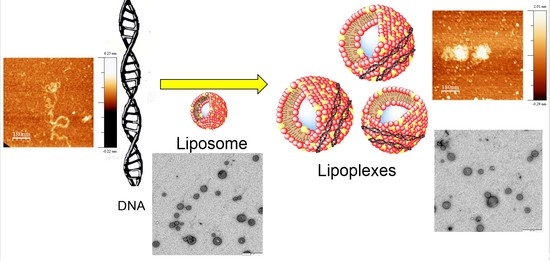
Cationic Single-Chained Surfactants with a Functional Group at the End of the Hydrophobic Tail DNA Compacting Efficiency
Abstract
1. Introduction
2. Materials and Methods
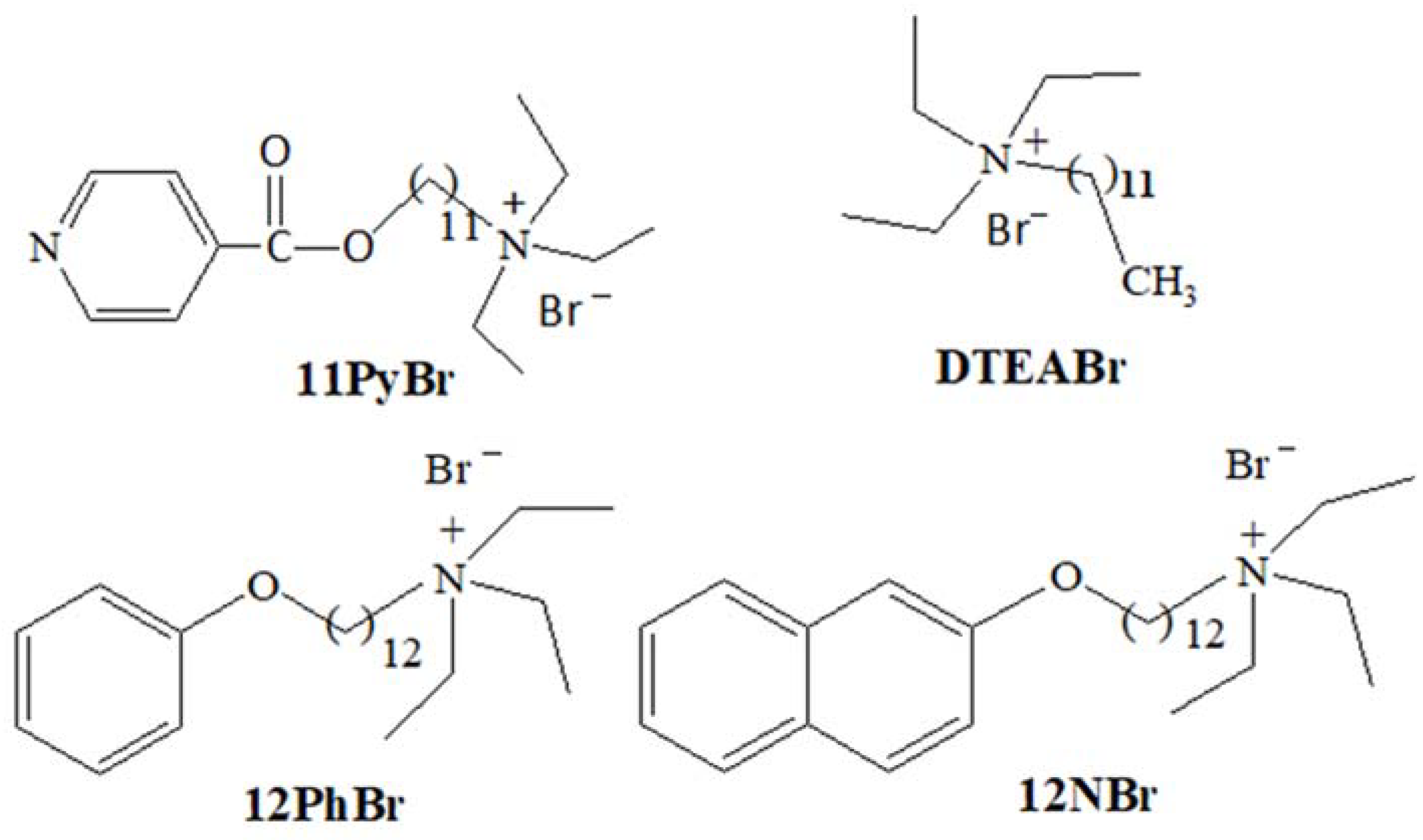
2.1. Materials
2.2. Preparation of Surfactant/ctDNA Solutions
2.3. Preparation of Lipoplexes
2.4. UV-Visible Spectroscopy
2.5. Fluorescence Measurements
2.6. Zeta Potential Measurements
2.7. Dynamic Light Scattering (DLS) Measurements
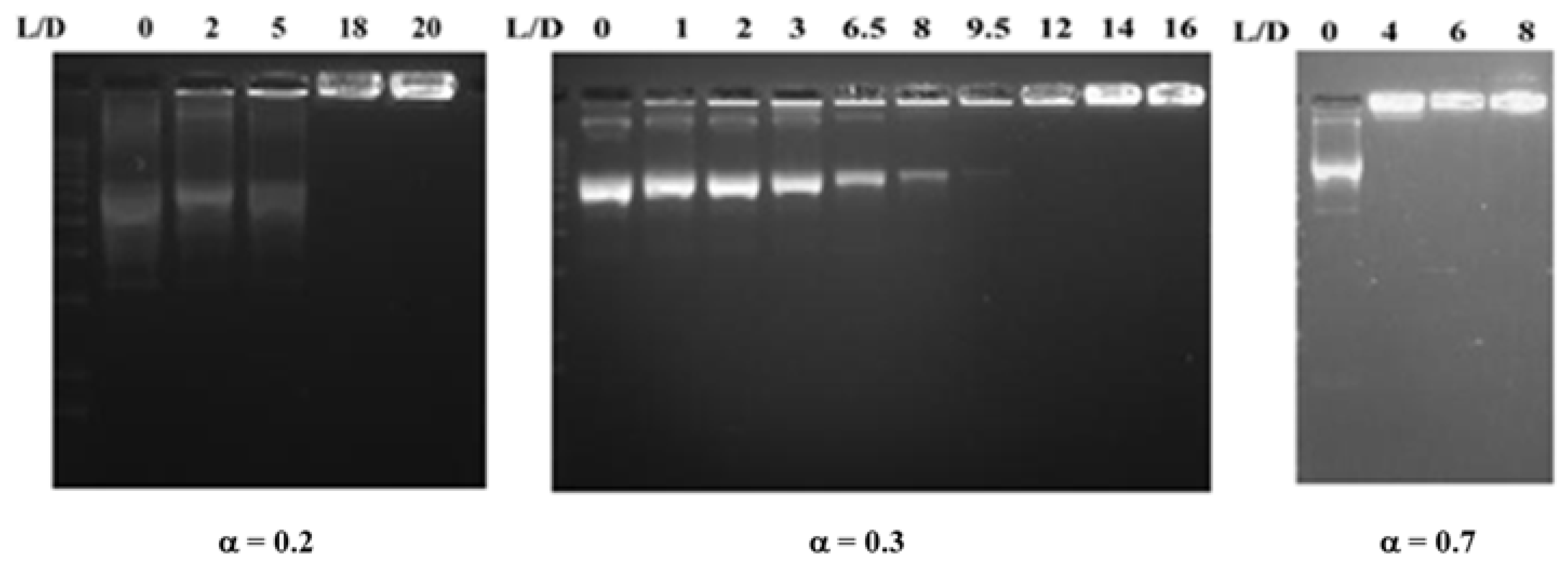
2.8. Agarose Gel Electrophoresis
2.9. Circular Dichroism Spectra
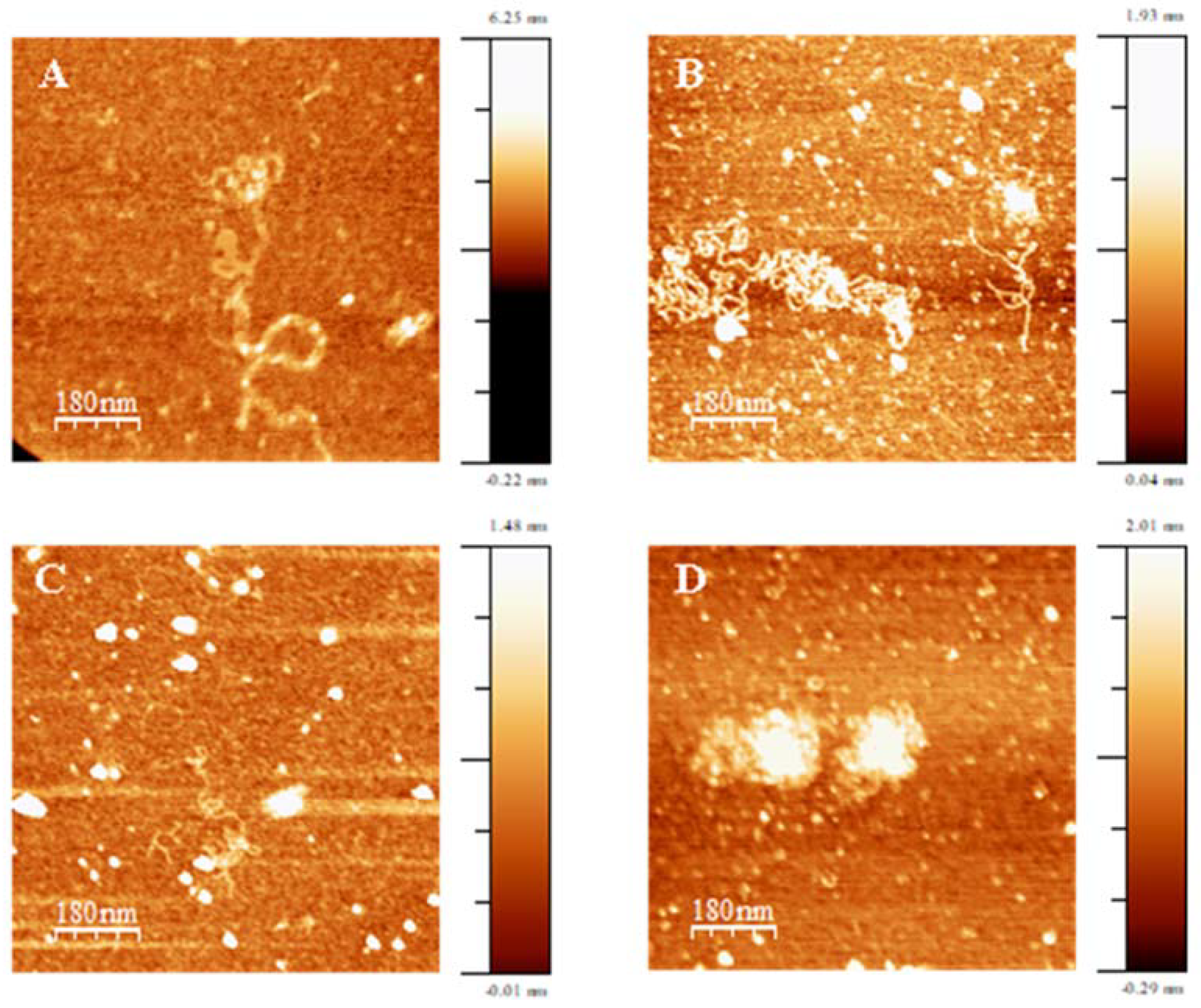
2.10. Atomic Force Microscopy, AFM
2.11. Electron Transmission Microscopy (TEM)
2.12. In Vitro Cytotoxicity Assays
2.13. Transfection Assays
2.14. Statistical Analysis
3. Results and Discussion
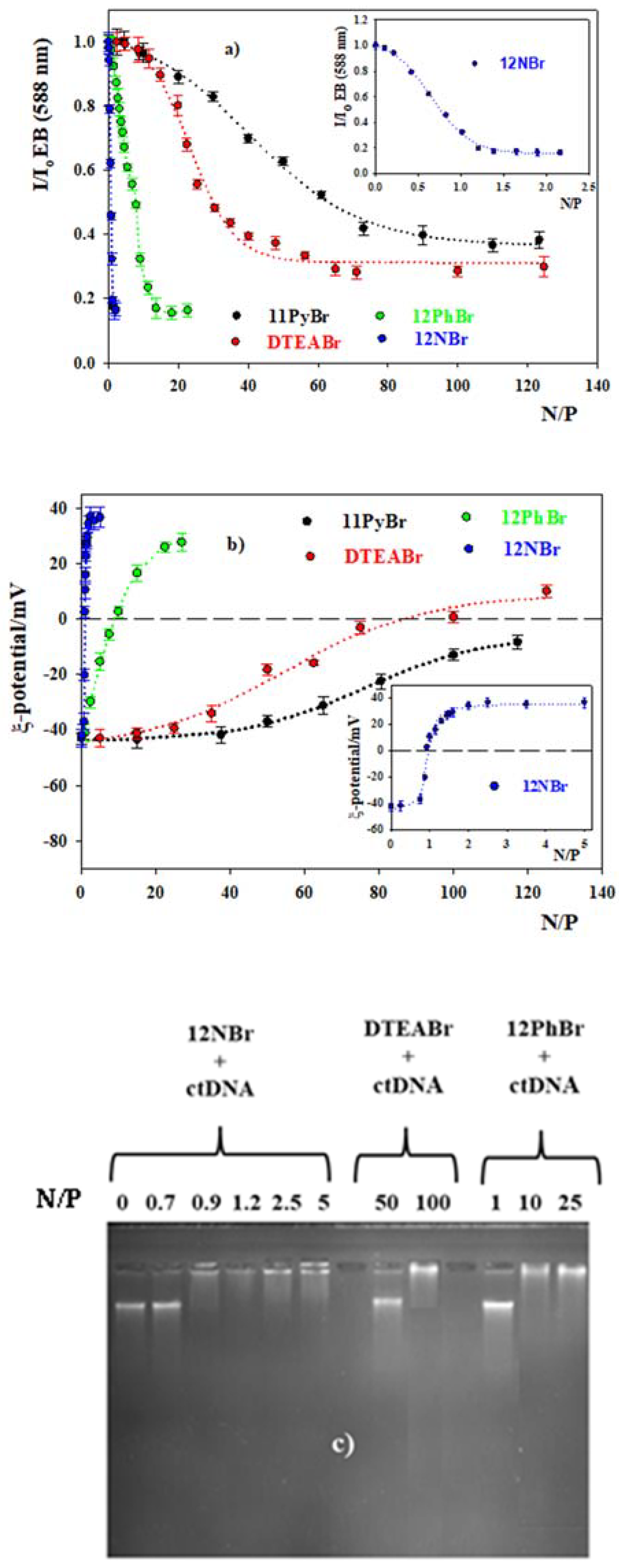
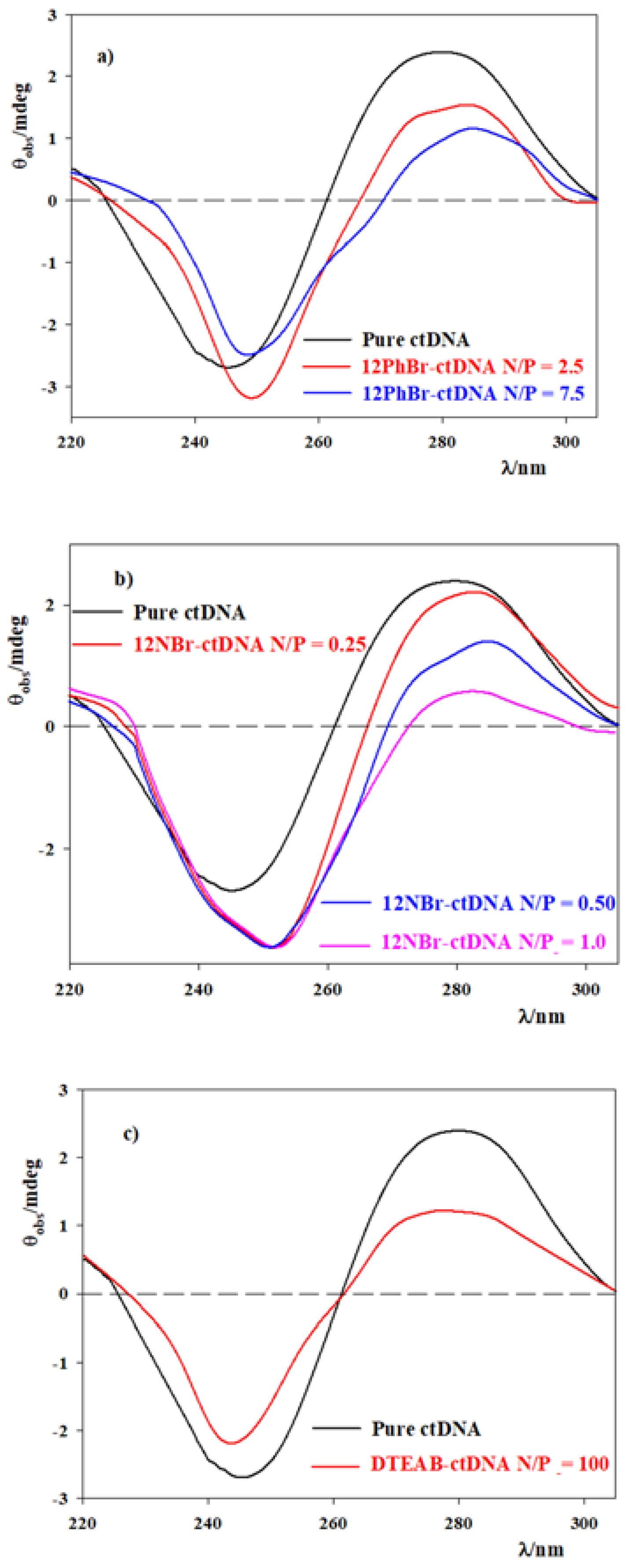
3.1. DNA Compacting Efficiency of the Surfactants
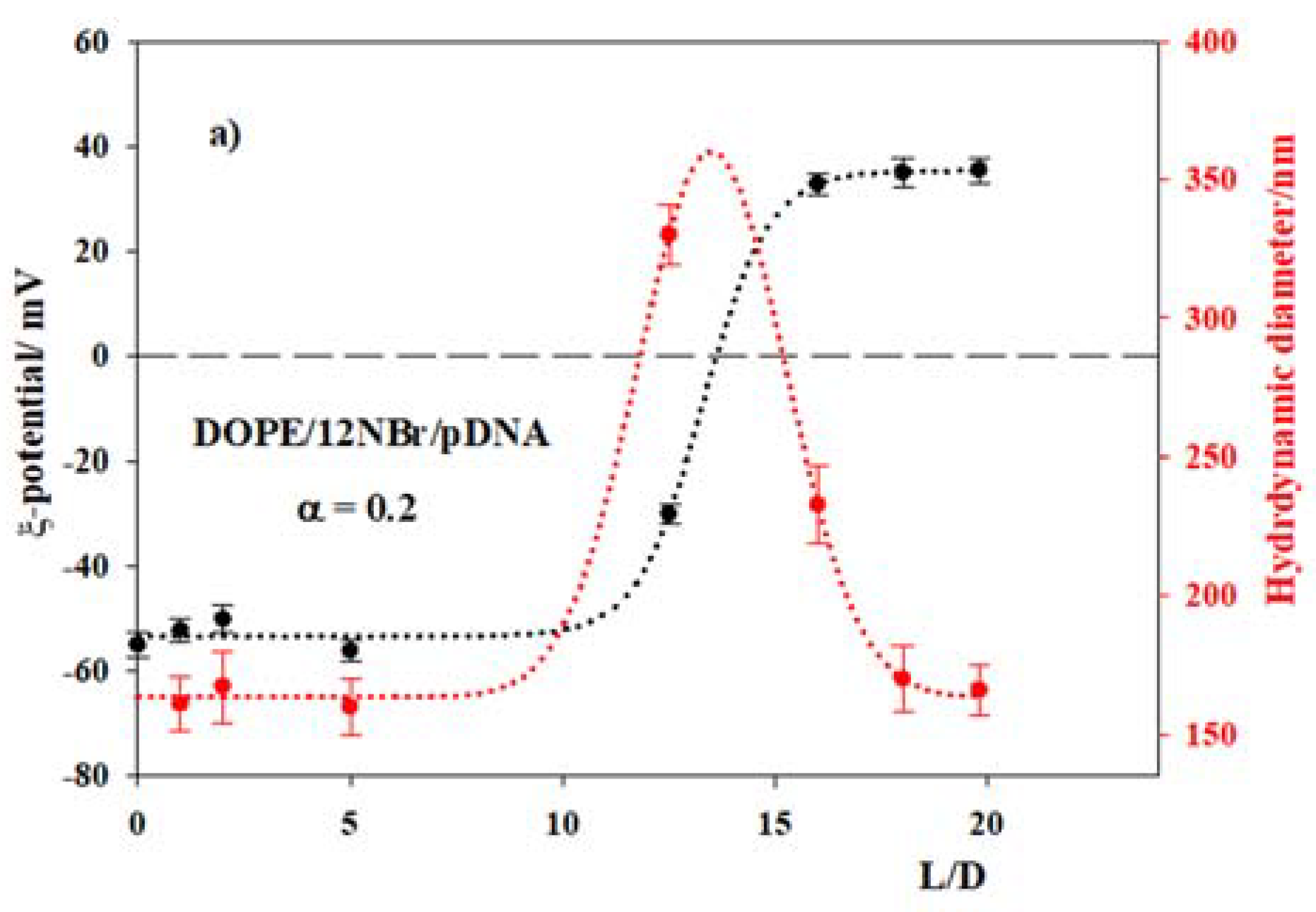
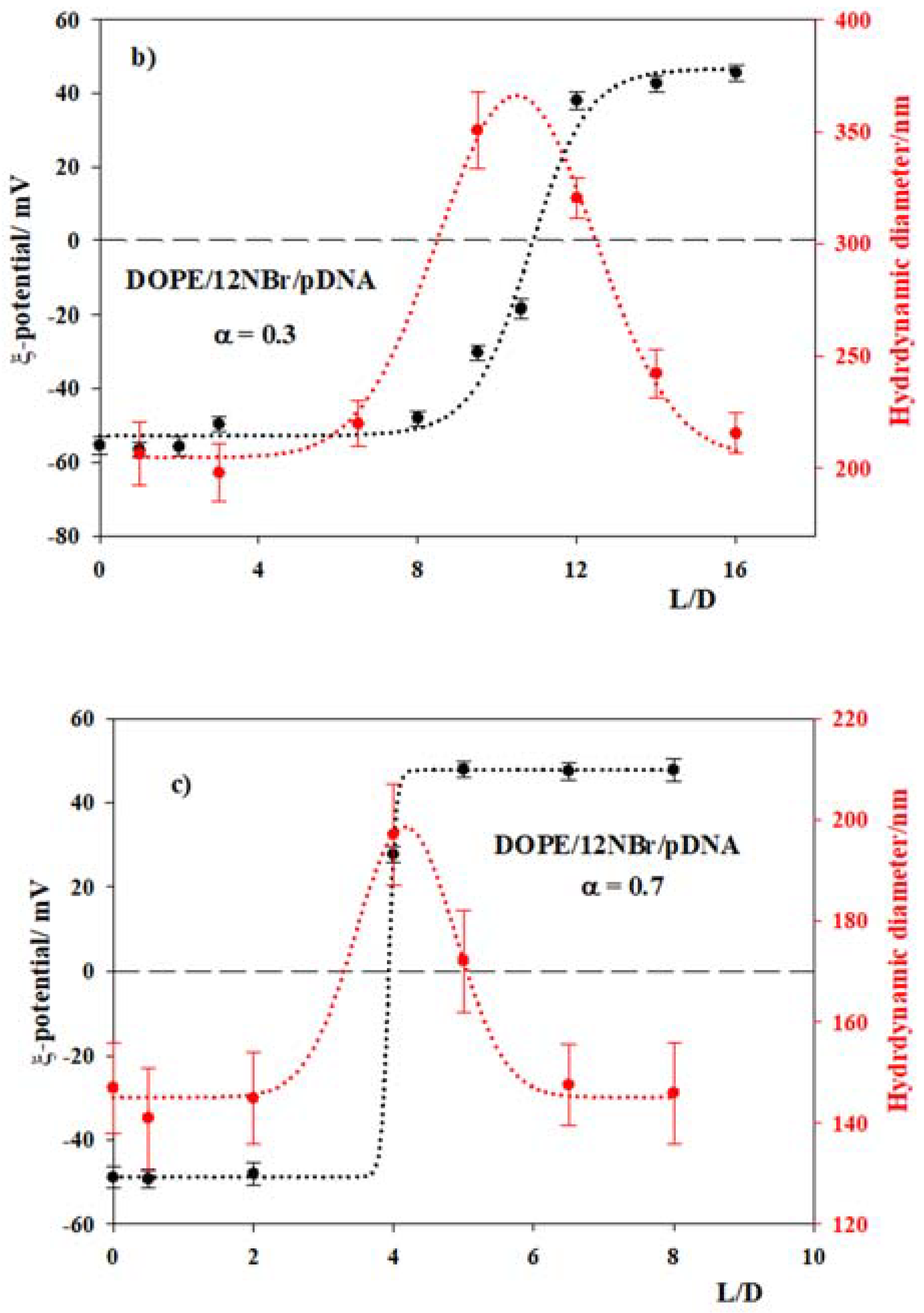
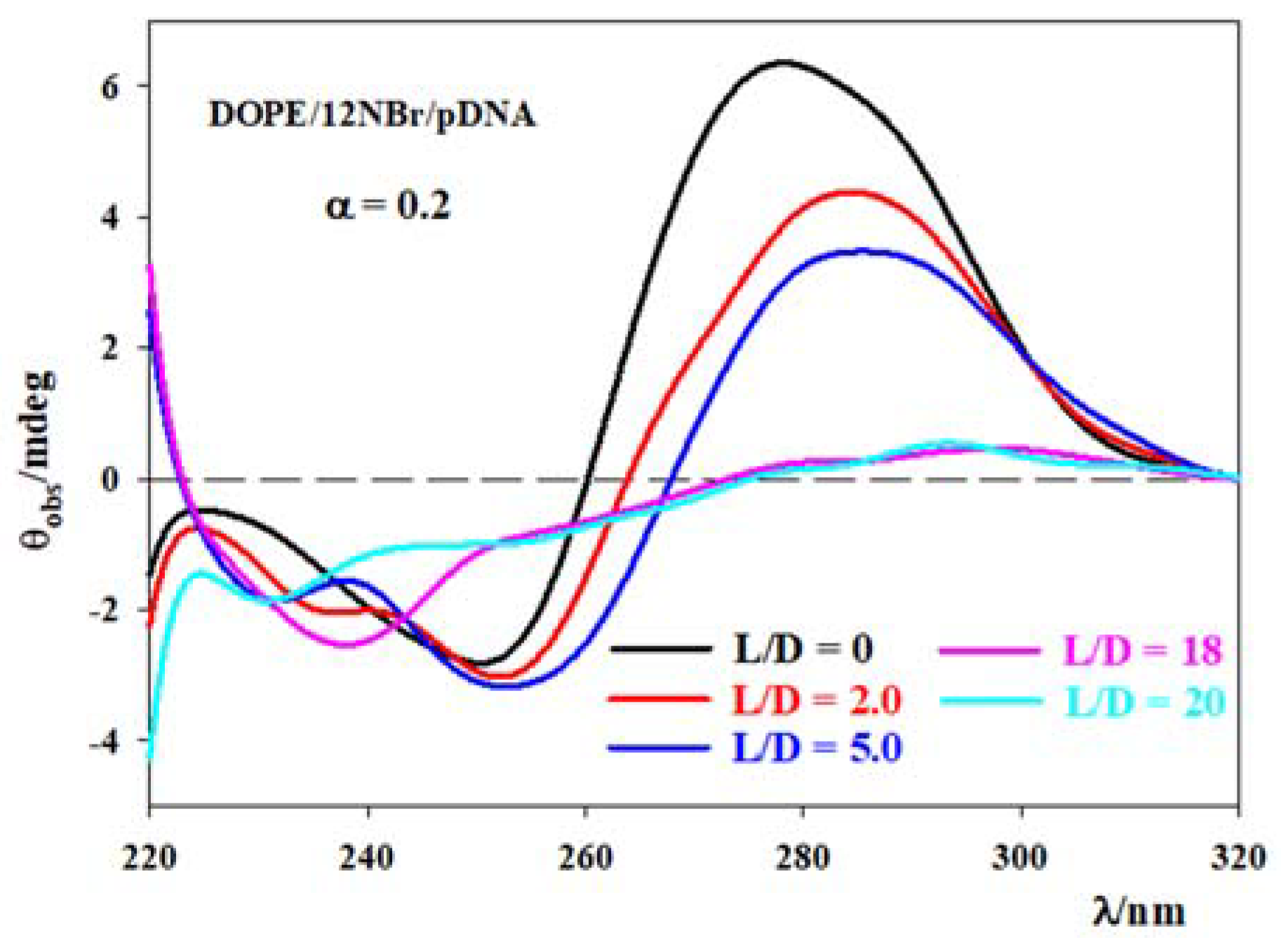
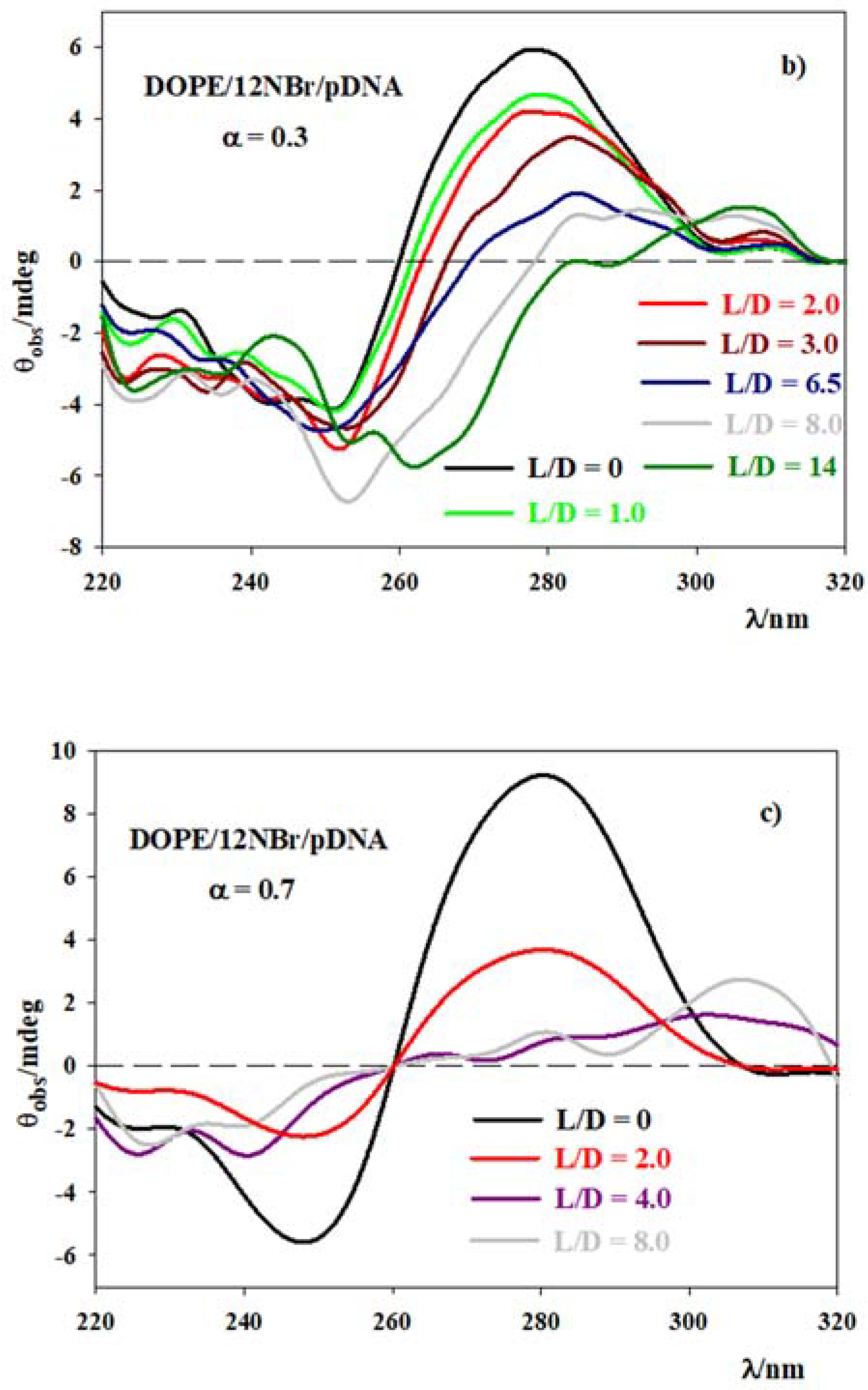
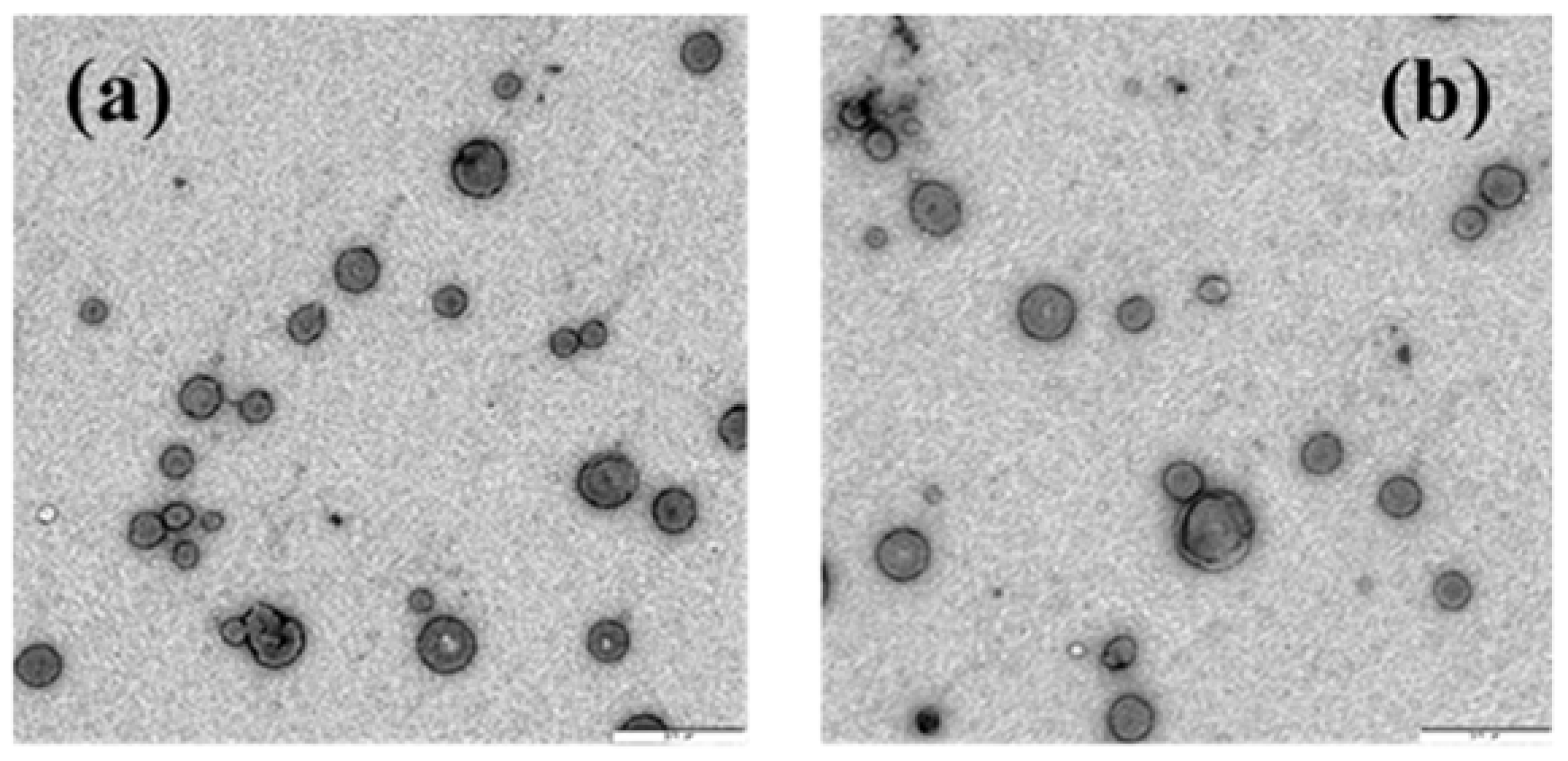
3.2. DOPE/12NBr/pDNA Lipoplexes
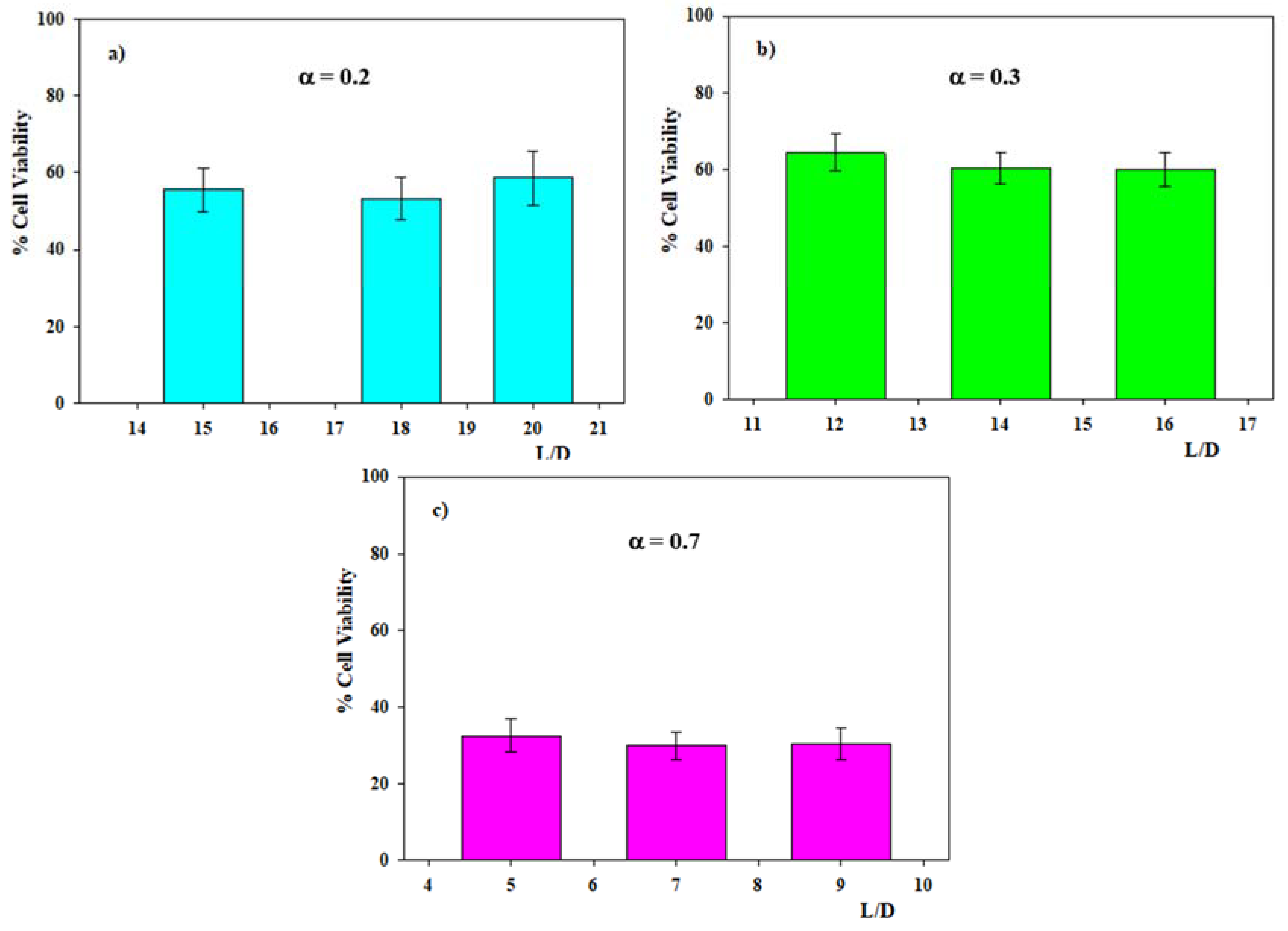
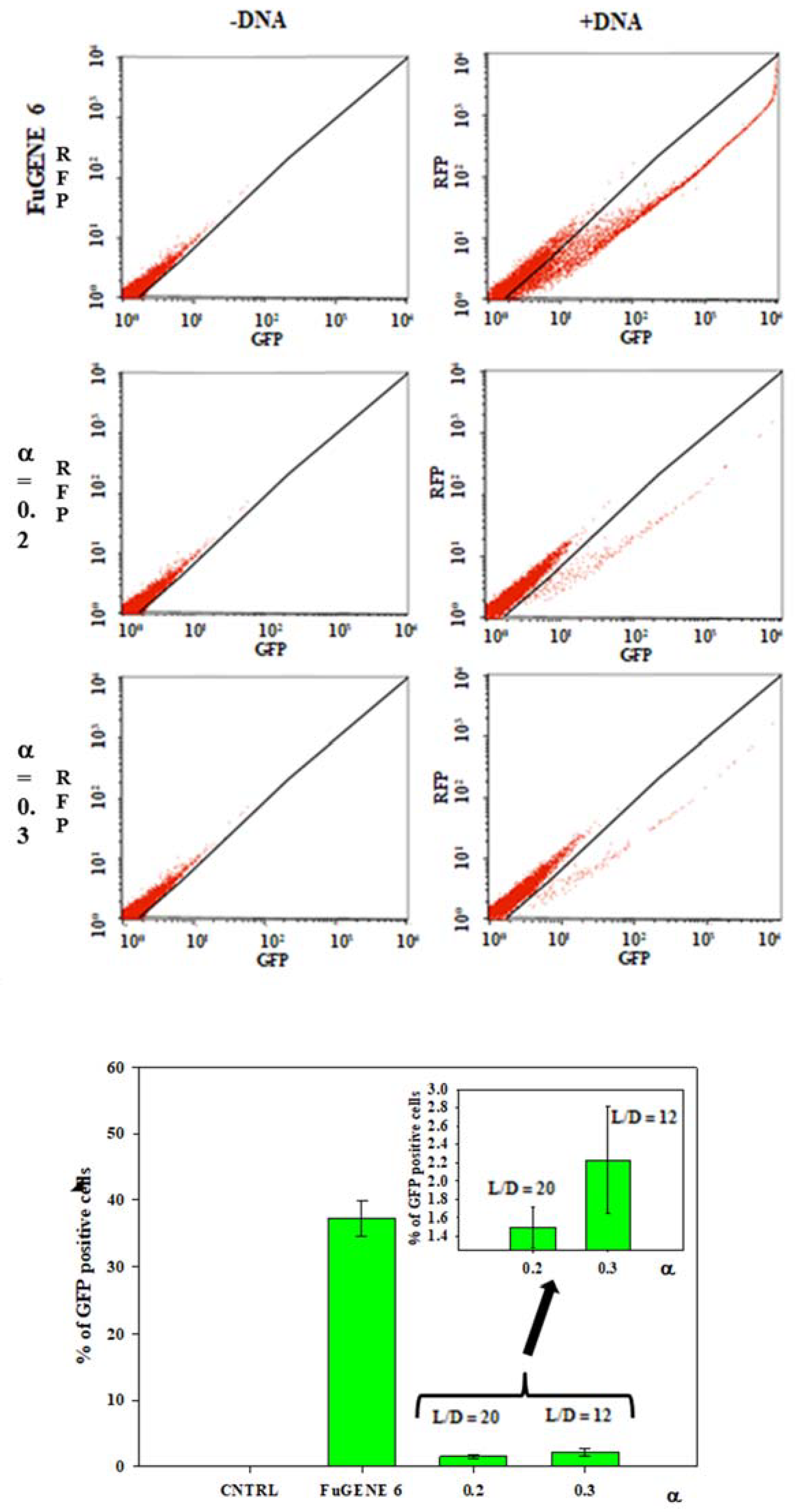
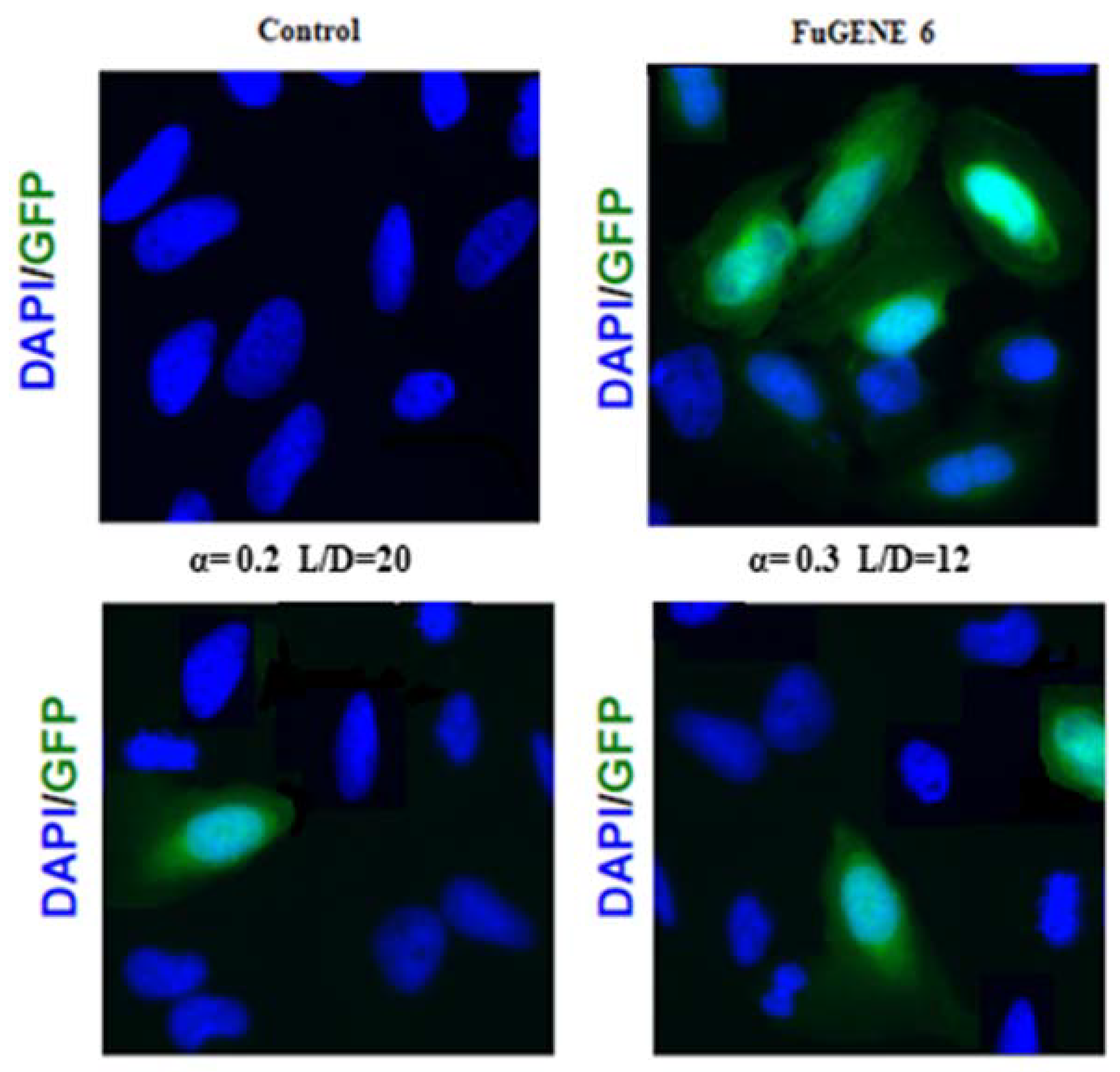
3.3. Transfection Efficiency of DOPE/12NBr/pDNA Lipoplexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kirby, A.J.; Camilleri, P.; Engberts, J.B.F.N.; Feiters, C.; Nolte, R.J.M.; Söderman, O.; Bergsma, M.; Bell, P.C.; Fielden, M.L.; García Rodríguez, C.L.; et al. Gemini surfactants: New synthetic vectors for gene transfection. Angew. Chem. Int. Ed. 2003, 42, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Zuhorn, I.; Engberts, J.B.F.N.; Hoekstra, C. Gene delivery by cationic lipid vectors: Overcoming cellular barriers. Eur. Biophys. J. 2007, 36, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and gemini surfactant: Impact on gene therapy: Part I. Nanomedicine 2016, 11, 289–306. [Google Scholar] [CrossRef]
- Ahmed, T.; Kamel, A.O.; Wettig, S.D. Interactions between DNA and gemini surfactant: Impact on gene therapy: Part II. Nanomedicine 2016, 11, 403–420. [Google Scholar] [CrossRef]
- Kim, K.; Chen, W.C.W.; Heo, Y.; Wang, Y. Polycations and their biomedical applications. Progr. Polym. Sci. 2016, 60, 18–50. [Google Scholar] [CrossRef]
- Zhi, D.; Bai, Y.; Yang, J.; Cui, S.; Zhao, Y.; Chen, H.; Zhang, S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 2018, 253, 117–140. [Google Scholar] [CrossRef]
