Molecular Modeling of Protein Corona Formation and Its Interactions with Nanoparticles and Cell Membranes for Nanomedicine Applications
Abstract
1. Introduction
2. Conformation, Binding Site and Strength of Plasma Proteins on the Nanoparticle
2.1. Gold Nanoparticles
2.2. Silver Nanoparticles
2.3. Carbon Nanomaterials
2.4. Polymer-Grafted Nanoparticles
2.5. Others
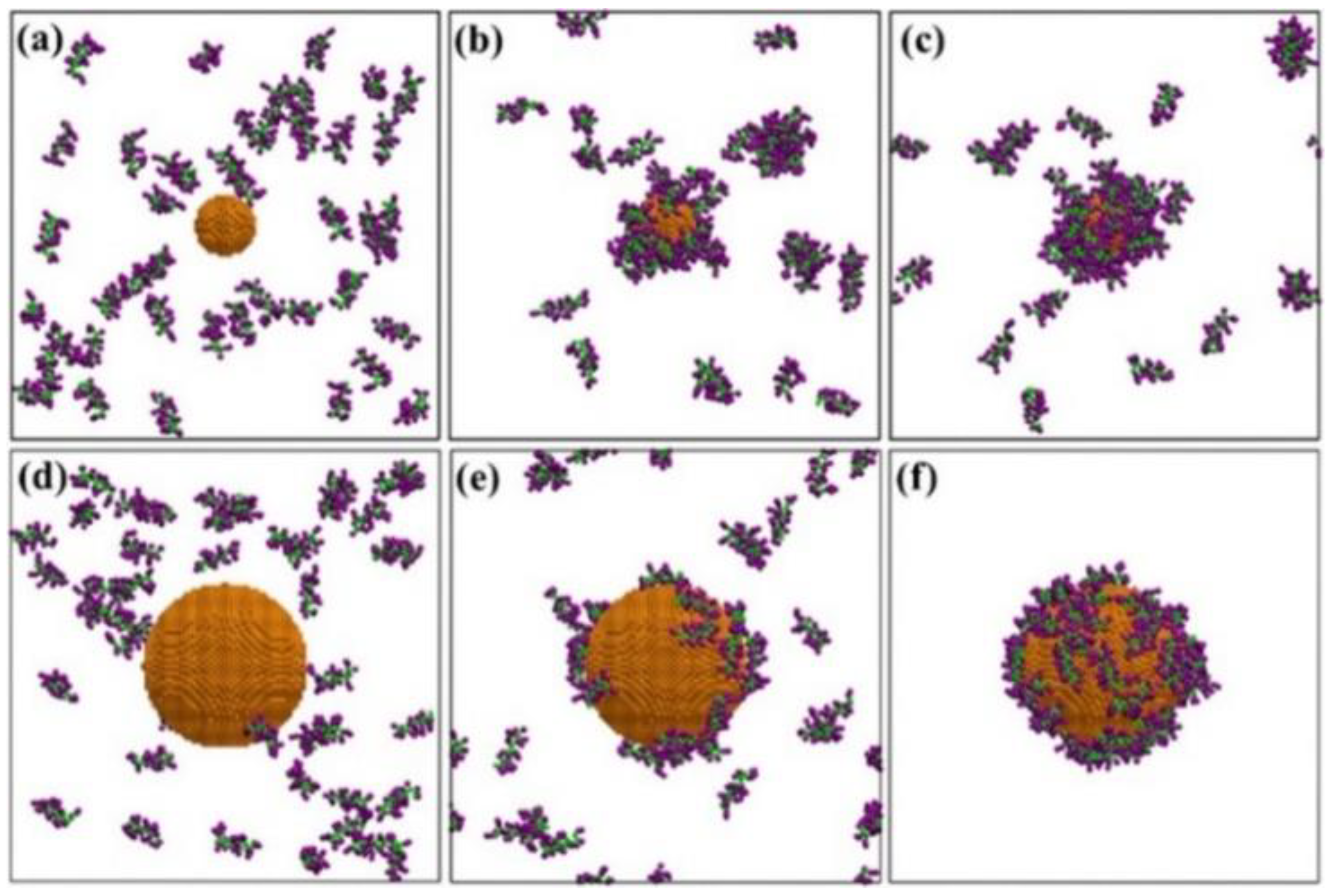
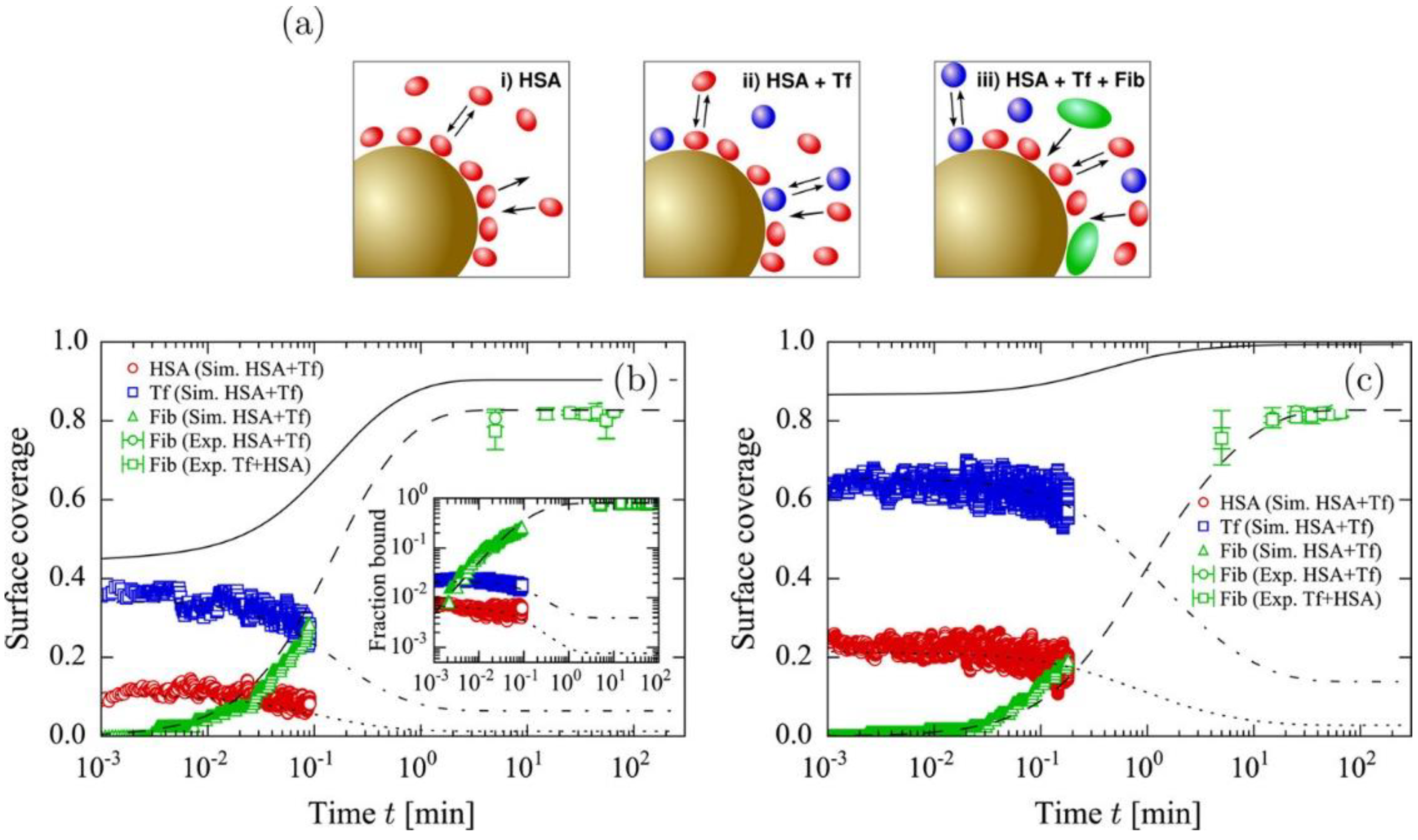
3. Competitive Adsorption and Desorption of Plasma Proteins on Nanoparticle Surfaces
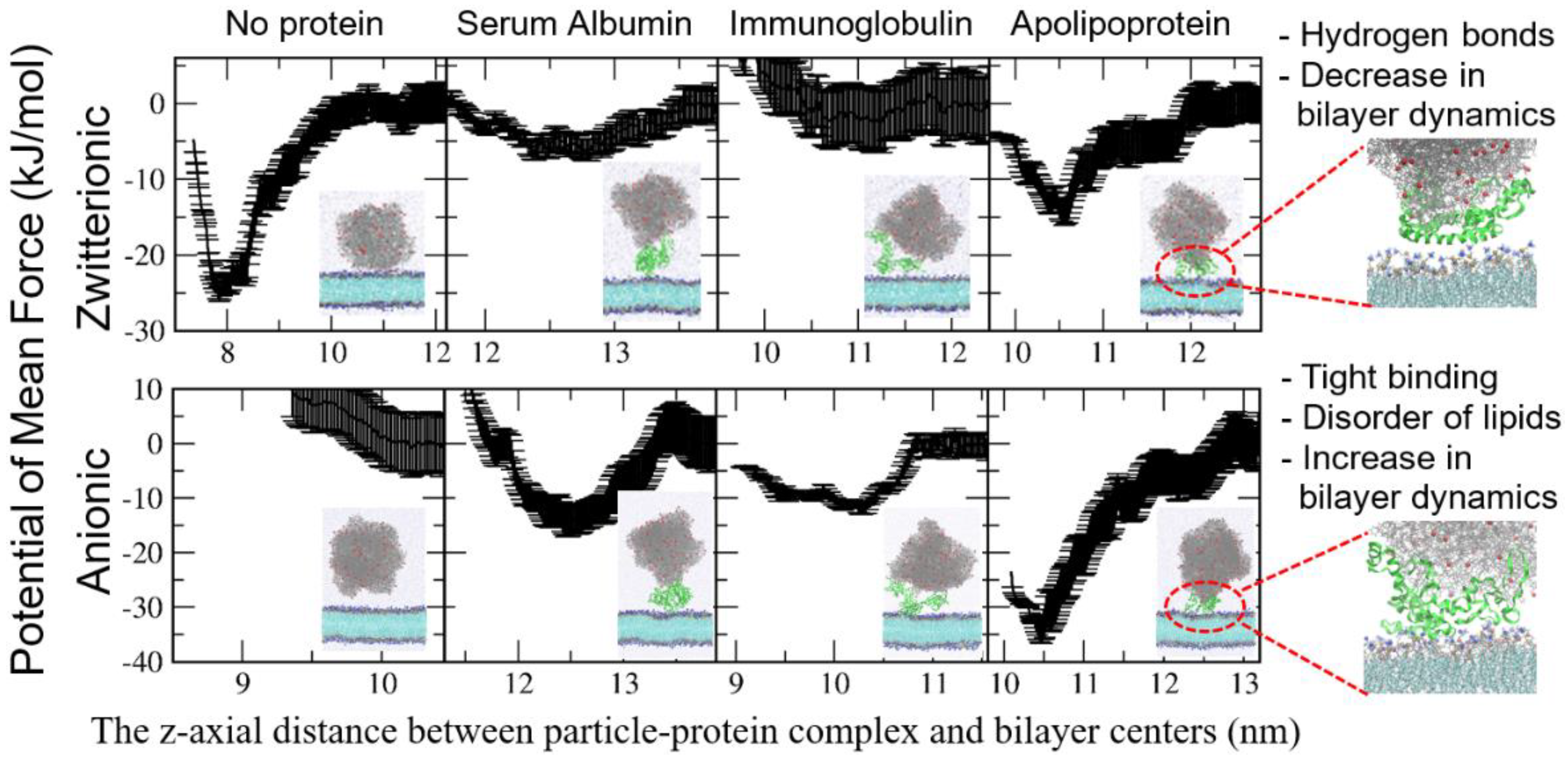
4. Interactions between the Protein Corona-Particle Complex and Lipid Membrane
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Segura, T.; Shea, L.D. Materials for non-viral gene delivery. Annu. Rev. Mater. Res. 2001, 31, 25–46. [Google Scholar] [CrossRef]
- Ferrari, M. Cancer nanotechnology: Opportunities and challenges. Nat. Rev. Cancer 2005, 5, 161–171. [Google Scholar] [CrossRef]
- Rolland, A.P. From genes to gene medicines: Recent advances in nonviral gene delivery. Crit. Rev. Ther. Drug Carr. Syst. 1998, 15, 143–198. [Google Scholar] [CrossRef]
- Davis, M.E. Non-viral gene delivery systems. Curr. Opin. Biotechnol. 2002, 13, 128–131. [Google Scholar] [CrossRef]
- Torchilin, V.P. Micellar nanocarriers: Pharmaceutical perspectives. Pharm. Res. 2007, 24, 1–16. [Google Scholar] [CrossRef]
- Nel, A.; Xia, T.; Madler, L.; Li, N. Toxic potential of materials at the nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef]
- Popielarski, S.R.; Pun, S.H.; Davis, M.E. A nanoparticle-based model delivery system to guide the rational design of gene delivery to the liver. 1. Synthesis and characterization. Bioconjugate Chem. 2005, 16, 1063–1070. [Google Scholar] [CrossRef]
- Luo, D.; Haverstick, K.; Belcheva, N.; Han, E.; Saltzman, W.M. Poly(ethylene glycol)-conjugated pamam dendrimer for biocompatible, high-efficiency DNA delivery. Macromolecules 2002, 35, 3456–3462. [Google Scholar] [CrossRef]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. Pegylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef]
