Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of 6S/nLNP and 6S/PLA NP
2.2.1. Lipid Extraction
2.2.2. Reassembly of 6S/nLNP and Construction of 6S/PLA NPs
2.3. Uptake of 6S/nLNP by E. coli
2.4. Microbiota Composition Analysis upon Oral Administration of 6S or 6S/nLNP
2.4.1. Animal
2.4.2. 16S rRNA Gene Sequencing
2.4.3. Microbiota Composition Data Analysis and Metagenome Prediction
2.5. Fecal Metabolites Extraction
2.6. Fecal Metabolic Profile
2.6.1. Ultra-High-Performance Liquid Chromatography Time-of-Flight Mass Spectrometry (UPLC-TOF-MS)
2.6.2. Metabolic Profile and Identification of Metabolites
2.7. In Vitro Anti-Inflammatory Assay of Fecal Metabolites
2.8. Electric Cell-Substrate Impedance Sensing (ECIS) Assay
2.9. In Vivo Efficacy Study
2.9.1. Animal
2.9.2. H&E Staining
2.10. Statistical Analysis
3. Results
3.1. Enteric Bacteria Effectively Take up 6S/nLNP
3.2. Oral Administration of 6S/nLNPs Accelerates 6S-Induced Changes in Gut Microbiota Composition
3.3. Changes in Microbiota Composition Produce Distinct Changes in Metabolic Profile
3.4. Gut Microbiota Composition Can Be Restored
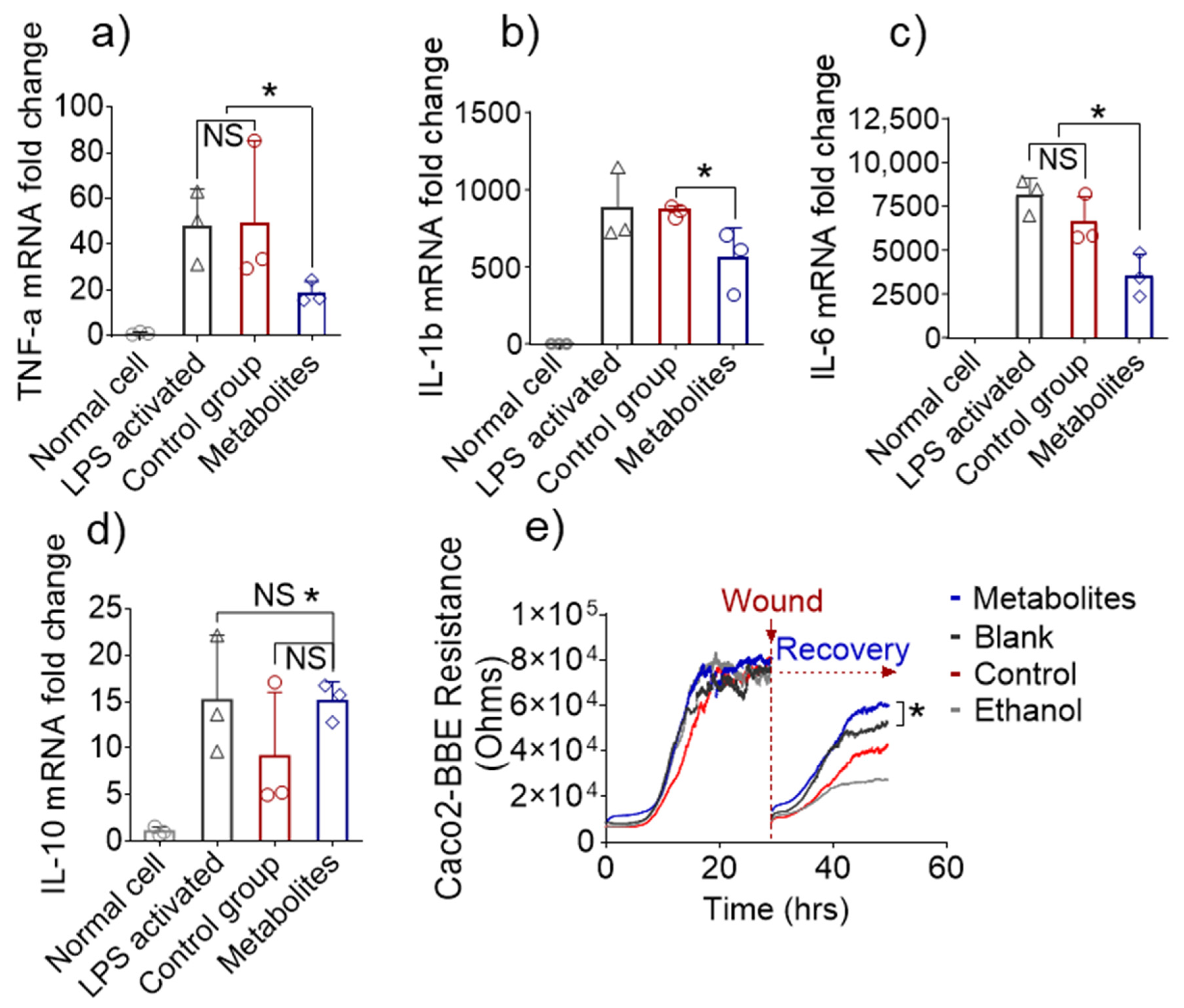
3.5. Altered Fecal Metabolites Exert Anti-Inflammatory Effects In Vitro
3.6. In Vivo Efficacy of Metabolites Derived from 6S/nLNP-Induced Changes in the Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danese, S.; Fiocchi, C. Ulcerative Colitis. N. Engl. J. Med. 2011, 365, 13. [Google Scholar] [CrossRef] [Green Version]
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Goel, A. MicroRNAs as Therapeutic Targets in Colitis and Colitis-Associated Cancer: Tiny Players with a Giant Impact. Gastroenterology 2015, 149, 859–861. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, P.B. Anti-adhesion molecules: Is gut specificity the key for a good safety profile? Curr. Drug Deliv. 2012, 9, 333–337. [Google Scholar] [CrossRef]
- Uchiyama, K.; Naito, Y.; Takagi, T. Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol. Ther. 2019, 199, 164–172. [Google Scholar] [CrossRef]
- Cani, P.D.; Delzenne, N.M. The gut microbiome as therapeutic target. Pharmacol. Ther. 2011, 130, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Roy, B.C.; Khan, S.A.; Septer, S.; Umar, S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms 2016, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Quagliariello, A.; Del Chierico, F.; Reddel, S.; Russo, A.; Onetti Muda, A.; D’Argenio, P.; Angelino, G.; Romeo, E.F.; Dall’Oglio, L.; De Angelis, P.; et al. Fecal Microbiota Transplant in Two Ulcerative Colitis Pediatric Cases: Gut Microbiota and Clinical Course Correlations. Microorganisms 2020, 8, 1486. [Google Scholar] [CrossRef]
