Development of Tailor-Made Dendrimer Ternary Complexes for Drug/Gene Co-Delivery in Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
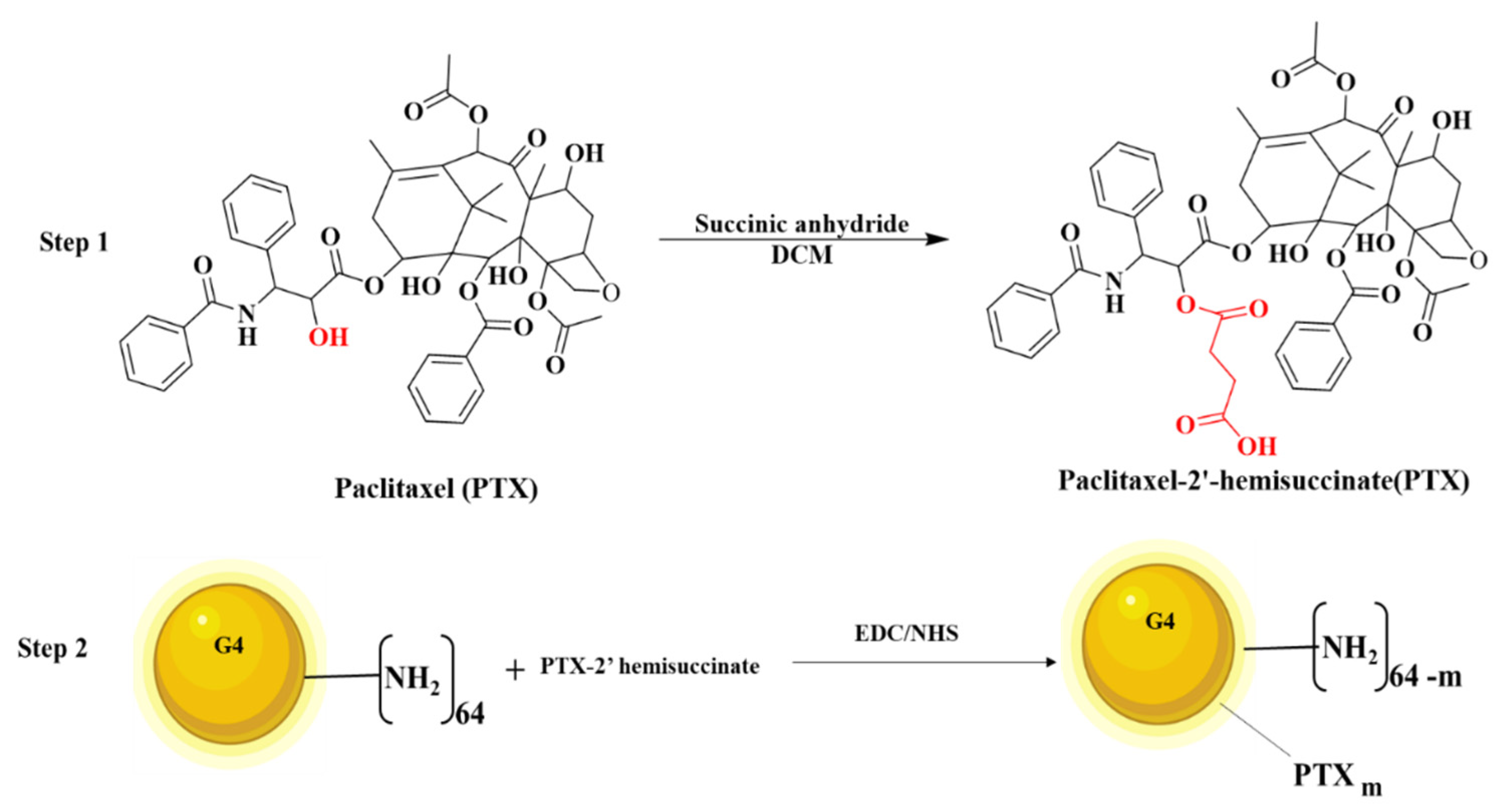
2.2.1. Synthesis of PTX-2-Hemisuccinate (PTX-SA)
2.2.2. Synthesis of PAMAM G4-PTX
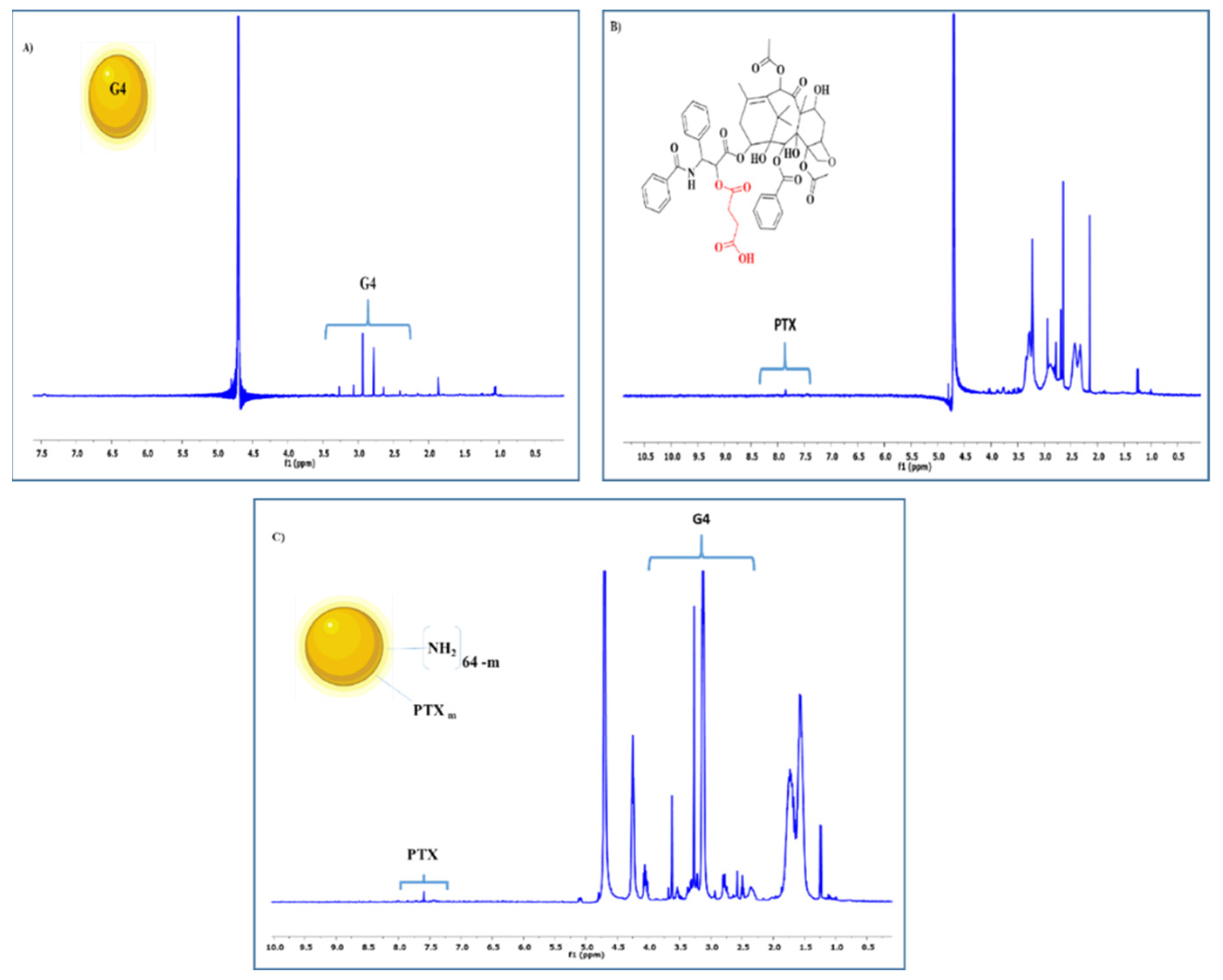
2.2.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.2.4. Preparation of PAMAM G4-PTX/pDNA and PAMAM G4-PTX/pDNA/PEI Complexes
2.2.5. Agarose Gel Electrophoresis
2.2.6. Characterization of Binary and Ternary Complexes
2.2.7. Cell Culture
2.2.8. Biocompatibility Study
2.2.9. FITC Plasmid Labeling
2.2.10. Cellular Uptake/Internalization
2.2.11. Determination of Cell Associated PTX
2.2.12. Protein Quantification
2.2.13. Measurement of Caspase-3 Activity
2.2.14. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Characterization of Multifunctional Dendrimer Conjugate
3.2. pDNA Complexation: Binary versus Ternary Systems
3.3. Evaluation of the Properties of Binary and Ternary Systems
3.4. Biocompatibility Evaluation
3.5. PTX and pDNA Cellular Internalization
3.6. Expression of p53 Protein
3.7. Detection of Apoptosis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pucci, C.; Martinelli, C.; Ciofani, G. Innovative approaches for cancer treatment: Current perspectives and new challenges. Ecancermedicalscience 2019, 13, 961. [Google Scholar] [CrossRef]
- Flotte, T.R.; Gao, G. 2020: Gene Therapy Enters Its Fourth Decade. Hum. Gene Ther. 2020, 31, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Belete, T.M. The Current Status of Gene Therapy for the Treatment of Cancer. Biol. Targets Ther. 2021, 15, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zawacka-Pankau, J.E. The Undervalued Avenue to Reinstate Tumor Suppressor Functionality of the p53 Protein Family for Improved Cancer Therapy-Drug Repurposing. Cancers 2020, 12, 2717. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Prives, C. p53: Master of life, death, and the epigenome. Genes Dev. 2017, 31, 955–956. [Google Scholar] [CrossRef]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal. Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Faria, R.; Sousa, Â.; Neves, A.R.; Queiroz, J.A.; Costa, D. Methotrexate-plasmid DNA polyplexes for cancer therapy: Characterization, cancer cell targeting ability and tuned in vitro transfection. J. Mol. Liq. 2019, 292, 111391. [Google Scholar] [CrossRef]
- Neves, A.R.; Sousa, A.; Faria, R.; Albuquerque, T.; Queiroz, J.A.; Costa, D. Cancer gene therapy mediated by RALA/plasmid DNA vectors: Nitrogen to phosphate groups ratio (N/P) as a tool for tunable transfection efficiency and apoptosis. Colloids Surf. B Biointerfaces 2020, 185, 110610. [Google Scholar] [CrossRef]