- Heinz, H.; Pramanik, C.; Heinz, O.; Ding, Y.; Mishra, R.K.; Marchon, D.; Flatt, R.J.; Estrela-Lopis, I.; Llop, J.; Moya, S.; et al. Nanoparticle decoration with surfactants: Molecular interactions, assembly, and applications. Surf. Sci. Rep. 2017, 72, 1–58. [Google Scholar] [CrossRef]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quntify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A decade of the protein corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef] [PubMed]
- Schöttler, S.; Becker, G.; Winzen, S.; Steinbach, T.; Mohr, K.; Landfester, K.; Mailänder, V.; Wurm, F.R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. [Google Scholar] [CrossRef]
- Schöttler, S.; Landfester, K.; Mailänder, V. Controlling the stealth effect of nanocarriers through understanding the protein corona. Angew. Chem. Int. Ed. 2016, 55, 8806–8815. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Lee, B.J. Protein corona: A new approach for nanomedicine design. Int. J. Nanomed. 2017, 12, 3137–3151. [Google Scholar] [CrossRef]
- Walkey, C.D.; Chan, W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef]
- Hellstrand, E.; Lynch, I.; Andersson, A.; Drakenberg, T.; Dahlbäck, B.; Dawson, K.A.; Linse, S.; Cedervall, T. Complete high-density lipoproteins in nanoparticle corona. FEBS J. 2009, 276, 3372–3381. [Google Scholar] [CrossRef]
- Liu, N.; Tang, M.; Ding, J. The interaction between nanoparticles-protein corona complex and cells and its toxic effect on cells. Chemosphere 2020, 245, 125624. [Google Scholar] [CrossRef]
- Zhdanov, V.P. Formation of a protein corona around nanoparticles. Curr. Opin. Colloid Interface Sci. 2019, 41, 95–103. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Pan, J.; Jiang, X.; Ji, Y.; Li, Y.; Qu, Y.; Zhao, Y.; Wu, X.; Chen, C. Revealing the binding structure of the protein corona on gold nanorods using synchrotron radiation-based techniques: Understanding the reduced damage in cell membranes. J. Am. Chem. Soc. 2013, 135, 17359–17368. [Google Scholar] [CrossRef]
- Ramezani, F.; Rafii-Tabar, H. An in-depth view of human serum albumin corona on gold nanoparticles. Mol. Biosyst. 2015, 11, 454–462. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. A closer look into the ubiquitin corona on gold nanoparticles by computational studies. New J. Chem. 2015, 39, 2474–2482. [Google Scholar] [CrossRef]