- Kumar, V.; Palazzolo, S.; Bayda, S.; Corona, G.; Toffoli, G.; Rizzolio, F. DNA nanotechnology for cancer therapy. Theranostics 2016, 6, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, T.; Pan, A.; Das, S.; Moulik, S.P. Ratiometric interactions of Anionic Surfactants with Calf Thymus DNA Bound cationic surfactants: Study II. J. Surfactants Deterg. 2018, 21, 127–137. [Google Scholar] [CrossRef]
- Ren, W.; Liu, H.; Yang, W.; Fan, Y.; Yang, L.; Wang, Y.; Liu, C.; Li, Z. A cytometric bead assay for sensitive DNA detection based on-enzyme-free signal amplification of hybridization chain reaction. Biosens. Bioelectron. 2013, 49, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Lopez-López, M.; López-Cornejo, P.; Martín, V.I.; Ostos, F.J.; Checa-Rodríguez, C.; Prados-Carvajal, R.; Lebron, J.A.; Huertas, P.; Moya, M.L. Importance of hydrophobic interactions in the single-chained cationic surfactant-DNA complexation. J. Colloid Interface Sci. 2018, 521, 197–205. [Google Scholar] [CrossRef]
- Li, X.; Sun, D.; Chen, Y.; Wang, K.; He, Q.; Wang, G. Studying compaction-decompaction of DNA molecules induced by surfactants. Biochem. Biophys. Res. Commun. 2018, 495, 2559–2565. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, H.; He, C.; Qiao, F.; Wang, S.; Wang, Y. DNA Condensation Induced by a Star-Shaped Hexameric Cationic Surfactant. ACS Appl. Mater. Interfaces 2017, 9, 23333–23341. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Bryant, Z.; Smith, S.B. Ten years of tension: Single-molecule DNA mechanics. Nature 2003, 421, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Ritort, F.; Mihardja, S.; Smith, S.B.; Bustamante, C. Condensation transition in DNA-polyaminoamide dendrimer fibers studied using optical tweezers. Phys. Rev. Lett. 2006, 96, 118301. [Google Scholar] [CrossRef]
- Husale, S.; Grange, W.; Karle, M.; Bürgi, S.; Hegner, M. Interaction of cationic surfactants with DNA: A single-molecule study. Nucleic Acids Res. 2008, 36, 1443–1449. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xu, L.; Dong, S.; Hao, J. Thermo-reversible capture and release of DNA by zwitterionic surfactants. Soft Matter 2016, 12, 7495–7504. [Google Scholar] [CrossRef]
- Hao, B.; Wang, K.; Zhou, Y.; Sui, C.; Wang, L.; Bai, R.; Yang, Z. Label-free detecting of the compaction and decompaction of ctDNA molecules induced by surfactants with SERS based on a nanoPAA-ZnCl2-AuLs solid substrate. ACS Omega 2020, 5, 1109–1119. [Google Scholar] [CrossRef]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The head group evolution of cationic lipids for gene delivery. Bioconj. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef]
