. In contrast, lamellar (LαsiRNA) DOTAP/DOPC–siRNA complexes (circles) show low KT at high ΦNL. The increased KNS at low ΦNL, when both formulations form lamellar complexes because of their high content of DOTAP (C0 ≈ 0), indicates an undesirable onset of vector toxicity. Reprinted with permission from [15]. Copyright 2010 American Chemical Society.
8. From In Vitro to In Vivo
An early major challenge to successful drug delivery with liposomes in vivo was their rapid clearance from circulation by a component of the immune system called the mononuclear phagocytic system [159,160,161]. A solution to this challenge are so-called STEALTH® liposomes, whose surface is covered by poly(ethylene glycol) (PEG), a hydrophilic polymer, typically at a molecular weight (MW) of 2000 or 5000 [162,163,164,165,166,167,168,169,170]. To anchor it to the surface of the liposome, the PEG is attached to a lipid moiety, creating a PEG-lipid (Figure 3). These lipids were evolved from glycosphingolipids with sialic acid groups [171,172], used in the earliest attempts to develop long-circulating liposomes by mimicking red blood cells.
The PEG-lipid sterically stabilizes STEALTH liposomes, meaning that the shell of hydrophilic polymer acts as a physical barrier to objects approaching the liposomes. The utility of this repulsive interaction (with a range on the scale of the size of the polymer chain [173,174,175]) is broad: it counteracts both electrostatic adhesion of oppositely charged particles to cationic or anionic liposomes [176] and the nonspecific adhesion of, e.g., proteins or other liposomes due to van der Waals interactions [160,162,163]. Accordingly, PEGylated liposomes exhibit extended in vivo circulation times (suggesting that PEGylation limits adhesion of blood plasma opsonins, a necessary event for removal by immune cells), and multiple formulations are now FDA-approved medicines. A prominent example is Doxil®, a liposomal formulation of doxorubicin, for cancer chemotherapy [177].
As mentioned earlier, complexation with NAs completely disrupts and rearranges the structure of CLs, independent of whether bilayer- (e.g., LαC) or nonbilayer-type (e.g., HIIC or HIC) CL–NA complexes form. These complexes are prone to aggregation in salt solution (e.g., cell culture medium). However, it is possible to use PEG-lipids to sterically stabilize CL–NA assemblies to sizes around 50–200 nm [55,126,178,179,180,181,182,183]. One way of achieving this is to use PEGylated CLs for complexation with NAs in an aqueous environment, where the specifics of the preparation method control important characteristics of the resulting CL–NA nanoparticles (NPs) [126,178,184]. Another preparation method combines an ethanolic solution of lipids with an aqueous solution of NA that is at a pH at which the ionizable CLs used are protonated, preferably using a microfluidic mixer. This method yields CL–NA NPs that have been termed LNPs after a dialysis step to remove residual ethanol and adjust the pH of the LNP suspension to physiological pH [55,181,182,183]. This is the method used for the preparation of patisiran and the CL-based COVID-19 vaccines.
The internal structure of CL–NA NPs again depends on the lipid composition: the formation of lamellar structures has been observed with SAXS and cryo-EM [126,178], as has the absence of periodic ordering (which makes structure determination with SAXS difficult) [126], in particular for LNP formulations that contain cationic lipids with a propensity for forming nonbilayer HII phases [185] and large amounts of cholesterol [183,186].
To avoid opsonization and nonspecific attachment to cells (essential for targeted in vivo applications) the surface of CL–NA NPs requires a PEG-lipid concentration that is high enough to ensure that the PEG chains are in the brush regime (i.e., stretched-out chains; e.g., 10 mol% PEG2000-lipid) [170,173,174,175,187]. Unfortunately, this extent of PEGylation strongly reduces the transfection efficiency of the CL–DNA NPs by increasing the barriers for both cell attachment and endosomal escape [179,180,184]. It is therefore important to devise strategies that can counter these undesirable effects of PEGylation.
Below, we describe two complementary strategies that can be used for partial recovery of the TE of PEGylated CL–DNA NPs (Figure 13): (1) low pH-induced dePEGylation to promote endolysosomal escape (Figure 14) [179,188,189] and (2) conjugating affinity targeting ligands to the PEG to favor uptake in target cells (Figure 15) [180].
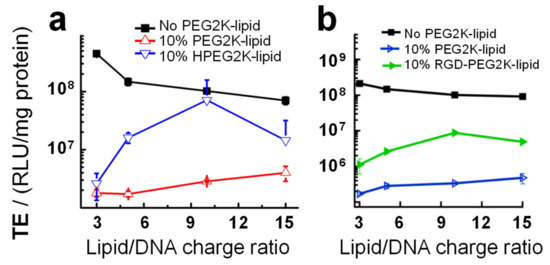
Figure 13.
Two strategies to enhance transfection efficiency (TE) of PEGylated CL–DNA complexes. TE in murine CCL-1 cells is plotted versus ρ (lipid/DNA charge ratio) for DOTAP/DOPC/PEG-lipid–DNA complexes (80 mol% DOTAP(1+)) and control complexes without PEG-lipid. TE drops strongly upon the inclusion of 10 mol% PEG-lipid. However, complexes containing RGD-PEG2K-lipid or HPEG2K-lipid instead show partial recovery of TE, which is due to distinct mechanisms as discussed in the text. (a) Comparison of the TE of complexes without PEG2K-lipid (black), with 10% PEG2K-lipid (red), and with 10 mol% acid-labile HPEG2K-lipid (blue). Adapted from [179], Copyright 2012, with permission from Elsevier. (b) Comparison of the TE of complexes without PEG2K-lipid (black), with 10% PEG2K-lipid (blue), and with 10 mol% RGD-PEG2K-lipid (green). Adapted from [180], Copyright 2014, with permission from Elsevier.
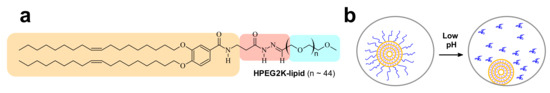
Figure 14.
(a) Structure of the HPEG2K-lipid. The acid-labile acylhydrazone moiety is underlain in red, the lipophilic tails in tan, and PEG in blue. (b) Schematic depiction of the proposed mechanism of TE recovery by the low-pH-sensitive HPEG2K-lipid. During the maturation of endosomes, acidification cleaves the PEG chains from the lipid tails. This unmasks the positive charge of the CL–DNA NP, allowing electrostatically mediated recruitment to, and fusion with, the negatively charged endosomal membrane, facilitating endosomal escape [179].
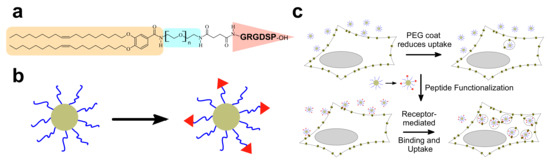
Figure 15.
(a) Structure of the RGD-PEG2K-lipid as an example of a ligand-PEG-lipid. The peptide ligand is highlighted with a red triangle, the lipophilic tails in beige, and PEG in blue. (b) Schematic depiction of ligand-tagging of CL–NA NPs. (c) PEGylation reduces cellular uptake of NPs, reducing efficacy. Functionalization of the distal end of a PEG-lipid with an appropriate ligand induces receptor-mediated binding and increases cellular uptake (and thus efficacy) in cells expressing the peptide’s receptor [180].
8.1. Low pH-Induced dePEGylation
The tail length affects the rate of desorption of PEG-lipids from CL–NA NPs, and careful choice of tails can tune NP stability in vivo [190]. However, other strategies are required at high PEG densities. One such strategy involves incorporating acid-labile bonds between the tails and the PEG moiety of the PEG-lipids, to induce dePEGylation at the low pH (≈5) of late endosomes. The acidification that is part of the maturation process of the endosome cleaves the PEG chains from the lipid tails and thus removes the PEG-coat of the NP. This enhances TE because it “switches on” the electrostatic attractions required for endosomal escape via membrane charge density-promoted activated fusion [17] by “unmasking” the charge of the NP (Figure 14b). The chemistry of the acid-labile bond is a crucial aspect of this strategy because it allows fine-tuning the pH ranges where cleavage occurs [191]. A successful example is a PEG-lipid (termed HPEG2K-lipid; PEG MW 2000 Da) bearing an acid-labile acylhydrazone bond between the lipid tails and the PEG chain (Figure 14a) [179]. The HPEG2K-lipid is stable at pH = 7, but the PEG chain is cleaved from the lipid tails at pH = 5. Figure 13 shows that complexes containing 10% HPEG2K-lipid (green diamonds) exhibit partial recovery of TE when compared to complexes containing 10% PEG2K-lipid (blue triangles) [179].
8.2. Affinity Targeting of PEGylated CL–DNA Complexes
PEGylation strongly reduces nonspecific attractive interactions of CLs and CL–NA NPs with the cell surface. This reduces efficacy but also opens the opportunity of specific targeting with affinity ligands such as peptides, antibodies, and aptamers. For example, uptake of PEGylated CL–DNA complexes in cultured cells can be increased by tagging the NPs with RGD peptides [192,193,194,195] that bind to the cell surface integrins expressed on the target cells (Figure 15). Figure 15a shows the GRGDSP-PEG2K-lipid [180,196], a PEG-lipid with a distally attached linear RGD peptide that shows high affinity for αvβ3 and αvβ5 integrins. Incorporating this peptide-PEG-lipid into NPs instead of the DOB-mPEG2000 lipid (Figure 3) results in NPs that are decorated with the peptide ligand (Figure 15b). While highly PEGylated NPs are sparsely taken up by cells (Figure 15c, top), the ligand-tagged NPs enter integrin-positive cells in greater number by receptor-mediated endocytosis. Live-cell imaging with quantitative colocalization analysis directly confirmed this increased NP uptake [180]. The increased internalization was paralleled by improved cellular delivery of the DNA payload: transfection experiments showed an increase in TE (Figure 13b).
8.3. Organ- and Disease-Specific Targeting Peptides
In addition to increasing efficacy by promoting uptake in cultured cells, peptide ligands may be used to target specific healthy and diseased tissues in vivo [197,198]. The systemically accessible molecular landscape on endothelial cells bears tissue type- and status-dependent signatures. This vascular heterogeneity can be explored in an unbiased manner by in vivo screening of phage libraries that display random, genetically encoded peptide sequences [199]. This screening process consists of injecting the phage library into the systemic circulation, rescuing the phage from the target tissue, and repeating the process to derive a phage pool with the ability to home to the intended target tissue [200]. The in vivo phage display approach has yielded a variety of homing peptides specific for normal organs as well as for tumor vasculature and tumor cells [198,201,202,203,204,205,206,207,208,209,210]. Recent advances in the approach, most notably the application of high-throughput sequencing and development of bioinformatics tools to analyze the resulting data, allow for streamlined, robust, and reproducible identification of homing peptides [211,212] and mapping of the vascular diversity.