- Shao, Q.; Hall, C.K. Protein adsorption on nanoparticles: Model development using computer simulation. J. Phys. Condens. Matter 2016, 28, 414019. [Google Scholar] [CrossRef]
- Shao, Q.; Hall, C.K. Allosteric effects of gold nanoparticles on human serum albumin. Nanoscale 2017, 9, 380–390. [Google Scholar] [CrossRef]
- Yang, J.; Wang, B.; You, Y.; Chang, W.J.; Tang, K.; Wang, Y.C.; Zhang, W.; Ding, F.; Gunasekaran, S. Probing the modulated formation of gold nanoparticles-beta-lactoglobulin corona complexes and their applications. Nanoscale 2017, 9, 17758–17769. [Google Scholar] [CrossRef] [PubMed]
- Tollefson, E.J.; Allen, C.R.; Chong, G.; Zhang, X.; Rozanov, N.D.; Bautista, A.; Cerda, J.J.; Pedersen, J.A.; Murphy, C.J.; Carlson, E.E.; et al. Preferential binding of cytochrome c to anionic ligand-coated gold nanoparticles: A complementary computational and experimental approach. ACS Nano 2019, 13, 6856–6866. [Google Scholar] [CrossRef] [PubMed]
- Power, D.; Rouse, I.; Poggio, S.; Brandt, E.; Lopez, H.; Lyubartsev, A.; Lobaskin, V. A multiscale model of protein adsorption on a nanoparticle surface. Model. Simul. Mater. Sci. Eng. 2019, 27, 084003. [Google Scholar] [CrossRef]
- Lu, X.; Xu, P.; Ding, H.M.; Yu, Y.S.; Huo, D.; Ma, Y.Q. Tailoring the component of protein corona via simple chemistry. Nat. Commun. 2019, 10, 4520. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Lee, M.J. Influence of the alanine side-chain methyl group on the peptide-gold nanoparticles interactions. J. Mol. Liq. 2020, 302, 112528. [Google Scholar] [CrossRef]
- Jahan Sajib, M.S.; Sarker, P.; Wei, Y.; Tao, X.; Wei, T. Protein corona on gold nanoparticles studied with coarse-grained simulations. Langmuir 2020, 36, 13356–13363. [Google Scholar] [CrossRef] [PubMed]
- Käkinen, A.; Ding, F.; Chen, P.; Mortimer, M.; Kahru, A.; Ke, P.C. Interaction of firefly luciferase and silver nanoparticles and its impact on enzyme activity. Nanotechnology 2013, 24, 345101. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Radic, S.; Chen, R.; Chen, P.; Geitner, N.K.; Brown, J.M.; Ke, P.C. Direct observation of a single nanoparticle-ubiquitin corona formation. Nanoscale 2013, 5, 9162–9169. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Chen, R.; Chen, P.; Wen, Y.; Ke, P.C.; Cho, S.S. Computational and experimental characterizations of silver nanoparticle-apolipoprotein biocorona. J. Phys. Chem. B 2013, 117, 13451–13456. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Seabrook, S.A.; Nedumpully-Govindan, P.; Chen, P.; Yin, H.; Waddington, L.; Chandana Epa, V.; Winkler, D.A.; Kirby, J.K.; Ding, F.; et al. Thermostability and reversibility of silver nanoparticle-protein binding. Phys. Chem. Chem. Phys. 2015, 17, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Nayak, P.S.; Borah, S.M.; Gogoi, H.; Asthana, S.; Bhatnagar, R.; Jha, A.N.; Jha, S. Lactoferrin adsorption onto silver nanoparticle interface: Implications of corona on protein conformation, nanoparticle cytotoxicity and the formulation adjuvanticity. Chem. Eng. J. 2019, 361, 470–484. [Google Scholar] [CrossRef]
- Ge, C.; Du, J.; Zhao, L.; Wang, L.; Liu, Y.; Li, D.; Yang, Y.; Zhou, R.; Zhao, Y.; Chai, Z.; et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. USA 2011, 108, 16968–16973. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, B.; Gregory, W.E.; Zhu, J.; Dasetty, S.; Karakaya, M.; Brown, J.M.; Rao, A.M.; Barrows, J.K.; Sarupria, S.; Podila, R. Influence of carbon nanomaterial defects on the formation of protein corona. RSC Adv. 2015, 5, 82395–82402. [Google Scholar] [CrossRef]
- Lee, H. Adsorption of plasma proteins onto pegylated single-walled carbon nanotubes: The effects of protein shape, peg size and grafting density. J. Mol. Graph. Model. 2017, 75, 1–8. [Google Scholar] [CrossRef]
- Lee, H.; Larson, R.G. Adsorption of plasma proteins onto pegylated lipid bilayers: The effect of peg size and grafting density. Biomacromolecules 2016, 17, 1757–1765. [Google Scholar] [CrossRef]
- Settanni, G.; Zhou, J.; Suo, T.; Schöttler, S.; Landfester, K.; Schmid, F.; Mailänder, V. Protein corona composition of poly(ethylene glycol)-and poly (phosphoester)-coated nanoparticles correlates strongly with the amino acid composition of the protein surface. Nanoscale 2017, 9, 2138–2144. [Google Scholar] [CrossRef]
- Settanni, G.; Schäfer, T.; Muhl, C.; Barz, M.; Schmid, F. Poly-sarcosine and poly(ethylene-glycol) interactions with proteins investigated using molecular dynamics simulations. Comput. Struct. Biotechnol. J. 2018, 16, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Lopez, H.; Lobaskin, V. Coarse-grained model of adsorption of blood plasma proteins onto nanoparticles. J. Chem. Phys. 2015, 143. [Google Scholar] [CrossRef] [PubMed]