- Shi, L.; Feng, H.; Li, Z.; Shi, J.; Jin, L.; Li, J. Co-Delivery of Paclitaxel and siRNA with pH-Responsive Polymeric Micelles for Synergistic Cancer Therapy. J. Biomed. Nanotechnol. 2021, 17, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Shang, F.; Chen, D.; Cao, T.; Wang, X.; Jiao, J.; He, S.; Liang, X. Protein liposomes-mediated targeted acetylcholinesterase gene delivery for effective liver cancer therapy. J. Nanobiotechnol. 2021, 19, 31. [Google Scholar] [CrossRef]
- Montaño-Samaniego, M.; Bravo-Estupiñan, D.M.; Méndez-Guerrero, O.; Alarcón-Hernández, E.; Ibáñez-Hernández, M. Strategies for Targeting Gene Therapy in Cancer Cells with Tumor-Specific Promoters. Front. Oncol. 2020, 10, 2671. [Google Scholar] [CrossRef] [PubMed]
- Santana-Armas, M.L.; Tros de Ilarduya, C. Strategies for cancer gene-delivery improvement by non-viral vectors. Int. J. Pharm. 2021, 596, 120291. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Chen, S.; Hu, Y.; Yang, F.; Huo, Q.; Xie, N. Tumour-Targeted and Redox-Responsive Mesoporous Silica Nanoparticles for Controlled Release of Doxorubicin and an siRNA Against Metastatic Breast Cancer. Int. J. Nanomed. 2021, 16, 1961–1976. [Google Scholar] [CrossRef]
- Costa, D.; Valente, A.J.M.; Queiroz, J. Stimuli-responsive polyamine-DNA blend nanogels for co-delivery in cancer therapy. Colloids Surf. B Biointerfaces 2015, 132, 194–201. [Google Scholar] [CrossRef]
- Li, G.; Meng, F.; Lu, T.; Wei, L.; Pan, X.; Nong, Z.; Wei, M.; Liao, C.; Li, X. Functionalised molybdenum disulfide nanosheets for co-delivery of doxorubicin and siRNA for combined chemo/gene/photothermal therapy on multidrug-resistant cancer. J. Pharm. Pharmacol. 2021, 73, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Nasab, S.H.; Amani, A.; Ebrahimi, H.A.; Hamidi, A.A. Design and preparation of a new multi-targeted drug delivery system using multifunctional nanoparticles for co-delivery of siRNA and paclitaxel. J. Pharm. Anal. 2021, 11, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, S.; Zhang, Z.; Wang, L.; You, Y. Cationic micelle: A promising nanocarrier for gene delivery with high transfection efficiency. J. Gene Med. 2019, 21, e3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhammad, K.; Zhao, J.; Gao, B.; Feng, Y. Polymeric nano-carriers for on-demand delivery of genes via specific responses to stimuli. J. Mater. Chem. B 2020, 8, 9621–9641. [Google Scholar] [CrossRef]