In vivo phage display and chemical optimization of the discovered peptides for increased stability and affinity have greatly expanded the spectrum of peptide ligands and potentiated their binding since the initial discovery of the RGD-motif [192,193,194,213]. An example of chemical optimization is head-to-tail cyclic RGD peptides (e.g., c(RGDfK) [214,215]) with high affinity (up to 1000× larger than linear RGD [216,217,218]) for the αvβ3 integrin overexpressed in many cancer cell lines in vitro and in malignant lesions in vivo. Another example is the development of stabilized versions of the fibrin-targeting CREKA peptide containing nonproteinogenic amino acids [219].
Importantly, and of translational relevance, the homing peptides are typically not species-specific—most of the homing peptides bind the corresponding human epitopes as well. Whereas homing peptides typically use docking interactions for accumulation in endothelial cells of target tissues, some homing peptides are tissue penetrating: they specifically recognize the endothelium of target vessels, extravasate, and penetrate deep into the extravascular tissue [220,221,222,223,224]. This class of homing peptides is particularly useful for targeting cells in the tissue stroma. One such tumor penetrating peptide, iRGD (sequence: disulfide-cyclized CRGDKGPDC) [221,222,223,225], is undergoing clinical testing for precision delivery of paclitaxel albumin nanoparticles (Abraxane) and gemcitabine to pancreatic cancer lesions (ClinicalTrials.gov Identifier: NCT03517176, “CEND-1 in combination with Nab-paclitaxel and Gemcitabine in Metastatic Pancreatic Cancer”).
Another example of a tumor penetrating peptide is TT1 (sequence: disulfide-cyclized CKRGARSTC) and its linear version linTT1 (AKRGARSTA). These peptides target p32 (HABP1/gC1qR), a protein that is upregulated and aberrantly expressed on the surface of malignant cells and activated macrophages in solid tumors and in atherosclerotic plaques [226,227,228,229,230,231,232,233].
In light of the current interest in CL–NA NPs for vaccine development, it is worth mentioning another recently identified targeting peptide, mUNO (sequence: CSPGAK). This peptide targets CD206 (known as the mannose receptor) on mouse and human cells when coated on polymeric nanoparticles [234,235,236]. The mannose receptor is widely used for delivering material to antigen-presenting dendritic cells for vaccine applications [237,238,239].
Receptors, i.e., the binding partners of homing peptides, can be identified using different biochemical approaches, such as affinity chromatography and proximity biotin ligation-based assays. Expression and accessibility of peptide receptors vary not only between different tissues and organs but also between healthy and diseased tissues [201,202,203,204] and between particular cell populations within the target tissue [234]. Examples include certain growth factor receptors (e.g., VEGFR2, neuropilin-1), cell adhesion molecules, and intracellular proteins (e.g., p32, nucleolin) aberrantly expressed at the cell surface in tumor vessels and malignant cells, CD206 expressed on M2 macrophages [234] and certain tumor-associated extracellular matrix isoforms [240,241].
8.4. Peptide Ligands Promote Tumor Targeting and Penetration of CL–DNA NPs In Vivo
Recent work used flow cytometry to screen peptides and compositional parameters of CL–DNA NPs (such as the lipid/DNA charge ratio and peptide coverage) for optimal in vivo performance (Figure 16a,b) [242]. The study investigated vectors tagged with linear GRGDSP (“RGD”), the cyclic iRGD tumor penetrating peptide (above), cRGD (c(RGDfK), above)), and RPARPAR. RPARPAR is a prototypical example of peptides containing an active C-end Rule (CendR) motif, R/KXXR/K. This motif, which has to be exposed at the C-terminus for activity, is shared between all tumor penetrating peptides [221]. Interestingly, intermediate peptide coverage (e.g., 5 mol% peptide-PEG-lipid + 5 mol% PEG-lipid rather than 10 mol% peptide-PEG-lipid) was optimal for in vitro binding and internalization in receptor-positive cells [242]. At a low lipid/DNA charge ratio, binding and internalization were moderately high for NPs tagged with iRGD and RPARPAR and highest for those tagged with cRGD (Figure 16a,b) in multiple cell lines in vitro. However, the higher positive charge of RPARPAR resulted in higher nonspecific attachment, showing that ligand charge is an additional important parameter to consider when designing targeted NPs.
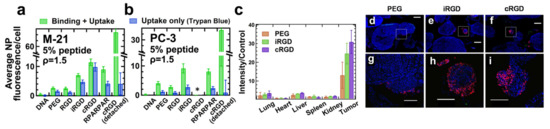
Figure 16.
Peptide-tagging for specific targeting of CL–DNA NPs in vitro and in vivo. NPs were formulated at a lipid/DNA charge ratio of 1.5 and a molar ratio of 10/70/10/5/5 of MVL5/DOPC/cholesterol/PEG-lipid/x, where x = PEG-lipid (control) or peptide-PEG-lipid. (a,b) Fluorescence from bound and internalized NPs containing labeled DNA in two cell lines (M21 and PC-3) measured by flow cytometry. The graphs compare several tagged (peptide-PEG-lipid) and untagged (PEG-lipid only) NPs with free DNA (no lipid) as a control. Binding and uptake were differentiated by the addition of Trypan Blue, a membrane-impermeable dye that quenches the fluorescence of NPs outside the cells. NPs tagged with cRGD detached a large number of cells from the substrate. These cells were measured separately. *: too few cells remained attached to allow measurement. (c) In vivo biodistribution of intraperitoneally (i.p.) administered CL–DNA NPs. Mice bearing intraperitoneal MKN-45P tumors were i.p. injected with either PBS (control) or ~0.5 mg of CL–DNA NPs. After 24 h the tumors and organs of interest were excised and the fluorescent signal from the Cy5-labeled DNA was imaged (inset) and quantified (bars; normalized to control; n = 3). The vast majority of the fluorescent DNA is found in the tumor, and peptide-tagged NPs show higher selectivity for the tumor than untagged NPs. (d–i) Confocal microscopy images showing CL–DNA NPs in sections of the tumor nodules. Parts (g–i) are enlarged views of the marked areas in parts (d–f), respectively. Cy5 (DNA-label, i.e., NPs): red, DAPI (cell nuclei): blue. Tumor nodules from mice treated with untagged (control) PEG2000-lipid NPs (d,g) show NPs on the nodule surface, while iRGD- (e,h) and cRGD-tagged NPs (f,i) penetrated into the tissue of smaller tumor nodules (diameter ~300 μm). Scale bars: 500 μm (d–f) and 200 μm (g–i). Adapted from [242], Copyright 2018, with permission from Elsevier.
In vivo biodistribution experiments, in which NPs tagged with iRGD and cRGD peptides were systemically administered to mice bearing peritoneally disseminated human gastric cancer (with a PEG-lipid control) revealed that both peptide-tagged NPs homed to the tumors at higher levels than the control NPs (Figure 16c). Importantly, in tumors, the peptide-tagged NPs also extravasated and penetrated into malignant tissue, in particular into small nodules (which are harder to remove surgically and thus a desirable target for therapeutics) (Figure 16e,f,h,i) [242].
9. Cationic Liposomes for The Delivery of Hydrophobic Drugs
Aside from NA delivery, cationic liposomes have received intense interest as carriers for other classes of therapeutics, in particular hydrophobic cancer drugs (Figure 1) [243,244,245,246,247,248,249]. Many of the principles underlying structure–activity relationships of CL–NA assemblies (such as the effect of lipid shape on assembly structure) that have been outlined in the previous sections are also relevant in this context, even as some intriguing differences exist (e.g., with respect to the effect of PEGylation, see below).
A prominent example of a hydrophobic cancer drug is paclitaxel (PTX), which is among the most widely used chemotherapy drugs to treat ovarian, breast, lung, pancreatic, and other cancers [250,251,252,253,254,255,256,257,258]. Because of their hydrophobicity, these drugs reside within the lipid bilayer rather than in the aqueous interior of the liposome (Figure 1, red spheres). This means that they will remain associated with the lipid membranes upon formation of CL–NA complexes, resulting in the exciting potential to combine them with NAs into therapeutics with dual action [259].
Because of their low water solubility, hydrophobic drugs such as PTX are not effective unless formulated with a carrier. Currently used formulations of PTX (such as Taxol® and Abraxane®); frequently cause hypersensitivity reactions and/or deliver PTX non-discriminately throughout the body [260,261,262]. Therefore, the development of liposomal formulations of PTX with high efficacy is an extremely active field of research [263,264,265,266,267], including in clinical trials [249,267]. Cationic liposomes have been shown to target tumor neovasculature [244,245,246,268,269] and are readily internalized by cells. This makes cationic liposomes desirable vectors for hydrophobic drugs because molecules such as PTX have to reach the inside of cells to unfold their activity.
A common feature of liposomal formulations of PTX is their relatively low loading capacity, at 3 mol% of the lipid content. Improving the PTX loading capacity of liposomal carriers requires enhancing PTX solubilization within the membrane [270]. Recent work showed that there is great potential for achieving this by modifying the lipid tails [109]. PTX membrane solubility in cationic liposomes with tails containing two cis double bonds (linoleoyl tails) was significantly increased compared to tails with one (oleoyl tails) double bond: 8 mol% PTX in the former remained soluble for approximately as long as 3 mol% PTX (the above-mentioned membrane solubility limit) in the latter (Figure 17). Comparison with DOPE-containing formulations suggests that the increase in solubility is likely not caused by the structural change from bilayers to inverse cylindrical micelles (Figure 4) but rather by the enhanced molecular affinity between lipid tails and PTX. Importantly, the efficacy of the PTX-loaded CLs was unaffected by changing the lipid tails in one cell line; in another cell line, the efficacy was even increased two-fold. These findings demonstrate the potential of chemical modifications of the lipid tails: liposomal PTX carriers with increased PTX solubility, at maintained or even increased efficacy, reduce side effects and costs because they require less lipid to deliver a given amount of PTX.
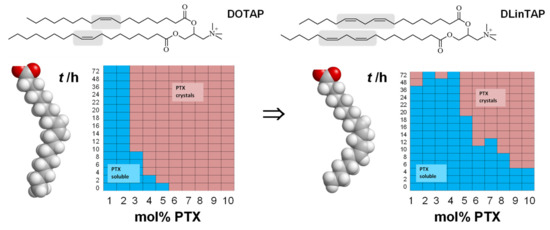
Figure 17.