- Lopez, H.; Brandt, E.G.; Mirzoev, A.; Zhurkin, D.; Lyubartsev, A.; Lobaskin, V. Multiscale Modelling of Bionano Interface. Adv. Exp. Med. Biol. 2017, 947, 173–206. [Google Scholar] [PubMed]
- Yu, G.; Zhou, J. Understanding the curvature effect of silica nanoparticles on lysozyme adsorption orientation and conformation: A mesoscopic coarse-grained simulation study. Phys. Chem. Chem. Phys. 2016, 18, 23500–23507. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Ahlstrom, L.S.; Brooks, C.L. Exploring protein–nanoparticle interactions with coarse-grained protein folding models. Small 2017, 13, 1603748. [Google Scholar] [CrossRef]
- Pilkington, E.H.; Xing, Y.; Wang, B.; Kakinen, A.; Wang, M.; Davis, T.P.; Ding, F.; Ke, P.C. Effects of protein corona on iapp amyloid aggregation, fibril remodelling, and cytotoxicity. Sci. Rep. 2017, 7, 2455. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, M.; Zhang, D.; Yang, Q.; Liu, T.; Lei, R.; Zhu, S.; Zhao, Y.; Chen, C. Probing adsorption behaviors of bsa onto chiral surfaces of nanoparticles. Small 2018, 14, 1703982. [Google Scholar] [CrossRef]
- Tavakol, M.; Montazeri, A.; Naghdabadi, R.; Hajipour, M.J.; Zanganeh, S.; Caracciolo, G.; Mahmoudi, M. Disease-related metabolites affect protein-nanoparticle interactions. Nanoscale 2018, 10, 7108–7115. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.; Davis, T.P.; Ke, P.C.; Wu, Y.; Ding, F. Understanding effects of pamam dendrimer size and surface chemistry on serum protein binding with discrete molecular dynamics simulations. ACS Sustain. Chem. Eng. 2018, 6, 11704–11715. [Google Scholar] [CrossRef]
- Fardanesh, A.; Zibaie, S.; Shariati, B.; Attar, F.; Rouhollah, F.; Akhtari, K.; Shahpasand, K.; Saboury, A.A.; Falahati, M. Amorphous aggregation of tau in the presence of titanium dioxide nanoparticles: Biophysical, computational, and cellular studies. Int. J. Nanomed. 2019, 14, 901–911. [Google Scholar] [CrossRef]