- Martín, V.I.; Sarrión, B.; López-López, M.; López-Cornejo, P.; Robina, I.; Moyá, M.L. Reversibility of the interactions between a novel surfactant derived from lysine and biomolecules. Colloids Surf. B 2015, 135, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Zinchenko, A.; Pyshkina, O.; Lezov, A.; Sergeyev, O.; Yoshikawa, K. Single DNA Molecules: Compaction and Decompaction. In DNA Interactions with Polymers and Surfactants; Dias, R.S., Lindman, B., Eds.; Wiley: Hoboken, NJ, USA, 2008; pp. 59–88. [Google Scholar] [CrossRef]
- Cai, K.; Cheng, R.; Wang, C.; Xia, Y.; Xu, T.; Gan, C. Interactions with ctDNA of novel sugar-based gemini cationic surfactants. Int. J. Biol. Macro. 2020, 156, 805–811. [Google Scholar] [CrossRef]
- Zakharova, L.A.; Kaupova, G.I.; Gabdrakhmanov, D.R.; Gaynanova, G.A.; Ermakova, E.A.; Mukhitov, A.R.; Galkina, I.V.; Cheresiz, S.V.; Pokrovsky, A.G.; Skvortsova, P.V.; et al. Alkyl triphenylphosphonium surfactants as nucleic acid carriers: Complexation efficacy toward DNA decamers, interaction with lipid bilayers and cytotoxicity studie. Phys. Chem. Chem. Phys. 2019, 21, 16706–16717. [Google Scholar] [CrossRef] [PubMed]
- Piecuch, A.; Lamch, L.; Paluch, E.; Obłazk, E.; Wilk, K.A. Biofilm prevention by dicephalic cationic surfactants and their interactions with DNA. J. Appl. Microbiol. 2016, 121, 682–692. [Google Scholar] [CrossRef]
- Lebrón, J.A.; Ostos, F.J.; López-López, M.; Moyá, M.L.; Kardell, O.; Sánchez, A.; Carrasco, C.J.; García-Calderón, M.; García-Calderón, C.B.; Rosado, I.V.; et al. Preparation and characterization of metallomicelles of Ru(II): Cytotoxic activity and use as vectors. Colloid Surf. B 2019, 175, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Bhar, R.; Kaur, G.; Mehta, S.K. Exploring interactions of copper hybrid surfactants with calf thymus-DNA. J. Mol. Liq. 2017, 241, 715–721. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, Y.; Zhang, Z.; Guo, X. Micellization of lactosylammonium surfactants with different counter ions and their interaction with DNA. J. Chem. Eng. Data 2016, 61, 2969–2978. [Google Scholar] [CrossRef]
- Fisicaro, E.; Compari, E.; Bacciottini, F.; Contardi, L.; Pongiluppi, E.; Barbero, N.; Viscardi, G.; Quagliotto, P.; Donofrio, G.; Krafft, M.P. Nonviral gene-delivery by highly fluorinated gemini bispyridinium surfactant-based DNA nanoparticles. J. Colloid Interface Sci. 2017, 487, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dong, S.; Hao, J. Recent progress of magnetic surfactants: Self-assembly, properties and functions. Curr. Opin. Colloid Interface Sci. 2018, 35, 81–90. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, Z.; Qiao, H.; Liu, M.; Shen, M.; Yuan, T.; Chen, J.; Dionysiou, D.D. Spectroscopic study on interaction between three cationic surfactants with different alkyl chain lengths and DNA. Spectrochim. Acta A 2015, 151, 237–246. [Google Scholar] [CrossRef]