Top: Structure of DOTAP, with oleoyl (C18 with a single cis double bond) tails, and the corresponding lipid DLinTAPwith linoleoyl tails (with an additional cis double bond). Bottom: Space-filling molecular models of the ground-state structure of the lipid tails and PTX solubility kinetic phase diagrams for the corresponding DOTAP/DOPC and DLinTAP/DLinPC formulations. Formulations of increasing PTX content (x-axis) were monitored over time (y-axis) for PTX crystallization (red color). Blue color indicates the absence of PTX crystals. See Figure 3 for the structures of DOPC and DLinPC. Solubility phase diagram data reprinted with permission from [109].
As mentioned earlier, PEGylation of CL-based NA carriers is necessary for in vivo applications but diminishes efficacy. Surprisingly, recent work showed that PEGylation of CL-based carriers of PTX enhances, rather than diminishes, delivery efficacy and cytotoxicity against human cancer cells (Figure 18a,b). This unexpected enhancement occurs even at low PEG-lipid content, when the PEG chains are in the transition regime between the mushroom and brush conformations. Cryogenic TEM showed that PEGylation leads to a mixture of nanometer-scale vesicles and anisotropic micelles (Figure 18a,c). The formation of disk-shaped micelles at sub-monolayer concentrations of the PEG-coat was unexpected; it had previously been assumed that vesicles would incorporate such relatively small amounts of PEG-lipid without modifying their shape. Confocal microscopy and flow cytometry revealed significantly enhanced cellular uptake of PTX for PEGylated (vesicles and disks) compared to bare PTX-loaded liposomes. This suggests that steric stabilization can facilitate NP entry into cells via distinct size-dependent endocytic pathways that are inaccessible to larger liposomes and aggregates. This study highlighted the value of understanding how PEGylation alters NP shape and structure, and thus NP efficacy, to design next-generation stealth drug carriers. Importantly, these PEGylated liposomes will allow integrating active cell-targeting strategies for delivery in vivo.
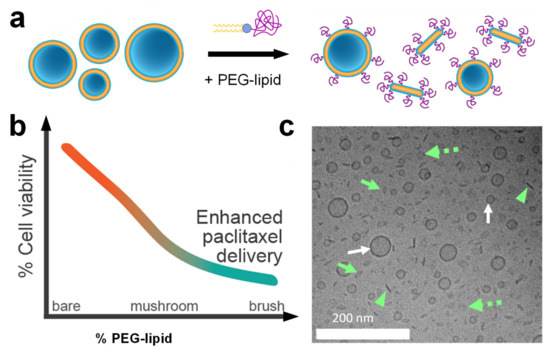
Figure 18.
(a) Schematic illustration of the structural transitions observed upon PEGylation of PTX-carrying CLs. Unilamellar vesicles of varying sizes are replaced by small vesicles and discoidal micelles (bicelles). (b) Cytotoxicity of PTX-carrying CLs as a function of increasing PEGylation (at a constant amount of PTX). The efficacy of the CLs against cancer cells increased (cell viability decreased) with the extent of PEGylation. (c) Cryogenic electron microscopy image of a formulation of DOTAP/DOPC/PEG2000-lipid/PTX at a molar ratio of 50/37/10/3. Small vesicles of varying size (white arrows) and edge-on (green arrowhead), tilted (green arrow), and top-down (green dashed arrow) views of discoidal micelles are discernible. Adapted with permission from [112]. Copyright 2020 American Chemical Society.
10. Concluding Remarks
As the range of applications of cationic liposomes in the delivery of therapeutics further matures and grows, it becomes ever more important to develop a fundamental science base that can inform the rational design of these self-assemblies. We have shown in this review that it is essential to consider principles from soft matter and biophysics and lipid and colloid science when trying to understand the structure–activities of the cationic and neutral lipids, because the lipids exert their function as an assembly of molecules. The results of ongoing studies investigating the barriers to targeted delivery and the mechanisms of intracellular uptake, transport, and release of these vectors and their interactions with cell components will continue to inform the design and synthesis of optimal lipid carriers of NAs.
Even as the medical applications are just beginning, cationic liposome-based vectors of nucleic acids for gene delivery, gene silencing, and gene editing have found myriad applications in fundamental and applied biological research. Further advances in the delivery systems will only add to these applications, be it in studies of chromosome structure and function through the development of efficient vectors for very large DNA constructs, or in molecular biology studies through improvements in the ability to efficiently deliver NAs into hard-to-transfect cell lines such as T-cells, macrophages and dendritic cells. Other important frontiers of the field are selectively targeting tissues beyond the liver (patisiran) and local delivery (mRNA vaccines). Finally, for cancer therapeutics, the ability of CLs to simultaneously deliver both NAs and hydrophobic drugs in combination therapies holds great promise.
Author Contributions
All authors equally contributed to writing of this review. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Institutes of Health under Award R01GM130769. Partial support was further provided by the National Science Foundation (NSF) under award DMR-1807327. VS was supported by the NSF Graduate Research Fellowship Program under grant no. DGE 1144085. The work utilized the Shared Experimental Facilities of the Materials Research Laboratory at UCSB. These facilities are supported by the MRSEC Program of the NSF under award DMR 1720256; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org accessed on 10 July 2021). P. Scodeller was supported by Eesti Teadusagentuur Starting Grant PUT-PSG38. T. Teesalu was supported by the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012) and by the Estonian Research Council (grants PRG230 and EAG79).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
P.S. and T.T. are inventors of patents on mUNO peptide. In addition, T.T. is inventor of iRGD and CendR peptides and a shareholder of Cend Therapeutics Inc., a company that holds a license for the mUNO, iRGD and CendR peptides. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Israelachvili, J.N. Intermolecular and Surface Forces, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Bangham, A. Model for biological membranes. New Sci. 1971, 49, 63–64. [Google Scholar]
- Gregoriadis, G.; Leathwood, P.D.; Ryman, B.E. Enzyme entrapment in liposomes. FEBS Lett. 1971, 14, 95–99. [Google Scholar] [CrossRef]
- Gregoriadis, G.; Ryman, B.E. Fate of Protein-Containing Liposomes Injected into Rats. Eur. J. Biochem. 1972, 24, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Gregoriadis, G. The Carrier Potential of Liposomes in Biology and Medicine (Part One). N. Engl. J. Med. 1976, 295, 704–710. [Google Scholar] [CrossRef]
- Gregoriadis, G. The Carrier Potential of Liposomes in Biology and Medicine (Part Two). N. Engl. J. Med. 1976, 295, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081. [Google Scholar] [CrossRef] [PubMed]
- Mislick, K.A.; Baldeschwieler, J.D. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc. Natl. Acad. Sci. USA 1996, 93, 12349–12354. [Google Scholar] [CrossRef]
- Sternberg, B.; Sorgi, F.L.; Huang, L. New structures in complex formation between DNA and cationic liposomes visualized by freeze—fracture electron microscopy. FEBS Lett. 1994, 356, 361–366. [Google Scholar] [CrossRef]
- Rädler, J.O.; Koltover, I.; Salditt, T.; Safinya, C.R. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997, 275, 810–814. [Google Scholar] [CrossRef]
- Koltover, I.; Salditt, T.; Rädler, J.O.; Safinya, C.R. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 1998, 281, 78–81. [Google Scholar] [CrossRef]
- Ewert, K.K.; Evans, H.M.; Zidovska, A.; Bouxsein, N.F.; Ahmad, A.; Safinya, C.R. A columnar phase of dendritic lipid-based cationic liposome-DNA complexes for gene delivery: Hexagonally ordered cylindrical micelles embedded in a DNA honeycomb lattice. J. Am. Chem. Soc. 2006, 128, 3998–4006. [Google Scholar] [CrossRef]
- Leal, C.; Bouxsein, N.F.; Ewert, K.K.; Safinya, C.R. Highly Efficient Gene Silencing Activity of siRNA Embedded in a Nanostructured Gyroid Cubic Lipid Matrix. J. Am. Chem. Soc. 2010, 132, 16841–16847. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.J.; Slack, N.L.; Ahmad, A.; George, C.X.; Samuel, C.E.; Safinya, C.R. Three-dimensional imaging of lipid gene-carriers: Membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophys. J. 2003, 84, 3307–3316. [Google Scholar] [CrossRef]
- Ahmad, A.; Evans, H.M.; Ewert, K.; George, C.X.; Samuel, C.E.; Safinya, C.R. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: Lipid-DNA complexes for gene delivery. J. Gene Med. 2005, 7, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Bouxsein, N.F.; McAllister, C.S.; Ewert, K.K.; Samuel, C.E.; Safinya, C.R. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry 2007, 46, 4785–4792. [Google Scholar] [CrossRef] [PubMed]
- Ponti, F.; Campolungo, M.; Melchiori, C.; Bono, N.; Candiani, G. Cationic lipids for gene delivery: Many players, one goal. Chem. Phys. Lipids 2021, 235, 105032. [Google Scholar] [CrossRef]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Koynova, R.; Tenchov, B.; Wang, L.; MacDonald, R.C. Hydrophobic Moiety of Cationic Lipids Strongly Modulates Their Transfection Activity. Mol. Pharm. 2009, 6, 951–958. [Google Scholar] [CrossRef]