- McErlean, E.M.; Ziminska, M.; McCrudden, C.M.; McBride, J.W.; Loughran, S.P.; Cole, G.; Mulholland, E.J.; Kett, V.; Buckley, N.E.; Robson, T.; et al. Rational design and characterisation of a linear cell penetrating peptide for non-viral gene delivery. J. Control. Release Off. J. Control. Release Soc. 2021, 330, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Pengnam, S.; Plianwong, S.; Patrojanasophon, P.; Radchatawedchakoon, W.; Yingyongnarongkul, B.E.; Opanasopit, P.; Charoensuksai, P. Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids. Pharmaceutics 2021, 13, 550. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Yi, J.; Peng, J.; Zhang, X.; Lei, L.; Zhao, D.; Lei, Z.; Nie, H. Characterization of synergistic anti-tumor effects of doxorubicin and p53 via graphene oxide-polyethyleneimine nanocarriers. J. Mater. Sci. Technol. 2017, 33, 807–814. [Google Scholar] [CrossRef]
- Lin, J.T.; Chen, H.; Wang, D.; Xiong, L.; Li, J.Z.; Chen, G.H.; Chen, G.B. Nuclear-targeted p53 and DOX co-delivery of chitosan derivatives for cancer therapy in vitro and in vivo. Colloids Surf. B Biointerfaces 2019, 183, 110440. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Dehshahri, A.; Sagar Madamsetty, V.; Zahmatkeshan, M.; Tavakol, S.; Makvandi, P.; Khorsandi, D.; Pardakhty, A.; Ashrafizadeh, M.; Ghasemipour Afshar, E.; et al. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Control. Release Off. J. Control. Release Soc. 2020, 325, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, H.; Kiran Rompicharla, S.V.; Ghosh, B.; Torchilin, V.; Biswas, S. Transferrin/α-tocopherol modified poly(amidoamine) dendrimers for improved tumor targeting and anticancer activity of paclitaxel. Nanomedicine 2019, 14, 3159–3176. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Xiao, Y.; Ouyang, Z.; Shen, M.; Shi, X. Efficient co-delivery of microRNA 21 inhibitor and doxorubicin to cancer cells using core–shell tecto dendrimers formed via supramolecular host–guest assembly. J. Mater. Chem. B 2020, 8, 2768–2774. [Google Scholar] [CrossRef]
- Sandoval-Yañez, C.; Castro Rodriguez, C. Dendrimers: Amazing Platforms for Bioactive Molecule Delivery Systems. Materials 2020, 13, 570. [Google Scholar] [CrossRef] [Green Version]
- Tarach, P.; Janaszewska, A. Recent Advances in Preclinical Research Using PAMAM Dendrimers for Cancer Gene Therapy. Int. J. Mol. Sci. 2021, 22, 2912. [Google Scholar] [CrossRef]