- Pietralik, Z.; Kumita, J.R.; Dobson, C.M.; Kozak, M. The influence of novel gemini surfactants containing cycloalkylside-chains on the structural phases of DNA in solution. Colloid Surf. B 2015, 131, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Aicart, E.; Junquera, E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid Interface Sci. 2016, 233, 161–175. [Google Scholar] [CrossRef]
- Kumar, K.; Barrán-Berdón, L.A.; Datta, S.; Múñoz-Úbeda, M.; Aicart-Ramos, C.; Kondaiah, P.; Junquera, E.; Aicart, E. A delocalizable cationic headgroup together with an oligooxyethylene spacer in gemini cationic lipids improves their biological activity as vectors of plasmid DNA. J. Mater. Chem. 2015, 2, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Grueso, E.; Roldan, E.; Perez-Tejeda, P.; Kuliszewska, E.; Molero, B.; Brecker, L.; Giraldez-Perez, R.M. Reversible DNA compaction induced by partial intercalation of 16-Ph-16 gemini surfactants: Evidence of triple helix formation. Phys. Chem. Chem. Phys. 2018, 20, 24902–24914. [Google Scholar] [CrossRef] [PubMed]
- Barbero, N.; Magistris, C.; Quagliotto, P.; Bonandini, L.; Barolo, C.; Buscaino, R.; Compari, C.; Contardi, L.; Fisicaro, E.; Viscardi, G. Synthesis, physicochemical characterization, and interaction with DNA of long-alkyl-chain gemini pyridinium surfactants. ChemPlusChem 2015, 80, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Wettig, S.D.; Verrall, R.D.; Foldvari, M. Gemini surfactants: A new family of building blocks for non-viral gene delivery systems. Curr. Gene Ther. 2008, 8, 9–23. [Google Scholar] [CrossRef]
- Sarrión, B.; Bernal, E.; Martín, V.I.; López-López, M.; López-Cornejo, P.; García-Calderón, M.; Moyá, M.L. Binding of 12-s-12 dimeric surfactants to calf thymus DNA: Evaluation of the spacer length influence. Colloid Surf. B 2016, 144, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, L.; van Eijk, M.C.P.; Söderman, P. Compaction of DNA by gemini surfactants: Effects of surfactant architecture. J. Colloid Interface Sci. 2002, 252, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.G.; Oliveira, I.S.; do Vale, M.L.C.; Marques, E.F. Serine-based gemini surfactants with different spacer linkages: From self-assembly to DNA compaction. Soft Matter 2014, 46, 9352–9361. [Google Scholar] [CrossRef]
- Bombelli, C.; Borocci, S.; Diociaiuti, M.; Galantini, L.; Luciani, P.; Mancini, G.; Sacco, M.G. Role of the spacer of cationic gemini amphiphiles in the condensation of DNA. Langmuir 2005, 21, 10271–10274. [Google Scholar] [CrossRef] [PubMed]