- Tranchant, I.; Thompson, B.; Nicolazzi, C.; Mignet, N.; Scherman, D. Physicochemical optimisation of plasmid delivery by cationic lipids. J. Gene Med. 2004, 6, S24–S35. [Google Scholar] [CrossRef] [PubMed]
- Labas, R.; Beilvert, F.; Barteau, B.; David, S.; Chèvre, R.; Pitard, B. Nature as a source of inspiration for cationic lipid synthesis. Genetica 2010, 138, 153–168. [Google Scholar] [CrossRef]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The Headgroup Evolution of Cationic Lipids for Gene Delivery. Bioconjug. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef]
- Wölk, C.; Janich, C.; Bakowsky, U.; Langner, A.; Brezesinski, G. Malonic acid based cationic lipids—The way to highly efficient DNA-carriers. Adv. Colloid Interface Sci. 2017, 248, 20–34. [Google Scholar] [CrossRef]
- Miller, A.D. Cationic Liposomes for Gene Therapy. Angew. Chem. Int. Ed. Engl. 1998, 37, 1768–1785. [Google Scholar] [CrossRef]
- Carrière, M.; Tranchant, I.; Niore, P.-A.; Byk, G.; Mignet, N.; Escriou, V.; Scherman, D.; Herscovici, J. Optimization of Cationic Lipid Mediated Gene Transfer: Structure-Function, Physico-Chemical, and Cellular Studies. J. Liposome Res. 2002, 12, 95–106. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Bajaj, A. Advances in gene delivery through molecular design of cationic lipids. Chem. Commun. 2009, 31, 4632–4656. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.; Slack, N.L.; Ahmad, A.; Evans, H.M.; Lin, A.J.; Samuel, C.E.; Safinya, C.R. Cationic lipid-DNA complexes for gene therapy: Understanding the relationship between complex structure and gene delivery pathways at the molecular level. Curr. Med. Chem. 2004, 11, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.K.; Evans, H.M.; Bouxsein, N.F.; Safinya, C.R. Dendritic cationic lipids with highly charged headgroups for efficient gene delivery. Bioconjug. Chem. 2006, 17, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Maier, M.A.; Jayaraman, M.; Matsuda, S.; Liu, J.; Barros, S.; Querbes, W.; Tam, Y.K.; Ansell, S.M.; Kumar, V.; Qin, J.; et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther. 2013, 21, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Chesnoy, S.; Huang, L. Structure and function of lipid-DNA complexes for gene delivery. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 27–47. [Google Scholar] [CrossRef]
- Ewert, K.; Evans, H.M.; Ahmad, A.; Slack, N.L.; Lin, A.J.; Martin-Herranz, A.; Safinya, C.R. Lipoplex Structures and Their Distinct Cellular Pathways. In Advances in Genetics, Vol. 53: Non-Viral Vectors for Gene Therapy, 2nd ed.; Huang, L., Hung, M.C., Wagner, E., Eds.; Elsevier; Academic Press: San Diego, CA, USA, 2005; pp. 119–155. [Google Scholar] [CrossRef]
- Ewert, K.; Ahmad, A.; Evans, H.; Safinya, C. Cationic lipid-DNA complexes for non-viral gene therapy: Relating supramolecular structures to cellular pathways. Expert Opin. Biol. Ther. 2005, 5, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Bielke, W.; Erbacher, C. (Eds.) Nucleic Acid Transfection; Springer: Berlin, Germany, 2010. [Google Scholar]
- Dan, N.; Danino, D. Structure and kinetics of lipid–nucleic acid complexes. Adv. Colloid Interface Sci. 2014, 205, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.K.; Zidovska, A.; Ahmad, A.; Bouxsein, N.F.; Evans, H.M.; McAllister, C.S.; Samuel, C.E.; Safinya, C.R. Cationic Liposome–Nucleic Acid Complexes for Gene Delivery and Silencing: Pathways and Mechanisms for Plasmid DNA and siRNA. Top. Curr. Chem. 2010, 296, 191–226. [Google Scholar] [CrossRef] [PubMed]
- Safinya, C.R.; Ewert, K.K.; Majzoub, R.N.; Leal, C. Cationic liposome-nucleic acid complexes for gene delivery and gene silencing. New J. Chem. 2014, 38, 5164–5172. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Huang, L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012, 45, 971–979. [Google Scholar] [CrossRef]
- Huang, L.; Hung, M.C.; Wagner, E. (Eds.) Non-Viral Vectors for Gene Therapy, 2nd ed.; Elsevier; Academic Press: San Diego, CA, USA, 2005. [Google Scholar]
- Huang, L.; Hung, M.-C.; Wagner, E. (Eds.) Nonviral Vectors for Gene Therapy; Academic Press: San Diego, CA, USA, 1999. [Google Scholar]
- Huang, L.; Liu, D.; Wagner, E. (Eds.) Nonviral Vectors for Gene Therapy: Lipid- and Polymer-Based Gene Transfer; Academic Press: New York, NY, USA, 2014. [Google Scholar]
- Lin, P.J.C.; Tam, Y.Y.C.; Hafez, I.; Sandhu, A.; Chen, S.; Ciufolini, M.A.; Nabi, I.R.; Cullis, P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine 2013, 9, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Foldvari, M.; Chen, D.W.; Nafissi, N.; Calderon, D.; Narsineni, L.; Rafiee, A. Non-viral gene therapy: Gains and challenges of non-invasive administration methods. J. Control. Release 2016, 240, 165–190. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021. [Google Scholar] [CrossRef]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef] [PubMed]
- The Journal of Gene Medicine: Gene Therapy Clinical Trials Worldwide. Available online: https://a873679.fmphost.com/fmi/webd/GTCT (accessed on 9 June 2021).
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Cogoni, C.; Macino, G. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 2000, 10, 638–643. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef]
- Caplen, N.J.; Parrish, S.; Imani, F.; Fire, A.; Morgan, R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 2001, 98, 9742–9747. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. Therapeutic siRNAs. Trends Pharmacol. Sci. 2004, 25, 22–28. [Google Scholar] [CrossRef]
- Karagiannis, T.C.; El-Osta, A. RNA interference and potential therapeutic applications of short interfering RNAs. Cancer Gene Ther. 2005, 12, 787–795. [Google Scholar] [CrossRef]
- Wan, C.; Allen, T.M.; Cullis, P.R. Lipid nanoparticle delivery systems for siRNA-based therapeutics. Drug Deliv. Transl. Res. 2014, 4, 74–83. [Google Scholar] [CrossRef]
- Spagnou, S.; Miller, A.D.; Keller, M. Lipidic Carriers of siRNA: Differences in the Formulation, Cellular Uptake, and Delivery with Plasmid DNA. Biochemistry 2004, 43, 13348–13356. [Google Scholar] [CrossRef]
- Ozcan, G.; Ozpolat, B.; Coleman, R.L.; Sood, A.K.; Lopez-Berestein, G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015, 87, 108–119. [Google Scholar] [CrossRef]
- Lares, M.R.; Rossi, J.J.; Ouellet, D.L. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010, 28, 570–579. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Pineda, M.; Moghadam, F.; Ebrahimkhani, M.R.; Kiani, S. Engineered CRISPR Systems for Next Generation Gene Therapies. ACS Synth. Biol. 2017, 6, 1614–1626. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J.; et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Ashwanikumar, N.; Robinson, E.; DuRoss, A.; Sun, C.; Murphy-Benenato, K.E.; Mihai, C.; Almarsson, Ö.; Sahay, G. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017, 17, 5711–5718. [Google Scholar] [CrossRef] [PubMed]
- Weissman, D.; Karikó, K. mRNA: Fulfilling the Promise of Gene Therapy. Mol. Ther. 2015, 23, 1416–1417. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Karikó, K.; Türeci, Ö. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine into mRNA Yields Superior Nonimmunogenic Vector with Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, T.S.; Lee, A.C.H.; Akinc, A.; Bramlage, B.; Bumcrot, D.; Fedoruk, M.N.; Harborth, J.; Heyes, J.A.; Jeffs, L.B.; John, M.; et al. RNAi-mediated gene silencing in non-human primates. Nature 2006, 441, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, R.; Coelho, F.; Silva, B.F.B. Lipid-Nucleic Acid Complexes: Physicochemical Aspects and Prospects for Cancer Treatment. Molecules 2020, 25, 5006. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv 2020. [Google Scholar] [CrossRef]
- United States Food and Drug Administration: Pfizer-BioNTech COVID-19 Vaccine Emergency Use Authorization Fact Sheet for Healthcare Providers Administering Vaccine. Available online: https://www.fda.gov/media/144413/download (accessed on 10 June 2021).
- Leventis, R.; Silvius, J.R. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochim. Biophys. Acta Biomembr. 1990, 1023, 124–132. [Google Scholar] [CrossRef]
- Ewert, K.; Ahmad, A.; Evans, H.M.; Schmidt, H.-W.; Safinya, C.R. Efficient synthesis and cell-transfection properties of a new multivalent cationic lipid for nonviral gene delivery. J. Med. Chem. 2002, 45, 5023–5029. [Google Scholar] [CrossRef]
- Heyes, J.; Palmer, L.; Bremner, K.; MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 2005, 107, 276–287. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Raper, S.E.; Chirmule, N.; Lee, F.S.; Wivel, N.A.; Bagg, A.; Gao, G.-p.; Wilson, J.M.; Batshaw, M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003, 80, 148–158. [Google Scholar] [CrossRef]
- Wilson, J.M. Lessons learned from the gene therapy trial for ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2009, 96, 151–157. [Google Scholar] [CrossRef]
- Kouprina, N.; Tomilin, A.N.; Masumoto, H.; Earnshaw, W.C.; Larionov, V. Human artificial chromosome-based gene delivery vectors for biomedicine and biotechnology. Expert Opin. Drug Deliv. 2014, 11, 517–535. [Google Scholar] [CrossRef]