- Hirten, R.P.; Grinspan, A.; Fu, S.C.; Luo, Y.; Suarez-Farinas, M.; Rowland, J.; Contijoch, E.J.; Mogno, I.; Yang, N.; Luong, T.; et al. Microbial Engraftment and Efficacy of Fecal Microbiota Transplant for Clostridium Difficile in Patients With and Without Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2019, 25, 969–979. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Strukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, D.; Jiang, H.; Zhang, S.; Pang, X.; Gao, S.; Zhang, H.; Zhang, S.; Xiao, Q.; Chen, L.; et al. Gut Microbiota Variation With Short-Term Intake of Ginger Juice on Human Health. Front. Microbiol. 2020, 11, 576061. [Google Scholar] [CrossRef]
- Nonaka, K.; Bando, M.; Sakamoto, E.; Inagaki, Y.; Naruishi, K.; Yumoto, H.; Kido, J.I. 6-Shogaol Inhibits Advanced Glycation End-Products-Induced IL-6 and ICAM-1 Expression by Regulating Oxidative Responses in Human Gingival Fibroblasts. Molecules 2019, 24, 3705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bischoff-Kont, I.; Furst, R. Benefits of Ginger and Its Constituent 6-Shogaol in Inhibiting Inflammatory Processes. Pharmaceuticals 2021, 14, 571. [Google Scholar] [CrossRef]
- Pan, M.H.; Hsieh, M.C.; Kuo, J.M.; Lai, C.S.; Wu, H.; Sang, S.; Ho, C.T. 6-Shogaol induces apoptosis in human colorectal carcinoma cells via ROS production, caspase activation, and GADD 153 expression. Mol. Nutr. Food Res. 2008, 52, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xu, C.; Liu, D.; Han, M.K.; Wang, L.; Merlin, D. Oral Delivery of Nanoparticles Loaded With Ginger Active Compound, 6-Shogaol, Attenuates Ulcerative Colitis and Promotes Wound Healing in a Murine Model of Ulcerative Colitis. J. Crohns Colitis 2018, 12, 217–229. [Google Scholar] [CrossRef]
- Zhang, M.; Viennois, E.; Prasad, M.; Zhang, Y.; Wang, L.; Zhang, Z.; Han, M.K.; Xiao, B.; Xu, C.; Srinivasan, S.; et al. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials 2016, 101, 321–340. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhang, M.; Lama, S.; Wang, L.; Merlin, D. Natural-lipid nanoparticle-based therapeutic approach to deliver 6-shogaol and its metabolites M2 and M13 to the colon to treat ulcerative colitis. J. Control. Release 2020, 323, 293–310. [Google Scholar] [CrossRef]