- Hubčík, L.; Galliková, D.; Pullmannová, P.; Lacinová, L.; Sulová, Z.; Hanulová, M.; Funari, S.S.; Devínsky, F.; Uhríková, D. DNA-DOPE-gemini surfactants complexes at low surface charge density: From structure to transfection efficiency. Gen. Physiol. Biophys. 2018, 37, 57–69. [Google Scholar] [CrossRef] [PubMed]
- López, A.; López-Cornejo, P.; López-López, M.; Lebrón, J.A.; Ostos, F.J.; Pérez-Alfonso, D.; Oviedo, J.; Laschewsky, A.; Moyá, M.L. Influence of the degree of oligomerization of surfactants on the DNA/surfactant interaction. Colloids Surf. B 2019, 182, 110399. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Nohara, A.; Negishi, Y.; Hawakami, H. Alkylimidazolium end-modified poly(ethylene glycol) to form the mono-ion complex with plasmid DNA for in vivo gene delivery. Biomacromolecules 2014, 15, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Chao, H.; Tan, L.F.; Yuan, L.X.; Ji, L.N. Ruthenium(II) mixed-ligand complexes containing 2-(6-methyl-3-chromonyl)-imidazol[4,5-f][1,10]-phenanthroline: Synthesis, DNA-binding and photoclavage studies. Inorg. Chim. Acta 2006, 359, 3807–3814. [Google Scholar] [CrossRef]
- Secco, F.; Venturini, M.; Biver, T.; Sanchez, T.; Prado-Gotor, R.; Grueso, E. Solvent effects on the kinetics of the interaction of 1-pyrenecarboxaldehyde with calf thymus DNA. J. Phys. Chem. B 2010, 114, 4686–4691. [Google Scholar] [CrossRef] [PubMed]
- Ostos, F.J. Use of Calixarene-Derived Macrocyclic Compounds as Drug Nanocarriers. Ph.D. Thesis, University of Seville, Sevilla, Spain, 2020. [Google Scholar]
- Martín, M.V.; Ostos, F.J.; Ángulo, M.; Márquez, A.; López-Cornejo, P.; López-López, M.; Carmona, M.T.; Moyá, M.L. Host-guest interactions between cyclodextrins and surfactants with functional groups at the end of the hydrophobic tail. J. Colloid Interface Sci. 2017, 491, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Martín, V.I.; Angulo, M.; López-Cornejo, P.; López-López, M.; Marchena, M.J.; Moyá, M.L. Stoppering/unstoppering of a rotaxane formed between an N-hetorycle ligand containing surfactant: β-cyclodextrin pseudorotaxane and pentacyanoferrate(II) ions. J. Colloid Interface Sci. 2017, 497, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Thin-Film hydration followed by extrusion method for liposome preparation. Methods Mol. Biol. 2017, 1522, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Moyá, M.L.; López-López, M.; Lebrón, J.A.; Ostos, F.J.; Pérez, D.; Camacho, V.; Beck, V.; Merino-Bohórquez, V.; Camean, M.; Madinabeitia, N.; et al. Preparation and characterization of new liposomes. Bactericidal activity of cefepime encapsulated into cationic liposomes. Pharmaceutics 2019, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell Sensitivity Assays: The MTT Assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar] [CrossRef]