- Gogulapati, N.M.; Manalan, B.V.; Nadendla, R.R. Poly (propylene imine) Dendrimer: Synthesis, characterization and applications in various drug delivery. Asian, J. Pharm. Pharmacol. 2020, 6, 190–203. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, J.; Zheng, Y.; Guo, R.; Wang, S.; Mignani, S.; Caminade, A.M.; Majoral, J.P.; Shi, X. Doxorubicin-Conjugated PAMAM Dendrimers for pH-Responsive Drug Release and Folic Acid-Targeted Cancer Therapy. Pharmaceutics 2018, 10, 162. [Google Scholar] [CrossRef] [Green Version]
- Palmerston Mendes, L.; Pan, J.; Torchilin, V.P. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tariq, I.; Ali, M.Y.; Sohail, M.F.; Amin, M.U.; Ali, S.; Bukhari, N.I.; Raza, A.; Pinnapireddy, S.R.; Schäfer, J.; Bakowsky, U. Lipodendriplexes mediated enhanced gene delivery: A cellular to pre-clinical investigation. Sci. Rep. 2020, 10, 21446. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, H.; Liang, S.; Li, X.; Wang, D. Synthesis and characterization of a PAMAM dendrimer nanocarrier functionalized by HA for targeted gene delivery systems and evaluation in vitro. J. Biomater. Sci. Polym. Ed. 2021, 32, 205–228. [Google Scholar] [CrossRef]
- Costa, D.; Briscoe, W.H.; Queiroz, J. Polyethylenimine coated plasmid DNA–surfactant complexes as potential gene delivery systems. Colloids Surf. B Biointerfaces 2015, 133, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Majoros, I.J.; Myc, A.; Thomas, T.; Mehta, C.B.; Baker, J.R., Jr. PAMAM dendrimer-based multifunctional conjugate for cancer therapy: Synthesis, characterization, and functionality. Biomacromolecules 2006, 7, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Sousa-Herves, A.; Würfel, P.; Wegner, N.; Khandare, J.; Licha, K.; Haag, R.; Welker, P.; Calderón, M. Dendritic polyglycerol sulfate as a novel platform for paclitaxel delivery: Pitfalls of ester linkage. Nanoscale 2015, 7, 3923–3932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, R.; Albuquerque, T.; Neves, A.R.; Bhatt, H.; Biswas, S.; Cardoso, A.M.; Pedroso de Lima, M.C.; Jurado, A.S.; Costa, D. Physicochemical characterization and targeting performance of triphenylphosphonium nano-polyplexes. J. Mol. Liq. 2020, 316, 113873. [Google Scholar] [CrossRef]
- Albuquerque, T.; Faria, R.; Sousa, Â.; Neves, A.R.; Queiroz, J.A.; Costa, D. Polymer-peptide ternary systems as a tool to improve the properties of plasmid DNA vectors in gene delivery. J. Mol. Liq. 2020, 309, 113157. [Google Scholar] [CrossRef]
- Srinageshwar, B.; Florendo, M.; Clark, B.; Johnson, K.; Munro, N.; Peruzzaro, S.; Antcliff, A.; Andrews, M.; Figacz, A.; Swanson, D.; et al. A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids. Pharmaceutics 2020, 12, 619. [Google Scholar] [CrossRef] [PubMed]