- Rieger, J.; Pelckmann, L.M.; Drewes, B. Preservation and Processing of Intestinal Tissue for the Assessment of Histopathology. Methods Mol. Biol. 2021, 2223, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. Methods Mol. Biol. 2017, 1522, 17–22. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Guijas, C.; Benton, H.P.; Warth, B.; Siuzdak, G. METLIN MS(2) molecular standards database: A broad chemical and biological resource. Nat. Methods 2020, 17, 953–954. [Google Scholar] [CrossRef] [PubMed]
- Altermann, E.; Klaenhammer, T.R. PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genom. 2005, 6, 60. [Google Scholar] [CrossRef] [Green Version]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castano-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Donia, M.S.; Fischbach, M.A. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science 2015, 349, 1254766. [Google Scholar] [CrossRef] [Green Version]
- Lopetuso, L.R.; Giorgio, M.E.; Saviano, A.; Scaldaferri, F.; Gasbarrini, A.; Cammarota, G. Bacteriocins and Bacteriophages: Therapeutic Weapons for Gastrointestinal Diseases? Int. J. Mol. Sci. 2019, 20, 183. [Google Scholar] [CrossRef] [Green Version]
- Guasch, J.F.; Enfedaque, J.; Ferrer, S.; Gargallo, D.; Regue, M. Bacteriocin 28b, a chromosomally encoded bacteriocin produced by most Serratia marcescens biotypes. Res. Microbiol. 1995, 146, 477–483. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of secondary metabolism in actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlin-Zhang, O.; Sung, J.; Viennois, E. In Vitro Intestinal Epithelial Wound-healing Assays Using Electric Cell-Substrate Impedance Sensing Instrument. Bio. Protoc. 2019, 9, e3351. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, T.; Rawla, P.; Ofosu, A.; Gaduputi, V. Fecal microbiota transplant—A new frontier in inflammatory bowel disease. J. Inflamm. Res. 2018, 11, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.Y.; Popov, J.; Pai, N. Fecal microbial transplant for the treatment of pediatric inflammatory bowel disease. World J. Gastroenterol. 2016, 22, 10304–10315. [Google Scholar] [CrossRef]
- Cho, S.; Spencer, E.; Hirten, R.; Grinspan, A.; Dubinsky, M.C. Fecal Microbiota Transplant for Recurrent Clostridium difficile Infection in Pediatric Inflammatory Bowel Disease. J. Pediatr. Gastroenterol. Nutr. 2019, 68, 343–347. [Google Scholar] [CrossRef]
- Goyal, A.; Yeh, A.; Bush, B.R.; Firek, B.A.; Siebold, L.M.; Rogers, M.B.; Kufen, A.D.; Morowitz, M.J. Safety, Clinical Response, and Microbiome Findings Following Fecal Microbiota Transplant in Children With Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2018, 24, 410–421. [Google Scholar] [CrossRef]