- Cosa, G.; Focsaneau, K.S.; Mc Lean, J.R.N.; Mac Namee, J.P.; Scaiano, J.C. Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solutions. Photochem. Photobiol. 2001, 73, 585–599. [Google Scholar] [CrossRef]
- Adjimatera, N.; Neal, A.P.; Blagbrough, I.S. Fluorescence techniques in non-viral gene therapy. In Fluorescence Spectroscopy in Biology Advanced Methods and Their Applications to Membranes, Proteins, DNA, and Cells; Hof, M., Hutterer, R., Fidler, V., Eds.; Springer: Berlin, Germany, 2005; Volume 3, pp. 201–228. [Google Scholar] [CrossRef]
- Geall, A.J.; Blagbrough, I.S. Rapid and sensitive ethidium bromide fluorescence quenching assay of polyamine conjugate-DNA interactions for the analysis of lipoplex formation in gene therapy. J. Pharm. Biomed. Anal. 2000, 22, 849–859. [Google Scholar] [CrossRef]
- Múñoz-Úbeda, M.; Misra, S.K.; Barrán-Berdón, A.L.; Aicart-Ramos, C.; Sierra, M.B.; Biswas, J.; Kondaiah, P.; Junquera, E.; Bhattacharya, S.; Aicart, E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy? J. Am. Chem. Soc. 2011, 133, 18014–18017. [Google Scholar] [CrossRef]
- Shklovskii, B.I. Screening of a micro Akon by multitalented ion: Correlation-induced inversion charge. Phys. Rev. E 1999, 60, 5802–5811. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Nucleic Acid Structure and Recognition; Oxford University Press: New York, NY, USA, 2002; pp. 89–138. [Google Scholar]
- Veeralakshmi, S.; Neru, S.; Sabapathi, G.; Arunachalam, S.; Venuvanaligam, P.; Kumar, P.; Anusha, C.; Ravikumar, V. Single and double chain surfactant-cobalt (III) complexes: The impact of hydrophobicity on the interaction with calf thymus DNA, and their biological activities. RSC Adv. 2015, 5, 31746–31758. [Google Scholar] [CrossRef]
- Camesano, A.; Nagarajan, R. Micelle formation and cmc of gemini surfactants: A thermodynamic model. Colloids Surf. A 2000, 167, 165–177. [Google Scholar] [CrossRef]
- Shim, G.; Kim, D.; Le, Q.-V.; Park, G.T.; Kwon, T.; Oh, Y.-K. Nonviral delivery systems for cancer gene therapy: Strategies and challenges. Curr. Gene Ther. 2018, 18, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Hidai, C.; Kitano, H. Nonviral gene therapy for cancer: A Review. Diseases 2018, 6, 57. [Google Scholar] [CrossRef]