- Bono, N.; Pennetta, C.; Bellucci, M.C.; Sganappa, A.; Malloggi, C.; Tedeschi, G.; Candiani, G.; Volonterio, A. Role of Generation on Successful DNA Delivery of PAMAM–(Guanidino)Neomycin Conjugates. ACS Omega 2019, 4, 6796–6807. [Google Scholar] [CrossRef] [Green Version]
- Dou, X.; Meints, G.A.; Sedaghat-Herati, R. New Insights into the Interactions of a DNA Oligonucleotide with mPEGylated-PAMAM by Circular Dichroism and Solution NMR. J. Phys. Chem. B 2019, 123, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Martins, I.; Tomás, H.; Lahoz, F.; Rodrigues, J. Engineered Fluorescent Carbon Dots and G4-G6 PAMAM Dendrimer Nanohybrids for Bioimaging and Gene Delivery. Biomacromolecules 2021, 22, 2436–2450. [Google Scholar] [CrossRef] [PubMed]
- Appelbe, O.K.; Kim, B.K.; Rymut, N.; Wang, J.; Kron, S.J.; Yeo, Y. Radiation-enhanced delivery of plasmid DNA to tumors utilizing a novel PEI polyplex. Cancer Gene Ther. 2018, 25, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Sousa, Â.; Faria, R.; Albuquerque, T.; Bhatt, H.; Biswas, S.; Queiroz, J.A.; Costa, D. Design of experiments to select triphenylphosphonium-polyplexes with suitable physicochemical properties for mitochondrial gene therapy. J. Mol. Liq. 2020, 302, 112488. [Google Scholar] [CrossRef]
- Utsuno, K.; Kono, H.; Tanaka, E.; Jouna, N.; Kojima, Y.; Uludağ, H. Low Molecular Weight Branched PEI Binding to Linear DNA. Chem. Pharm. Bull. 2016, 64, 1484–1491. [Google Scholar] [CrossRef] [Green Version]
- Costa, D.; Valente, A.J.M.; Queiroz, J.A.; Sousa, Â. Finding the ideal polyethylenimine-plasmid DNA system for co-delivery of payloads in cancer therapy. Colloids Surf. B Biointerfaces 2018, 170, 627–636. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.K.; Pedersen, J.N.; Marie, R. Size and surface charge characterization of nanoparticles with a salt gradient. Nat. Commun. 2020, 11, 2337. [Google Scholar] [CrossRef] [PubMed]
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef]
- Manzanares, D.; Ceña, V. Endocytosis: The Nanoparticle and Submicron Nanocompounds Gateway into the Cell. Pharmaceutics 2020, 12, 371. [Google Scholar] [CrossRef] [Green Version]
- Moore, T.L.; Rodriguez-Lorenzo, L.; Hirsch, V.; Balog, S.; Urban, D.; Jud, C.; Rothen-Rutishauser, B.; Lattuada, M.; Petri-Fink, A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 2015, 44, 6287–6305. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, B.; Yan, B. Enabling Anticancer Therapeutics by Nanoparticle Carriers: The Delivery of Paclitaxel. Int. J. Mol. Sci. 2011, 12, 4395–4413. [Google Scholar] [CrossRef]