- Jamot, S.; Raghunathan, V.; Patel, K.; Kelly, C.R.; Lim, S.H. Factors Associated with the Use of Fecal Microbiota Transplant in Patients with Recurrent Clostridium difficile Infections. Infect. Control. Hosp. Epidemiol. 2018, 39, 302–306. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, M.; Merlin, D. Advances in Plant-derived Edible Nanoparticle-based lipid Nano-drug Delivery Systems as Therapeutic Nanomedicines. J. Mater. Chem. B 2018, 6, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; San, K.Y.; Bennett, G.N. Improved succinate production from galactose-rich feedstocks by engineered Escherichia coli under anaerobic conditions. Biotechnol. Bioeng. 2020, 117, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lim, B.C.; Yeom, S.J.; Kim, Y.S.; Kim, H.J.; Lee, J.K.; Lee, S.H.; Kim, S.W.; Oh, D.K. Differential selectivity of the Escherichia coli cell membrane shifts the equilibrium for the enzyme-catalyzed isomerization of galactose to tagatose. Appl. Environ. Microbiol. 2008, 74, 2307–2313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugenholtz, F.; de Vos, W.M. Mouse models for human intestinal microbiota research: A critical evaluation. Cell Mol. Life Sci. 2018, 75, 149–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Name | Formula | CAS | METLIN ID | Mass | ΔPPM | After Treatment |
|---|---|---|---|---|---|---|
| Bacteriocin 28b | C12H11N3O2 | 148466-49-3 | 89961 | 229.0851 | 0 | ↑ |
| Undecylprodigiosin | C25H35N3O | 52340-48-4 | 69260 | 393.2780 | 1 | ↑ |
| NAc-DNP-Cys | C11H11N3O7S | 35897-25-7 | 69126 | 329.0318 | 4 | ↑ |
| Iprodione | C13H13Cl2N3O3 | 36734-19-7 | 68915 | 329.0334 | 0 | ↑ |
| 24 unknown metabolites | n/a | n/a | n/a | n/a | n/a | ↓ |
| 60 unknown metabolites | n/a | n/a | n/a | n/a | n/a | ↑ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Long, D.; Sung, J.; Alghoul, Z.; Merlin, D. Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome. Pharmaceutics 2021, 13, 1355. https://doi.org/10.3390/pharmaceutics13091355
Yang C, Long D, Sung J, Alghoul Z, Merlin D. Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome. Pharmaceutics. 2021; 13(9):1355. https://doi.org/10.3390/pharmaceutics13091355
Chicago/Turabian StyleYang, Chunhua, Dingpei Long, Junsik Sung, Zahra Alghoul, and Didier Merlin. 2021. "Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome" Pharmaceutics 13, no. 9: 1355. https://doi.org/10.3390/pharmaceutics13091355
APA StyleYang, C., Long, D., Sung, J., Alghoul, Z., & Merlin, D. (2021). Orally Administered Natural Lipid Nanoparticle-Loaded 6-Shogaol Shapes the Anti-Inflammatory Microbiota and Metabolome. Pharmaceutics, 13(9), 1355. https://doi.org/10.3390/pharmaceutics13091355