- Larin, Z.; Mejía, J.E. Advances in human artificial chromosome technology. Trends Genet. 2002, 18, 313–319. [Google Scholar] [CrossRef]
- Harrington, J.J.; Van Bokkelen, G.; Mays, R.W.; Gustashaw, K.; Willard, H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997, 15, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-D.; Huang, L. Non-viral is superior to viral gene delivery. J. Control. Release 2007, 123, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Gindy, M.E.; Leone, A.M.; Cunningham, J.J. Challenges in the pharmaceutical development of lipid-based short interfering ribonucleic acid therapeutics. Expert Opin. Drug Deliv. 2012, 9, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Ewert, K.; Zidovska, A.; Ahmad, A.; Bouxsein, N.; Evans, H.; McAllister, C.; Samuel, C.; Safinya, C. Cationic Liposome–Nucleic Acid Complexes for Gene Delivery and Silencing: Pathways and Mechanisms for Plasmid DNA and siRNA. In Nucleic Acid Transfection; Bielke, W., Erbacher, C., Eds.; Springer: Berlin, Germany, 2010; pp. 191–226. [Google Scholar] [CrossRef]
- Mahato, R.I.; Kim, S.W. Pharmaceutical Perspectives of Nucleic Acid-Based Therapy; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 2 1976, 72, 1525–1568. [Google Scholar] [CrossRef]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of lipid bilayers and vesicles. Biochim. Biophys. Acta Biomembr. 1977, 470, 185–201. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J.; Tilcock, C.P.S. Lipid polymorphism and the roles of lipids in membranes. Chem. Phys. Lipids 1986, 40, 127–144. [Google Scholar] [CrossRef]
- Tilcock, C.P.S. Lipid polymorphism. Chem. Phys. Lipids 1986, 40, 109–125. [Google Scholar] [CrossRef]
- Helfrich, W.Z. Elastic Properties of Lipid Bilayers: Theory and Possible Experiments. Z. Naturforsch. C J. Biosci. 1973, 28, 693–703. [Google Scholar] [CrossRef]
- Lipowsky, R.; Sackmann, E. (Eds.) Structure and Dynamics of Membranes; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Safran, S.A. Statistical Thermodynamics of Surfaces, Interfaces, and Membranes; Addison-Wesley: Reading, UK, 1994. [Google Scholar]
- Landau, L.D.; Lifshitz, E.M. Theory of Elasticity, Volume 7 of Course of Theoretical Physics; Pergamon Press: Oxford, UK, 1970. [Google Scholar]
- Szleifer, I.; Kramer, D.; Ben-Shaul, A.; Roux, D.; Gelbart, W.M. Curvature Elasticity of Pure and Mixed Surfactant Films. Phys. Rev. Lett. 1988, 60, 1966–1969. [Google Scholar] [CrossRef]
- Seddon, J.M. Structure of the inverted hexagonal (HII) phase, and non-lamellar phase transitions of lipids. Biochim. Biophys. Acta 1990, 1031, 1–69. [Google Scholar] [CrossRef]
- Gruner, S.M. Stability of lyotropic phases with curved interfaces. J. Phys. Chem. 1989, 93, 7562–7570. [Google Scholar] [CrossRef]
- Zhen, Y.; Ewert, K.K.; Fisher, W.S.; Steffes, V.M.; Li, Y.; Safinya, C.R. Paclitaxel loading in cationic liposome vectors is enhanced by replacement of oleoyl with linoleoyl tails with distinct lipid shapes. Sci. Rep. 2021, 11, 7311. [Google Scholar] [CrossRef] [PubMed]
- Behr, J.; Demeneix, B.; Loeffler, J.; Perez-Mutul, J. Efficient gene transfer into mammalian primary endocrine cells with lipopolyamine-coated DNA. Proc. Natl. Acad. Sci. USA 1989, 86, 6982–6986. [Google Scholar] [CrossRef] [PubMed]
- Zidovska, A.; Ewert, K.K.; Quispe, J.; Carragher, B.; Potter, C.S.; Safinya, C.R. Block liposome and nanotube formation is a general phenomenon of two-component membranes containing multivalent lipids. Soft Matter 2011, 7, 8363–8369. [Google Scholar] [CrossRef] [PubMed]
- Steffes, V.M.; Zhang, Z.; MacDonald, S.; Crowe, J.; Ewert, K.K.; Carragher, B.; Potter, C.S.; Safinya, C.R. PEGylation of Paclitaxel-Loaded Cationic Liposomes Drives Steric Stabilization of Bicelles and Vesicles thereby Enhancing Delivery and Cytotoxicity to Human Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, M.; Edwards, K. Phase Behavior and Aggregate Structure in Mixtures of Dioleoylphosphatidylethanolamine and Poly(Ethylene Glycol)-Lipids. Biophys. J. 2001, 80, 313–323. [Google Scholar] [CrossRef]
- Sandström, M.C.; Johansson, E.; Edwards, K. Structure of Mixed Micelles Formed in PEG-Lipid/Lipid Dispersions. Langmuir 2007, 23, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Zidovska, A.; Evans, H.M.; Ewert, K.K.; Quispe, J.; Carragher, B.; Potter, C.S.; Safinya, C.R. Liquid crystalline phases of dendritic lipid-DNA self-assemblies: Lamellar, hexagonal, and DNA bundles. J. Phys. Chem. B 2009, 113, 3694–3703. [Google Scholar] [CrossRef]
- Siegel, D.P. The Modified Stalk Mechanism of Lamellar/Inverted Phase Transitions and Its Implications for Membrane Fusion. Biophys. J. 1999, 76, 291–313. [Google Scholar] [CrossRef]
- Porte, G. Lamellar phases and disordered phases of fluid bilayer membranes. J. Phys. Condens. Matter 1992, 4, 8649–8670. [Google Scholar] [CrossRef]
- Yang, L.; Huang, H.W. Observation of a Membrane Fusion Intermediate Structure. Science 2002, 297, 1877–1879. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.E.; Ces, O.; Squires, A.M.; Mulet, X.; Winter, R.; Finet, S.M.; Templer, R.H.; Seddon, J.M. A pressure-jump time-resolved x-ray diffraction study of cubic-cubic transition kinetics in monoolein. Langmuir 2008, 24, 2331–2340. [Google Scholar] [CrossRef]
- Lindblom, G.; Larsson, K.; Johansson, L.; Fontell, K.; Forsen, S. The cubic phase of monoglyceride-water systems. Arguments for a structure based upon lamellar bilayer units. J. Am. Chem. Soc. 1979, 101, 5465–5470. [Google Scholar] [CrossRef]
- Leal, C.; Ewert, K.K.; Shirazi, R.S.; Bouxsein, N.F.; Safinya, C.R. Nanogyroids Incorporating Multivalent Lipids: Enhanced Membrane Charge Density and Pore Forming Ability for Gene Silencing. Langmuir 2011, 27, 7691–7697. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.S. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions I. Colligative Properties. J. Chem. Phys. 1969, 51, 924–933. [Google Scholar] [CrossRef]
- Piotrowski-Daspit, A.S.; Kauffman, A.C.; Bracaglia, L.G.; Saltzman, W.M. Polymeric vehicles for nucleic acid delivery. Adv. Drug Deliv. Rev. 2020, 156, 119–132. [Google Scholar] [CrossRef]
- Lasic, D.D.; Strey, H.; Stuart, M.C.A.; Podgornik, R.; Frederik, P.M. The Structure of DNA−Liposome Complexes. J. Am. Chem. Soc. 1997, 119, 832–833. [Google Scholar] [CrossRef]
- Huebner, S.; Battersby, B.J.; Grimm, R.; Cevc, G. Lipid-DNA Complex Formation: Reorganization and Rupture of Lipid Vesicles in the Presence of DNA as Observed by Cryoelectron Microscopy. Biophys. J. 1999, 76, 3158–3166. [Google Scholar] [CrossRef]
- Majzoub, R.N.; Ewert, K.K.; Jacovetty, E.L.; Carragher, B.; Potter, C.S.; Li, Y.; Safinya, C.R. Patterned Threadlike Micelles and DNA-Tethered Nanoparticles: A Structural Study of PEGylated Cationic Liposome–DNA Assemblies. Langmuir 2015, 31, 7073–7083. [Google Scholar] [CrossRef] [PubMed]
- Safinya, C.R.; Roux, D.; Smith, G.S.; Sinha, S.K.; Dimon, P.; Clark, N.A.; Bellocq, A.M. Steric Interactions in a Model Multimembrane System: A Synchrotron X-ray Study. Phys. Rev. Lett. 1986, 57, 2718–2721. [Google Scholar] [CrossRef]
- Roux, D.; Safinya, C.R. A Synchrotron X-ray study of Competing Undulation and Electrostatic Interlayer Interactions in Fluid Multimembrane Lyotropic Phases. J. Phys. 1988, 49, 307–318. [Google Scholar] [CrossRef]
- Safinya, C.R.; Sirota, E.B.; Roux, D.; Smith, G.S. Universality in interacting membranes: The effect of cosurfactants on the interfacial rigidity. Phys. Rev. Lett. 1989, 62, 1134–1137. [Google Scholar] [CrossRef]
- Lei, N.; Safinya, C.R.; Bruinsma, R.F. Discrete Harmonic Model for Stacked Membranes: Theory and Experiment. J. Phys. II 1995, 5, 1155–1163. [Google Scholar] [CrossRef]
- Koltover, I.; Salditt, T.; Safinya, C.R. Phase diagram, stability, and overcharging of lamellar cationic lipid-DNA self-assembled complexes. Biophys. J. 1999, 77, 915–924. [Google Scholar] [CrossRef]
- Safinya, C.R. Structures of lipid-DNA complexes: Supramolecular assembly and gene delivery. Curr. Opin. Struct. Biol. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Salditt, T.; Koltover, I.; Rädler, J.O.; Safinya, C.R. Two-dimensional smectic ordering of linear DNA chains in self-assembled DNA-cationic liposome mixtures. Phys. Rev. Lett. 1997, 79, 2582–2585. [Google Scholar] [CrossRef]
- Salditt, T.; Koltover, I.; Rädler, J.O.; Safinya, C.R. Self-assembled DNA--cationic-lipid complexes: Two-dimensional smectic ordering, correlations, and interactions. Phys. Rev. E 1998, 58, 889–904. [Google Scholar] [CrossRef]
- Zidovska, A.; Evans, H.M.; Ahmad, A.; Ewert, K.K.; Safinya, C.R. The Role of Cholesterol and Structurally Related Molecules in Enhancing Transfection of Cationic Liposome–DNA Complexes. J. Phys. Chem. B 2009, 113, 5208–5216. [Google Scholar] [CrossRef][Green Version]
- Bouxsein, N.F.; Leal, C.l.; McAllister, C.S.; Ewert, K.K.; Li, Y.; Samuel, C.E.; Safinya, C.R. Two-Dimensional Packing of Short DNA with Nonpairing Overhangs in Cationic Liposome–DNA Complexes: From Onsager Nematics to Columnar Nematics with Finite-Length Columns. J. Am. Chem. Soc. 2011, 133, 7585–7595. [Google Scholar] [CrossRef]
- Koynova, R.; Tenchov, B. Cationic Lipids: Molecular Structure/Transfection Activity Relationships and Interactions with Biomembranes. Top. Curr. Chem. 2010, 296, 51–93. [Google Scholar] [CrossRef] [PubMed]