- Aoubala, M.; Murray-Zmijewski, F.; Khoury, M.P.; Fernandes, K.; Perrier, S.; Bernard, H.; Prats, A.C.; Lane, D.P.; Bourdon, J.C. p53 directly transactivates Δ133p53α, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2011, 18, 248–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rompicharla, S.V.K.; Kumari, P.; Bhatt, H.; Ghosh, B.; Biswas, S. Biotin functionalized PEGylated poly(amidoamine) dendrimer conjugate for active targeting of paclitaxel in cancer. Int. J. Pharm. 2019, 557, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Rompicharla, S.V.K.; Kumari, P.; Ghosh, B.; Biswas, S. Octa-arginine modified poly(amidoamine) dendrimers for improved delivery and cytotoxic effect of paclitaxel in cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 847–859. [Google Scholar] [CrossRef]
- Biswas, S.; Deshpande, P.P.; Navarro, G.; Dodwadkar, N.S.; Torchilin, V.P. Lipid modified triblock PAMAM-based nanocarriers for siRNA drug co-delivery. Biomaterials 2013, 34, 1289–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulhari, H.; Pooja, D.; Shrivastava, S.; Kuncha, M.; Naidu, V.G.M.; Bansal, V.; Sistla, R.; Adams, D.J. Trastuzumab-grafted PAMAM dendrimers for the selective delivery of anticancer drugs to HER2-positive breast cancer. Sci. Rep. 2016, 6, 23179. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Pim, D.; Banks, L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999, 18, 7690–7700. [Google Scholar] [CrossRef] [Green Version]
- Teo, P.Y.; Cheng, W.; Hedrick, J.L.; Yang, Y.Y. Co-delivery of drugs and plasmid DNA for cancer therapy. Adv. Drug Deliv. Rev. 2016, 98, 41–63. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A Target for Anticancer Therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichani, K.; Li, J.; Suzuki, M.; Houston, J. Evaluation of Caspase-3 Activity During Apoptosis with Fluorescence Lifetime-Based Cytometry Measurements and Phasor Analyses. Cytometry 2020, 97, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Rooprai, H.K.; Lawrence, P.; Keshavarz, S.; Yashod, P.; Gullan, R.W.; Selway, R.P.; Davies, D. DRAQ7 as an Alternative to MTT Assay for Measuring Viability of Glioma Cells Treated with Polyphenols. Anticancer. Res. 2020, 40, 5427–5436. [Google Scholar] [CrossRef]






| PAMAM G4-PTX/pDNA/PEI N/P Ratio | Size (nm) | PdI | Zeta Potential (mV) | CC% |
|---|---|---|---|---|
| N/P 2:1:1 | 461 ± 35 | 0.63 ± 0.06 | −5.05 ± 0.09 | 34.33 ± 3.22 |