- Huang, Z.; Tomitaka, A.; Raymond, A.; Nair, M. Current application of CRISPR/Cas9 gene-editing technique to eradication of HIV/AIDS. Gene Ther. 2017, 24, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Sticca, R.P.; Holmes, L.M.; Burgin, K.E.; Li, J.; Williamson, J.; Evans, L.; Smith, S.J.; Stephenson, J.J.; Wagner, T.E. Dendritoma vaccination combined with low dose interleukin-2 in metastatic melanoma patients induced immunological and clinical responses. Int. J. Oncol. 2006, 28, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Blagbrough, I.S.; Metwally, A.A. siRNA and Gene Formulation for Efficient Gene Therapy. In Gene Therapy—Tools and Potential Applications; Martin Molina, F., Ed.; InTechOpen: Rijeka, Croatia, 2013; pp. 135–173. [Google Scholar] [CrossRef]
- Blagbrough, I.S.; Metwally, A.A.; Ahmed, O.A.A. Polyamine-based agents for gene and siRNA transfer. In Polyamine Drug Discovery; Woster, P.M., Casero, R.A., Jr., Eds.; RSC: Cambridge, UK, 2012; pp. 205–237. [Google Scholar] [CrossRef]
- Ghonaim, H.M.; Ahmed, O.A.A.; Pourzand, C.; Blagbrough, I.S. Varying the chain length in N4,N9-diacyl spermines: Non-viral lipopolyamine vectors for efficient plasmid DNA formulation. Mol. Pharm. 2008, 5, 1111–1121. [Google Scholar] [CrossRef]
- Metwally, A.A.; Pourzand, C.; Blagbrough, I.S. Efficient gene silencing by self-assembled complexes of siRNA and symmetrical fatty acid amides of spermine. Pharmaceutics 2011, 3, 125–140. [Google Scholar] [CrossRef]
- Gallego-Yerga, L.; Blanco-Fernández, L.; Martínez-Negro, M.; Vélaz, I.; Aicart, E.; Junquera, E.; Mellet, M.D.C.O.; De Ilarduya, C.T.; Fernández, J.M.M.G.; Benito, J.M. Plasmid-Templated Control of DNA Cyclodextrin nanoparticles morphology through Molecular Vector Design for Effective Gene Delivery. Chem. A Eur. J. 2018, 24, 3825–3835. [Google Scholar] [CrossRef]
- Blanco, J.L.J.; Benito, J.M.; Mellet, C.O.; García-Fernández, J.M. Molecular nanoparticle-based gene delivery systems. J. Drug Deliv. Sci. Technol. 2017, 42, 18–37. [Google Scholar] [CrossRef]
- Ostos, F.J.; Lebrón, J.A.; Moyá, M.L.; López-López, M.; Sánchez, A.; Clavero, A.; García-Calderón, C.B.; Rosado, I.; Cornejo, P.L. P-Sulfocalix[6]arene as nanocarrier for controlled delivery of doxorubicin. Chem. Asian J. 2017, 12, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Lin, W.; Liu, D.; He, C. Application of liposomal technologies for delivery of platinum analogs in oncology. Int. J. Nanomed. 2013, 8, 3309–3319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barron, L.G.; Uyechi, L.S.; Szoka, F.C.; Szoka, F.C., Jr. Cationic lipids are essential for gene delivery mediated by intravenous administration of lipoplexes. Gene Ther. 1999, 6, 1179–1183. [Google Scholar] [CrossRef]
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Hope, M.J. Lipid-based formulations of antisense oligonucleotides for systemic delivery applications. Method. Enzym. 2000, 313, 322–341. [Google Scholar] [CrossRef]