- Tenchov, B.G.; Wang, L.; Koynova, R.; MacDonald, R.C. Modulation of a membrane lipid lamellar-nonlamellar phase transition by cationic lipids: A measure for transfection efficiency. Biochim. Biophys. Acta Biomembr. 2008, 1778, 2405–2412. [Google Scholar] [CrossRef]
- Koynova, R.; Tenchov, B. Cationic phospholipids: Structure–transfection activity relationships. Soft Matter 2009, 5, 3187–3200. [Google Scholar] [CrossRef]
- Ziller, A.; Nogueira, S.S.; Hühn, E.; Funari, S.S.; Brezesinski, G.; Hartmann, H.; Sahin, U.; Haas, H.; Langguth, P. Incorporation of mRNA in Lamellar Lipid Matrices for Parenteral Administration. Mol. Pharm. 2018, 15, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Michanek, A.; Kristen, N.; Höök, F.; Nylander, T.; Sparr, E. RNA and DNA interactions with zwitterionic and charged lipid membranes—A DSC and QCM-D study. Biochim. Biophys. Acta Biomembr. 2010, 1798, 829–838. [Google Scholar] [CrossRef]
- Artzner, F.; Zantl, R.; Rapp, G.; Rädler, J.O. Observation of a Rectangular Columnar Phase in Condensed Lamellar Cationic Lipid-DNA Complexes. Phys. Rev. Lett. 1998, 81, 5015–5018. [Google Scholar] [CrossRef]
- Koynova, R.; MacDonald, R.C. Columnar DNA Superlattices in Lamellar o-Ethylphosphatidylcholine Lipoplexes: Mechanism of the Gel-Liquid Crystalline Lipid Phase Transition. Nano Lett. 2004, 4, 1475–1479. [Google Scholar] [CrossRef]
- McManus, J.J.; Rädler, J.O.; Dawson, K.A. Observation of a Rectangular Columnar Phase in a DNA−Calcium−Zwitterionic Lipid Complex. J. Am. Chem. Soc. 2004, 126, 15966–15967. [Google Scholar] [CrossRef]
- Bouxsein, N.F.; Leal, C.; McAllister, C.S.; Li, Y.; Ewert, K.K.; Samuel, C.E.; Safinya, C.R. 3D Columnar Phase of Stacked Short DNA Organized by Coherent Membrane Undulations. Langmuir 2019, 35, 11891–11901. [Google Scholar] [CrossRef] [PubMed]
- Golubović, L.; Golubović, M. Fluctuations of Quasi-Two-Dimensional Smectics Intercalated between Membranes in Multilamellar Phases of DNA-Cationic Lipid Complexes. Phys. Rev. Lett. 1998, 80, 4341–4344. [Google Scholar] [CrossRef]
- Golubović, L.; Lubensky, T.C.; O’Hern, C.S. Structural properties of the sliding columnar phase in layered liquid crystalline systems. Phys. Rev. E 2000, 62, 1069–1094. [Google Scholar] [CrossRef]
- O’Hern, C.S.; Lubensky, T.C. Sliding Columnar Phase of DNA-Lipid Complexes. Phys. Rev. Lett. 1998, 80, 4345–4348. [Google Scholar] [CrossRef]
- Kulkarni, C.V.; Wachter, W.; Iglesias-Salto, G.; Engelskirchen, S.; Ahualli, S. Monoolein: A magic lipid? Phys. Chem. Chem. Phys. 2011, 13, 3004–3021. [Google Scholar] [CrossRef]
- Kang, M.; Kim, H.; Leal, C. Self-organization of nucleic acids in lipid constructs. Curr. Opin. Colloid Interface Sci. 2016, 26, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Negro, M.; Kumar, K.; Barrán-Berdón, A.L.; Datta, S.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Efficient Cellular Knockdown Mediated by siRNA Nanovectors of Gemini Cationic Lipids Having Delocalizable Headgroups and Oligo-Oxyethylene Spacers. ACS Appl. Mater. Interfaces 2016, 8, 22113–22126. [Google Scholar] [CrossRef]
- Kim, H.; Leal, C. Cuboplexes: Topologically Active siRNA Delivery. ACS Nano 2015, 9, 10214–10226. [Google Scholar] [CrossRef]
- Kim, H.; Sung, J.; Chang, Y.; Alfeche, A.; Leal, C. Microfluidics Synthesis of Gene Silencing Cubosomes. ACS Nano 2018, 12, 9196–9205. [Google Scholar] [CrossRef] [PubMed]
- Leal, C.; Ewert, K.K.; Bouxsein, N.F.; Shirazi, R.S.; Li, Y.; Safinya, C.R. Stacking of short DNA induces the gyroid cubic-to-inverted hexagonal phase transition in lipid-DNA complexes. Soft Matter 2013, 9, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Farhood, H.; Serbina, N.; Huang, L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim. Biophys. Acta Biomembr. 1995, 1235, 289–295. [Google Scholar] [CrossRef]
- Degors, I.M.S.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Martens, T.F.; Remaut, K.; Demeester, J.; De Smedt, S.C.; Braeckmans, K. Intracellular delivery of nanomaterials: How to catch endosomal escape in the act. Nano Today 2014, 9, 344–364. [Google Scholar] [CrossRef]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef]
- Poste, G.; Bucana, C.; Raz, A.; Bugelski, P.; Kirsh, R.; Fidler, I.J. Analysis of the Fate of Systemically Administered Liposomes and Implications for Their Use in Drug Delivery. Cancer Res. 1982, 42, 1412–1422. [Google Scholar]
- Lasic, D.D. Liposomes: From Physics to Applications; Elsevier: San Diego, CA, USA, 1993. [Google Scholar]
- Yan, X.; Scherphof, G.L.; Kamps, J.A.A.M. Liposome Opsonization. J. Liposome Res. 2005, 15, 109–139. [Google Scholar] [CrossRef]
- Lasic, D.D.; Needham, D. The “Stealth” Liposome: A Prototypical Biomaterial. Chem. Rev. 1995, 95, 2601–2628. [Google Scholar] [CrossRef]
- Lasic, D.D.; Martin, F. (Eds.) Stealth Liposomes; CRC Press: Boca Raton, FL, USA, 1995. [Google Scholar]
- Woodle, M.C.; Newman, M.; Collins, L.; Redemann, C.; Martin, F. Improved long circulating (Stealth®) liposomes using synthetic lipids. Proc. Intern. Symp. Control. Rel. Bioact. Mater. 1990, 17, 77–78. [Google Scholar]
- Klibanov, A.L.; Maruyama, K.; Torchilin, V.P.; Huang, L. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990, 268, 235–237. [Google Scholar] [CrossRef]
- Blume, G.; Cevc, G. Liposomes for the sustained drug release in vivo. Biochim. Biophys. Acta Biomembr. 1990, 1029, 91–97. [Google Scholar] [CrossRef]
- Papahadjopoulos, D.; Allen, T.M.; Gabizon, A.; Mayhew, E.; Matthay, K.; Huang, S.K.; Lee, K.D.; Woodle, M.C.; Lasic, D.D.; Redemann, C. Sterically stabilized liposomes: Improvements in pharmacokinetics and antitumor therapeutic efficacy. Proc. Natl. Acad. Sci. USA 1991, 88, 11460–11464. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; Hansen, C.; Martin, F.; Redemann, C.; Yau-Young, A. Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo. Biochim. Biophys. Acta Biomembr. 1991, 1066, 29–36. [Google Scholar] [CrossRef]
- Woodle, M.C.; Lasic, D.D. Sterically stabilized liposomes. Biochim. Biophys. Acta 1992, 1113, 171–199. [Google Scholar] [CrossRef]
- Silvander, M. Steric stabilization of liposomes—A review. Prog. Colloid Polym. Sci. 2002, 120, 35–40. [Google Scholar] [CrossRef]
- Gabizon, A.; Papahadjopoulos, D. Liposome formulations with prolonged circulation time in blood and enhanced uptake by tumors. Proc. Natl. Acad. Sci. USA 1988, 85, 6949–6953. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; Hansen, C.; Rutledge, J. Liposomes with prolonged circulation times: Factors affecting uptake by reticuloendothelial and other tissues. Biochim. Biophys. Acta Biomembr. 1989, 981, 27–35. [Google Scholar] [CrossRef]
- De Gennes, P.-G. Scaling Concepts in Polymer Physics; Cornell University Press: New York, NY, USA, 1979. [Google Scholar]
- Kuhl, T.L.; Leckband, D.E.; Lasic, D.D.; Israelachvili, J.N. Modulation of interaction forces between bilayers exposing short-chained ethylene oxide headgroups. Biophys. J. 1994, 66, 1479–1488. [Google Scholar] [CrossRef]
- Kenworthy, A.K.; Hristova, K.; Needham, D.; McIntosh, T.J. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol). Biophys. J. 1995, 68, 1921–1936. [Google Scholar] [CrossRef]
- Hong, K.; Zheng, W.; Baker, A.; Papahadjopoulos, D. Stabilization of cationic liposome-plasmid DNA complexes by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Lett. 1997, 400, 233–237. [Google Scholar] [CrossRef]
- Barenholz, Y. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Silva, B.F.B.; Majzoub, R.N.; Chan, C.-L.; Li, Y.; Olsson, U.; Safinya, C.R. PEGylated cationic liposome–DNA complexation in brine is pathway-dependent. Biochim. Biophys. Acta Biomembr. 2014, 1838, 398–412. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.-L.; Majzoub, R.N.; Shirazi, R.S.; Ewert, K.K.; Chen, Y.-J.; Liang, K.S.; Safinya, C.R. Endosomal escape and transfection efficiency of PEGylated cationic liposome–DNA complexes prepared with an acid-labile PEG-lipid. Biomaterials 2012, 33, 4928–4935. [Google Scholar] [CrossRef] [PubMed]
- Majzoub, R.N.; Chan, C.-L.; Ewert, K.K.; Silva, B.F.B.; Liang, K.S.; Jacovetty, E.L.; Carragher, B.; Potter, C.S.; Safinya, C.R. Uptake and transfection efficiency of PEGylated cationic liposome–DNA complexes with and without RGD-tagging. Biomaterials 2014, 35, 4996–5005. [Google Scholar] [CrossRef]
- Belliveau, N.M.; Huft, J.; Lin, P.J.C.; Chen, S.; Leung, A.K.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther. Nucleic Acids 2012, 1, e37. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomedicine 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; van der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and siRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef] [PubMed]
- Martin-Herranz, A.; Ahmad, A.; Evans, H.M.; Ewert, K.; Schulze, U.; Safinya, C.R. Surface Functionalized Cationic Lipid-DNA Complexes for Gene Delivery: PEGylated Lamellar Complexes Exhibit Distinct DNA-DNA Interaction Regimes. Biophys. J. 2004, 86, 1160–1168. [Google Scholar] [CrossRef][Green Version]
- Hafez, I.M.; Maurer, N.; Cullis, P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001, 8, 1188–1196. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing siRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef]