- Moya, C.; Escudero, R.; Malaspina, D.C.; De La Mata, M.; Hernández-Saz, J.; Faraudo, J.; Roig, A. Insights into preformed human serum albumin corona on iron oxide nanoparticles: Structure, effect of particle size, impact on mri efficiency, and metabolization. ACS Appl. Bio Mater. 2019, 2, 3084–3094. [Google Scholar] [CrossRef]
- Derakhshankhah, H.; Hosseini, A.; Taghavi, F.; Jafari, S.; Lotfabadi, A.; Ejtehadi, M.R.; Shahbazi, S.; Fattahi, A.; Ghasemi, A.; Barzegari, E.; et al. Molecular interaction of fibrinogen with zeolite nanoparticles. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Dzubiella, J. Probing the protein corona around charged macromolecules: Interpretation of isothermal titration calorimetry by binding models and computer simulations. Colloid Polym. Sci. 2020, 298, 747–759. [Google Scholar] [CrossRef]
- Sanchez-Guzman, D.; Giraudon-Colas, G.; Marichal, L.; Boulard, Y.; Wien, F.; Degrouard, J.; Baeza-Squiban, A.; Pin, S.; Renault, J.P.; Devineau, S. In situ analysis of weakly bound proteins reveals molecular basis of soft corona formation. ACS Nano 2020, 14, 9073–9088. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, T.; Jing, C.; Liu, S.; Zhang, C.; Alvarez, P.J.J.; Chen, W. Nanocrystal facet modulation to enhance transferrin binding and cellular delivery. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Hassanian, M.; Aryapour, H.; Goudarzi, A.; Javan, M.B. Are zinc oxide nanoparticles safe? A structural study on human serum albumin using in vitro and in silico methods. J. Biomol. Struct. Dyn. 2021, 39, 330–335. [Google Scholar] [CrossRef]
- Lee, H. Effects of nanoparticle electrostatics and protein–protein interactions on corona formation: Conformation and hydrodynamics. Small 2020, 16, 1906598. [Google Scholar] [CrossRef]
- Vroman, L. Effect of adsorbed proteins on the wettability of hydrophilic and hydrophobic solids. Nature 1962, 196, 476–477. [Google Scholar] [CrossRef]
- Vilaseca, P.; Dawson, K.A.; Franzese, G. Understanding and modulating the competitive surface-adsorption of proteins through coarse-grained molecular dynamics simulations. Soft Matter 2013, 9, 6978–6985. [Google Scholar] [CrossRef]