- Srinivas, R.; Samanta, S.; Chaudhuri, A. Cationic amphiphiles: Promising carriers of genetic materials in gene therapy. Chem. Soc. Rev. 2009, 38, 3326–3338. [Google Scholar] [CrossRef] [PubMed]
- Mignet, N.; Vandermeulen, G.; Pembouong, G.; Largeau, C.; Thompson, B.; Spanedda, M.V.; Wasungu, L.; Rols, M.-P.; Bessodes, M.; Bureau, M.F.; et al. Cationic and anionic lipoplexes inhibit gene transfection by electroporation in vivo. J. Gene Med. 2010, 12, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Andrzejewska, W.; Wilkowska, P.; Cheabaszczewska, M.; Kozak, M. The study of complexation between dicationic surfactants and the DNA duplex using structural and spectroscopic methods. RSC Adv. 2017, 7, 26006–26018. [Google Scholar] [CrossRef]
- Xu, Y.; Szoka, F. Mechanism of DNA release from cationic liposomes/DNA complexes used in cell transfection. Biochemistry 1996, 35, 5616–5623. [Google Scholar] [CrossRef] [PubMed]
- Duangjit, S.; Pamornpathomkul, B.; Opanasopit, P.; Rojanarata, T.; Obata, Y.; Takayama, K.; Ngawhirunpat, T. Role of the charge, carbon chain length, and content of surfactant on the skin penetration of meloxicam-loaded liposomes. Int. J. Nanomed. 2014, 9, 2005–2017. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebrón, J.A.; López-Cornejo, P.; García-Dionisio, E.; Huertas, P.; García-Calderón, M.; Moyá, M.L.; Ostos, F.J.; López-López, M. Cationic Single-Chained Surfactants with a Functional Group at the End of the Hydrophobic Tail DNA Compacting Efficiency. Pharmaceutics 2021, 13, 589. https://doi.org/10.3390/pharmaceutics13040589
Lebrón JA, López-Cornejo P, García-Dionisio E, Huertas P, García-Calderón M, Moyá ML, Ostos FJ, López-López M. Cationic Single-Chained Surfactants with a Functional Group at the End of the Hydrophobic Tail DNA Compacting Efficiency. Pharmaceutics. 2021; 13(4):589. https://doi.org/10.3390/pharmaceutics13040589
Chicago/Turabian StyleLebrón, José Antonio, Pilar López-Cornejo, Elena García-Dionisio, Pablo Huertas, Margarita García-Calderón, María Luisa Moyá, Francisco José Ostos, and Manuel López-López. 2021. "Cationic Single-Chained Surfactants with a Functional Group at the End of the Hydrophobic Tail DNA Compacting Efficiency" Pharmaceutics 13, no. 4: 589. https://doi.org/10.3390/pharmaceutics13040589
APA StyleLebrón, J. A., López-Cornejo, P., García-Dionisio, E., Huertas, P., García-Calderón, M., Moyá, M. L., Ostos, F. J., & López-López, M. (2021). Cationic Single-Chained Surfactants with a Functional Group at the End of the Hydrophobic Tail DNA Compacting Efficiency. Pharmaceutics, 13(4), 589. https://doi.org/10.3390/pharmaceutics13040589