- Needham, D.; McIntosh, T.J.; Lasic, D.D. Repulsive interactions and mechanical stability of polymer-grafted lipid membranes. Biochim. Biophys. Acta Biomembr. 1992, 1108, 40–48. [Google Scholar] [CrossRef]
- Kong, L.; Campbell, F.; Kros, A. DePEGylation strategies to increase cancer nanomedicine efficacy. Nanoscale Horiz. 2019, 4, 378–387. [Google Scholar] [CrossRef]
- Romberg, B.; Hennink, W.E.; Storm, G. Sheddable Coatings for Long-Circulating Nanoparticles. Pharm. Res. 2008, 25, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Mui, B.L.; Tam, Y.K.; Jayaraman, M.; Ansell, S.M.; Du, X.; Tam, Y.Y.C.; Lin, P.J.C.; Chen, S.; Narayanannair, J.K.; Rajeev, K.G.; et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol. Ther. Nucleic Acids 2013, 2, e139. [Google Scholar] [CrossRef]
- Walker, G.F.; Fella, C.; Pelisek, J.; Fahrmeir, J.; Boeckle, S.; Ogris, M.; Wagner, E. Toward synthetic viruses: Endosomal pH-triggered deshielding of targeted polyplexes greatly enhances gene transfer in vitro and in vivo. Mol. Ther. 2005, 11, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Pierschbacher, M.D.; Hayman, E.G.; Ruoslahti, E. Location of the cell-attachment site in fibronectin with monoclonal antibodies and proteolytic fragments of the molecule. Cell 1981, 26, 259–267. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Pierschbacher, M.D.; Ruoslahti, E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. USA 1984, 81, 5985–5988. [Google Scholar] [CrossRef]
- Pytela, R.; Pierschbacher, M.D.; Ruoslahti, E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell 1985, 40, 191–198. [Google Scholar] [CrossRef]
- Ewert, K.K.; Kotamraju, V.R.; Majzoub, R.N.; Steffes, V.M.; Wonder, E.A.; Teesalu, T.; Ruoslahti, E.; Safinya, C.R. Synthesis of linear and cyclic peptide–PEG–lipids for stabilization and targeting of cationic liposome–DNA complexes. Bioorg. Med. Chem. Lett. 2016, 26, 1618–1623. [Google Scholar] [CrossRef]
- Ruoslahti, E.; Pierschbacher, M.D. Arg-Gly-Asp: A versatile cell recognition signal. Cell 1986, 44, 517–518. [Google Scholar] [CrossRef]
- Ruoslahti, E.; Bhatia, S.N.; Sailor, M.J. Targeting of drugs and nanoparticles to tumors. J. Cell Biol. 2010, 188, 759–768. [Google Scholar] [CrossRef]
- Clackson, T.; Lowmand, H.B. (Eds.) Phage Display: A Practical Approach; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Teesalu, T.; Sugahara, K.N.; Ruoslahti, E. Mapping of Vascular ZIP Codes by Phage Display. Meth. Enzymol. 2012, 503, 35–56. [Google Scholar] [CrossRef]
- Ruoslahti, E. Specialization of Tumour Vasculature. Nat. Rev. Cancer 2002, 2, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. Vascular zip codes in angiogenesis and metastasis. Biochem. Soc. Trans. 2004, 32, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Laakkonen, P.; Åkerman, M.E.; Biliran, H.; Yang, M.; Ferrer, F.; Karpanen, T.; Hoffman, R.M.; Ruoslahti, E. Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9381–9386. [Google Scholar] [CrossRef]
- Ruoslahti, E.; Duza, T.; Zhang, L. Vascular Homing Peptides with Cell-Penetrating Properties. Curr. Pharm. Des. 2005, 11, 3655–3660. [Google Scholar] [CrossRef]
- Arap, W.; Pasqualini, R.; Ruoslahti, E. Cancer Treatment by Targeted Drug Delivery to Tumor Vasculature in a Mouse Model. Science 1998, 279, 377–380. [Google Scholar] [CrossRef]
- Laakkonen, P.; Porkka, K.; Hoffman, J.A.; Ruoslahti, E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat. Med. 2002, 8, 751–755. [Google Scholar] [CrossRef]
- Park, J.-H.; von Maltzahn, G.; Xu, M.J.; Fogal, V.; Kotamraju, V.R.; Ruoslahti, E.; Bhatia, S.N.; Sailor, M.J. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Giraudo, E.; Hoffman, J.A.; Hanahan, D.; Ruoslahti, E. Lymphatic Zip Codes in Premalignant Lesions and Tumors. Cancer Res. 2006, 66, 5696–5706. [Google Scholar] [CrossRef]
- Lee, S.-M.; Lee, E.-J.; Hong, H.-Y.; Kwon, M.-K.; Kwon, T.-H.; Choi, J.-Y.; Park, R.-W.; Kwon, T.-G.; Yoo, E.-S.; Yoon, G.-S.; et al. Targeting Bladder Tumor Cells In vivo and in the Urine with a Peptide Identified by Phage Display. Mol. Cancer Res. 2007, 5, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Simberg, D.; Duza, T.; Park, J.H.; Essler, M.; Pilch, J.; Zhang, L.; Derfus, A.M.; Yang, M.; Hoffman, R.M.; Bhatia, S.; et al. Biomimetic amplification of nanoparticle homing to tumors. Proc. Natl. Acad. Sci. USA 2007, 104, 932–936. [Google Scholar] [CrossRef]
- Pleiko, K.; Põšnograjeva, K.; Haugas, M.; Paiste, P.; Tobi, A.; Kurm, K.; Riekstina, U.; Teesalu, T. In vivo phage display: Identification of organ-specific peptides using deep sequencing and differential profiling across tissues. Nucleic Acids Res. 2021, 49, e38. [Google Scholar] [CrossRef] [PubMed]
- Põsnograjeva, K.; Pleiko, K.; Haugas, M.; Teesalu, T. New tools for streamlined in vivo homing peptide identification. In Cell-Penetrating Peptides: Methods and Protocols, 3rd ed.; Langel, Ü., Ed.; Springer: New York, NY, USA, 2021. [Google Scholar]
- Scodeller, P.; Asciutto, E.K. Targeting Tumors Using Peptides. Molecules 2020, 25, 808. [Google Scholar] [CrossRef] [PubMed]
- Kessler, H.; Diefenbach, B.; Finsinger, D.; Geyer, A.; Gurrath, M.; Goodman, S.L.; Hölzemann, G.; Haubner, R.; Jonczyk, A.; Müller, G.; et al. Design of superactive and selective integrin receptor antagonists containing the RGD sequence. Lett. Pept. Sci. 1995, 2, 155–160. [Google Scholar] [CrossRef]
- Haubner, R.; Gratias, R.; Diefenbach, B.; Goodman, S.L.; Jonczyk, A.; Kessler, H. Structural and Functional Aspects of RGD-Containing Cyclic Pentapeptides as Highly Potent and Selective Integrin αVβ3 Antagonists. J. Am. Chem. Soc. 1996, 118, 7461–7472. [Google Scholar] [CrossRef]
- Mas-Moruno, C.; Rechenmacher, F.; Kessler, H. Cilengitide: The First Anti-Angiogenic Small Molecule Drug Candidate. Design, Synthesis and Clinical Evaluation. Anti-Cancer Agents Med. Chem. 2010, 10, 753–768. [Google Scholar] [CrossRef]
- Dechantsreiter, M.A.; Planker, E.; Mathä, B.; Lohof, E.; Hölzemann, G.; Jonczyk, A.; Goodman, S.L.; Kessler, H. N-Methylated Cyclic RGD Peptides as Highly Active and Selective αVβ3 Integrin Antagonists. J. Med. Chem. 1999, 42, 3033–3040. [Google Scholar] [CrossRef]
- Temming, K.; Schiffelers, R.M.; Molema, G.; Kok, R.J. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist. Updates 2005, 8, 381–402. [Google Scholar] [CrossRef]
- Zanuy, D.; Sayago, F.J.; Revilla-López, G.; Ballano, G.; Agemy, L.; Kotamraju, V.R.; Jiménez, A.I.; Cativiela, C.; Nussinov, R.; Sawvel, A.M.; et al. Engineering strategy to improve peptide analogs: From structure-based computational design to tumor homing. J. Comput. Aided Mol. Des. 2013, 27, 31–43. [Google Scholar] [CrossRef]
- Ruoslahti, E. Tumor penetrating peptides for improved drug delivery. Adv. Drug Deliv. Rev. 2017, 110, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-Penetrating Delivery of Compounds and Nanoparticles into Tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a Tumor-Penetrating Peptide Enhances the Efficacy of Cancer Drugs. Science 2010, 328, 1031–1035. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.N.; Ruoslahti, E. Tumor penetrating peptides. Front. Oncol. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Simón-Gracia, L.; Hunt, H.; Scodeller, P.; Gaitzsch, J.; Kotamraju, V.R.; Sugahara, K.N.; Tammik, O.; Ruoslahti, E.; Battaglia, G.; Teesalu, T. iRGD peptide conjugation potentiates intraperitoneal tumor delivery of paclitaxel with polymersomes. Biomaterials 2016, 104, 247–257. [Google Scholar] [CrossRef]
- Sharma, S.; Kotamraju, V.R.; Mölder, T.; Tobi, A.; Teesalu, T.; Ruoslahti, E. Tumor-Penetrating Nanosystem Strongly Suppresses Breast Tumor Growth. Nano Lett. 2017, 17, 1356–1364. [Google Scholar] [CrossRef]
- Hunt, H.; Simón-Gracia, L.; Tobi, A.; Kotamraju, V.R.; Sharma, S.; Nigul, M.; Sugahara, K.N.; Ruoslahti, E.; Teesalu, T. Targeting of p32 in peritoneal carcinomatosis with intraperitoneal linTT1 peptide-guided pro-apoptotic nanoparticles. J. Control. Release 2017, 260, 142–153. [Google Scholar] [CrossRef]
- Paasonen, L.; Sharma, S.; Braun, G.B.; Kotamraju, V.R.; Chung, T.D.Y.; She, Z.-G.; Sugahara, K.N.; Yliperttula, M.; Wu, B.; Pellecchia, M.; et al. New p32/gC1qR Ligands for Targeted Tumor Drug Delivery. ChemBioChem 2016, 17, 570–575. [Google Scholar] [CrossRef]
- Wannasarit, S.; Wang, S.; Figueiredo, P.; Trujillo, C.; Eburnea, F.; Simón-Gracia, L.; Correia, A.; Ding, Y.; Teesalu, T.; Liu, D.; et al. A Virus-Mimicking pH-Responsive Acetalated Dextran-Based Membrane-Active Polymeric Nanoparticle for Intracellular Delivery of Antitumor Therapeutics. Adv. Funct. Mater. 2019, 29, 1905352. [Google Scholar] [CrossRef]
- Säälik, P.; Lingasamy, P.; Toome, K.; Mastandrea, I.; Rousso-Noori, L.; Tobi, A.; Simón-Gracia, L.; Hunt, H.; Paiste, P.; Kotamraju, V.R.; et al. Peptide-guided nanoparticles for glioblastoma targeting. J. Control. Release 2019, 308, 109–118. [Google Scholar] [CrossRef]
- Simon-Gracia, L.; Savier, E.; Parizot, C.; Brossas, J.Y.; Loisel, S.; Teesalu, T.; Conti, F.; Charlotte, F.; Scatton, O.; Aoudjehane, L.; et al. Bifunctional Therapeutic Peptides for Targeting Malignant B Cells and Hepatocytes: Proof of Concept in Chronic Lymphocytic Leukemia. Adv. Ther. 2020, 3, 2000131. [Google Scholar] [CrossRef]