- Vilanova, O.; Mittag, J.J.; Kelly, P.M.; Milani, S.; Dawson, K.A.; Rädler, J.O.; Franzese, G. Understanding the kinetics of protein–nanoparticle corona formation. ACS Nano 2016, 10, 10842–10850. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Competitive binding of proteins to gold nanoparticles disclosed by molecular dynamics simulations. J. Phys. Chem. C 2015, 119, 22172–22180. [Google Scholar] [CrossRef]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Multiscale molecular dynamics simulation of multiple protein adsorption on gold nanoparticles. Int. J. Mol. Sci. 2019, 20, 3539. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Jiao, B.; Shi, X.; Valle, R.P.; Fan, Q.; Zuo, Y.Y. Physicochemical properties of nanoparticles regulate translocation across pulmonary surfactant monolayer and formation of lipoprotein corona. ACS Nano 2013, 7, 10525–10533. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.-M.; Ma, Y.-Q. Computer simulation of the role of protein corona in cellular delivery of nanoparticles. Biomaterials 2014, 35, 8703–8710. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Kang, S.G.; Tian, X.; Garate, J.A.; Zhao, L.; Ge, C.; Zhou, R. Protein corona mitigates the cytotoxicity of graphene oxide by reducing its physical interaction with cell membrane. Nanoscale 2015, 7, 15214–15224. [Google Scholar] [CrossRef]
- Lee, H. Effect of protein corona on nanoparticle–lipid membrane binding: The binding strength and dynamics. Langmuir 2021, 37, 3751–3760. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H. Molecular Modeling of Protein Corona Formation and Its Interactions with Nanoparticles and Cell Membranes for Nanomedicine Applications. Pharmaceutics 2021, 13, 637. https://doi.org/10.3390/pharmaceutics13050637
Lee H. Molecular Modeling of Protein Corona Formation and Its Interactions with Nanoparticles and Cell Membranes for Nanomedicine Applications. Pharmaceutics. 2021; 13(5):637. https://doi.org/10.3390/pharmaceutics13050637
Chicago/Turabian StyleLee, Hwankyu. 2021. "Molecular Modeling of Protein Corona Formation and Its Interactions with Nanoparticles and Cell Membranes for Nanomedicine Applications" Pharmaceutics 13, no. 5: 637. https://doi.org/10.3390/pharmaceutics13050637
APA StyleLee, H. (2021). Molecular Modeling of Protein Corona Formation and Its Interactions with Nanoparticles and Cell Membranes for Nanomedicine Applications. Pharmaceutics, 13(5), 637. https://doi.org/10.3390/pharmaceutics13050637