| N/P 5:1:1 | 427 ± 14 | 0.59 ± 0.05 | −4.53 ± 0.10 | 41.67 ± 2.52 |
| N/P 10:1:1 | 412 ± 32 | 0.53 ± 0.04 | −4.24 ± 0.07 | 41.33 ± 2.31 |
| N/P 20:1:1 | 409 ± 34 | 0.44 ± 0.03 | −4.03 ± 0.13 | 41.33 ± 1.53 |
| N/P 50:1:1 | 390 ± 26 | 0.38 ± 0.03 | −3.12 ± 0.04 | 43.33 ± 1.16 |
| N/P 100:1:1 | 384 ± 40 | 0.34 ± 0.03 | −2.71 ± 0.43 | 45.33 ± 3.06 |
| N/P 2:1.5:1 | 385 ± 24 | 0.38 ± 0.03 | −2.14 ± 0.04 | 45.33 ± 1.53 |
| N/P 5:1.5:1 | 383 ± 33 | 0.35 ± 0.02 | −2.24 ± 0.08 | 46.67 ± 1.16 |
| N/P 10:1.5:1 | 381 ± 42 | 0.34 ± 0.04 | −2.58 ± 0.27 | 46.67 ± 1.53 |
| N/P 20:1.5:1 | 373 ± 34 | 0.35 ± 0.03 | −1.92 ± 0.14 | 47.33 ± 1.16 |
| N/P 50:1.5:1 | 355 ± 23 | 0.33 ± 0.03 | −1.09 ± 0.05 | 52.67 ± 2.52 |
| N/P 100:1.5:1 | 334 ± 44 | 0.36 ± 0.04 | +1.31 ± 0.27 | 58.33 ± 0.58 |
| N/P 2:2:1 | 308 ± 23 | 0.38 ± 0.02 | +1.22 ± 0.04 | 66.33 ± 1.16 |
| N/P 5:2:1 | 306 ± 31 | 0.33 ± 0.03 | +2.25 ± 0.07 | 69.67 ± 2.08 |
| N/P 10:2:1 | 283 ± 33 | 0.36 ± 0.02 | +4.58 ± 0.03 | 77.67 ± 1.16 |
| N/P 20:2:1 | 259 ± 42 | 0.43 ± 0.03 | +4.94 ± 0.10 | 79.33 ± 2.52 |
| N/P 50:2:1 | 217 ± 31 | 0.35 ± 0.03 | +6.99 ± 0.21 | 85.33 ± 1.16 |
| N/P 100:2:1 | 194 ± 23 | 0.31 ± 0.03 | +10.05 ± 0.18 | 89.67 ± 2.08 |
| P53 Content (ng/mL) | ||
|---|---|---|
| HeLa | Control cells | 0.0 ± 0 |
| PAMAM G4-PTX/pDNA/PEI N/P 50:2:1 | 482.7 ± 0.943 | |
| N2a | Control cells | 40.9 ± 5.71 |
| PAMAM G4-PTX/pDNA/PEI N/P 50:2:1 | 403.2 ± 6.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, A.R.; Albuquerque, T.; Faria, R.; Paul, M.; Biswas, S.; Sousa, Â.; Costa, D. Development of Tailor-Made Dendrimer Ternary Complexes for Drug/Gene Co-Delivery in Cancer. Pharmaceutics 2021, 13, 1256. https://doi.org/10.3390/pharmaceutics13081256
Neves AR, Albuquerque T, Faria R, Paul M, Biswas S, Sousa Â, Costa D. Development of Tailor-Made Dendrimer Ternary Complexes for Drug/Gene Co-Delivery in Cancer. Pharmaceutics. 2021; 13(8):1256. https://doi.org/10.3390/pharmaceutics13081256
Chicago/Turabian StyleNeves, Ana Raquel, Tânia Albuquerque, Rúben Faria, Milan Paul, Swati Biswas, Ângela Sousa, and Diana Costa. 2021. "Development of Tailor-Made Dendrimer Ternary Complexes for Drug/Gene Co-Delivery in Cancer" Pharmaceutics 13, no. 8: 1256. https://doi.org/10.3390/pharmaceutics13081256
APA StyleNeves, A. R., Albuquerque, T., Faria, R., Paul, M., Biswas, S., Sousa, Â., & Costa, D. (2021). Development of Tailor-Made Dendrimer Ternary Complexes for Drug/Gene Co-Delivery in Cancer. Pharmaceutics, 13(8), 1256. https://doi.org/10.3390/pharmaceutics13081256