- Simón-Gracia, L.; Sidorenko, V.; Uustare, A.; Ogibalov, I.; Tasa, A.; Tshubrik, O.; Teesalu, T. Novel Anthracycline Utorubicin for Cancer Therapy. Angew. Chem. Int. Ed. Engl. 2021. [Google Scholar] [CrossRef]
- Simón-Gracia, L.; Scodeller, P.; Fuentes, S.S.; Gómez, V.; Vallejo, X.R.; San Sebastián, E.; Sidorenko, V.; Di Silvio, D.; Suck, M.; De Lorenzi, F.; et al. Application of polymersomes engineered to target p32 protein for detection of small breast tumors in mice. Oncotarget 2018, 9, 18682–18697. [Google Scholar] [CrossRef] [PubMed]
- Scodeller, P.; Simón-Gracia, L.; Kopanchuk, S.; Tobi, A.; Kilk, K.; Säälik, P.; Kurm, K.; Squadrito, M.L.; Kotamraju, V.R.; Rinken, A.; et al. Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide. Sci. Rep. 2017, 7, 14655. [Google Scholar] [CrossRef] [PubMed]
- Lepland, A.; Asciutto, E.K.; Malfanti, A.; Simón-Gracia, L.; Sidorenko, V.; Vicent, M.J.; Teesalu, T.; Scodeller, P. Targeting Pro-Tumoral Macrophages in Early Primary and Metastatic Breast Tumors with the CD206-Binding mUNO Peptide. Mol. Pharm. 2020, 17, 2518–2531. [Google Scholar] [CrossRef]
- Figueiredo, P.; Lepland, A.; Scodeller, P.; Fontana, F.; Torrieri, G.; Tiboni, M.; Shahbazi, M.A.; Casettari, L.; Kostiainen, M.A.; Hirvonen, J.; et al. Peptide-guided resiquimod-loaded lignin nanoparticles convert tumor-associated macrophages from M2 to M1 phenotype for enhanced chemotherapy. Acta Biomater. 2020. [Google Scholar] [CrossRef] [PubMed]
- Keler, T.; Ramakrishna, V.; Fanger, M.W. Mannose receptor-targeted vaccines. Expert Opin. Biol. Ther. 2004, 4, 1953–1962. [Google Scholar] [CrossRef]
- Conniot, J.; Scomparin, A.; Peres, C.; Yeini, E.; Pozzi, S.; Matos, A.I.; Kleiner, R.; Moura, L.I.F.; Zupančič, E.; Viana, A.S.; et al. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat. Nanotechnol. 2019, 14, 891–901. [Google Scholar] [CrossRef]
- Silva, J.M.; Zupancic, E.; Vandermeulen, G.; Oliveira, V.G.; Salgado, A.; Videira, M.; Gaspar, M.; Graca, L.; Préat, V.; Florindo, H.F. In vivo delivery of peptides and Toll-like receptor ligands by mannose-functionalized polymeric nanoparticles induces prophylactic and therapeutic anti-tumor immune responses in a melanoma model. J. Control. Release 2015, 198, 91–103. [Google Scholar] [CrossRef]
- Lingasamy, P.; Tobi, A.; Haugas, M.; Hunt, H.; Paiste, P.; Asser, T.; Rätsep, T.; Kotamraju, V.R.; Bjerkvig, R.; Teesalu, T. Bi-specific tenascin-C and fibronectin targeted peptide for solid tumor delivery. Biomaterials 2019, 219, 119373. [Google Scholar] [CrossRef]
- Lingasamy, P.; Tobi, A.; Kurm, K.; Kopanchuk, S.; Sudakov, A.; Salumäe, M.; Rätsep, T.; Asser, T.; Bjerkvig, R.; Teesalu, T. Tumor-penetrating peptide for systemic targeting of Tenascin-C. Sci. Rep. 2020, 10, 5809. [Google Scholar] [CrossRef]
- Wonder, E.; Simón-Gracia, L.; Scodeller, P.; Majzoub, R.N.; Kotamraju, V.R.; Ewert, K.K.; Teesalu, T.; Safinya, C.R. Competition of charge-mediated and specific binding by peptide-tagged cationic liposome–DNA nanoparticles in vitro and in vivo. Biomaterials 2018, 166, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Campbell, R.B.; Ying, B.; Kuesters, G.M.; Hemphill, R. Fighting cancer: From the bench to bedside using second generation cationic liposomal therapeutics. J. Pharm. Sci. 2009, 98, 411–429. [Google Scholar] [CrossRef]
- Strieth, S.; Eichhorn, M.E.; Sauer, B.; Schulze, B.; Teifel, M.; Michaelis, U.; Dellian, M. Neovascular targeting chemotherapy: Encapsulation of paclitaxel in cationic liposomes impairs functional tumor microvasculature. Int. J. Cancer 2004, 110, 117–124. [Google Scholar] [CrossRef]
- Strieth, S.; Nussbaum, C.F.; Eichhorn, M.E.; Fuhrmann, M.; Teifel, M.; Michaelis, U.; Berghaus, A.; Dellian, M. Tumor-selective vessel occlusions by platelets after vascular targeting chemotherapy using paclitaxel encapsulated in cationic liposomes. Int. J. Cancer 2008, 122, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Sody, M.; Strieth, S.; Krasnici, S.; Sauer, B.; Schulze, B.; Teifel, M.; Michaelis, U.; Naujoks, K.; Dellian, M. Neovascular Targeting Therapy: Paclitaxel Encapsulated in Cationic Liposomes Improves Antitumoral Efficacy. Clin. Cancer Res. 2003, 9, 2335–2341. [Google Scholar] [PubMed]
- Kunstfeld, R.; Wickenhauser, G.; Michaelis, U.; Teifel, M.; Umek, W.; Naujoks, K.; Wolff, K.; Petzelbauer, P. Paclitaxel Encapsulated in Cationic Liposomes Diminishes Tumor Angiogenesis and Melanoma Growth in a “Humanized” SCID Mouse Model. J. Investig. Dermatol. 2003, 120, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Fasol, U.; Frost, A.; Büchert, M.; Arends, J.; Fiedler, U.; Scharr, D.; Scheuenpflug, J.; Mross, K. Vascular and pharmacokinetic effects of EndoTAG-1 in patients with advanced cancer and liver metastasis. Ann. Oncol. 2012, 23, 1030–1036. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, Š.; Turánek, J. Liposomal paclitaxel formulations. J. Control. Release 2012, 163, 322–334. [Google Scholar] [CrossRef]
- Wani, M.C.; Taylor, H.L.; Wall, M.E.; Coggon, P.; McPhail, A.T. Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc. 1971, 93, 2325–2327. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Weaver, B.A. How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 2014, 25, 2677–2681. [Google Scholar] [CrossRef]
- Rowinsky, E.K.; Donehower, R.C. Paclitaxel (Taxol). N. Engl. J. Med. 1995, 332, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Markman, M.; Mekhail, T.M. Paclitaxel in cancer therapy. Expert Opin. Pharmacother. 2002, 3, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Belani, C.P. Paclitaxel for non-small cell lung cancer. Expert Opin. Pharmacother. 2004, 5, 1771–1780. [Google Scholar] [CrossRef]
- Hironaka, S.; Zenda, S.; Boku, N.; Fukutomi, A.; Yoshino, T.; Onozawa, Y. Weekly paclitaxel as second-line chemotherapy for advanced or recurrent gastric cancer. Gastric Cancer 2006, 9, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.; Matsui, T.; Kodera, Y. Paclitaxel chemotherapy for the treatment of gastric cancer. Gastric Cancer 2009, 12, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Moxley, K.M.; McMeekin, D.S. Endometrial Carcinoma: A Review of Chemotherapy, Drug Resistance, and the Search for New Agents. Oncologist 2010, 15, 1026–1033. [Google Scholar] [CrossRef]
- Teo, P.Y.; Cheng, W.; Hedrick, J.L.; Yang, Y.Y. Co-delivery of drugs and plasmid DNA for cancer therapy. Adv. Drug Deliv. Rev. 2016, 98, 41–63. [Google Scholar] [CrossRef] [PubMed]
- Dorr, R.T. Pharmacology and Toxicology of Cremophor EL Diluent. Ann. Pharmacother. 1994, 28, S11–S14. [Google Scholar] [CrossRef]
- Weiss, R.B.; Donehower, R.C.; Wiernik, P.H.; Ohnuma, T.; Gralla, R.J.; Trump, D.L.; Jr, J.R.B.; Echo, D.A.V.; Hoff, D.D.V.; Leyland-Jones, B. Hypersensitivity reactions from taxol. J. Clin. Oncol. 1990, 8, 1263–1268. [Google Scholar] [CrossRef]
- Gelderblom, H.; Verweij, J.; Nooter, K.; Sparreboom, A. Cremophor EL: The drawbacks and advantages of vehicle selection for drug formulation. Eur. J. Cancer 2001, 37, 1590–1598. [Google Scholar] [CrossRef]
- Sofias, A.M.; Dunne, M.; Storm, G.; Allen, C. The battle of “nano” paclitaxel. Adv. Drug Deliv. Rev. 2017, 122, 20–30. [Google Scholar] [CrossRef]
- Hong, S.-S.; Choi, J.Y.; Kim, J.O.; Lee, M.-K.; Kim, S.H.; Lim, S.-J. Development of paclitaxel-loaded liposomal nanocarrier stabilized by triglyceride incorporation. Int. J. Nanomed. 2016, 11, 4465–4477. [Google Scholar] [CrossRef]
- Zhou, R.; Mazurchuk, R.V.; Tamburlin, J.H.; Harrold, J.M.; Mager, D.E.; Straubinger, R.M. Differential Pharmacodynamic Effects of Paclitaxel Formulations in an Intracranial Rat Brain Tumor Model. J. Pharmacol. Exp. Ther. 2010, 332, 479–488. [Google Scholar] [CrossRef]
- Ait-Oudhia, S.; Mager, D.E.; Straubinger, R.M. Application of Pharmacokinetic and Pharmacodynamic Analysis to the Development of Liposomal Formulations for Oncology. Pharmaceutics 2014, 6, 137–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.A.; Anyarambhatla, G.; Ma, L.; Ugwu, S.; Xuan, T.; Sardone, T.; Ahmad, I. Development and characterization of a novel Cremophor® EL free liposome-based paclitaxel (LEP-ETU) formulation. Eur. J. Pharm. Biopharm. 2005, 59, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Dellian, M.; Yuan, F.; Trubetskoy, V.S.; Torchilin, V.P.; Jain, R.K. Vascular permeability in a human tumour xenograft: Molecular charge dependence. Br. J. Cancer 2000, 82, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, M.E.; Ischenko, I.; Luedemann, S.; Strieth, S.; Papyan, A.; Werner, A.; Bohnenkamp, H.; Guenzi, E.; Preissler, G.; Michaelis, U.; et al. Vascular targeting by EndoTAG™-1 enhances therapeutic efficacy of conventional chemotherapy in lung and pancreatic cancer. Int. J. Cancer 2010, 126, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, Š.; Turánek-Knötigová, P.; MaŠek, J.; Korvasová, Z.; Škrabalová, M.; Plocková, J.; Bartheldyová, E.; Turánek, J. Liposomes with high encapsulation capacity for paclitaxel: Preparation, characterisation and in vivo anticancer effect. J. Pharm. Sci. 2010, 99, 2309–2319. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).