Mechanistic Insights into Side Effects of Troglitazone and Rosiglitazone Using a Novel Inverse Molecular Docking Protocol
Abstract
1. Introduction
2. Materials and Methods
2.1. Inverse Molecular Docking
2.2. Binding Site Comparisons Using ProBiS Web Server
2.3. Method Validation
3. Results
3.1. Targets of Troglitazone in the Human Proteome
3.2. Targets of Rosiglitazone in the Human Proteome
3.3. Binding Site Comparison Using ProBiS Web Server
3.3.1. Troglitazone
3.3.2. Rosiglitazone
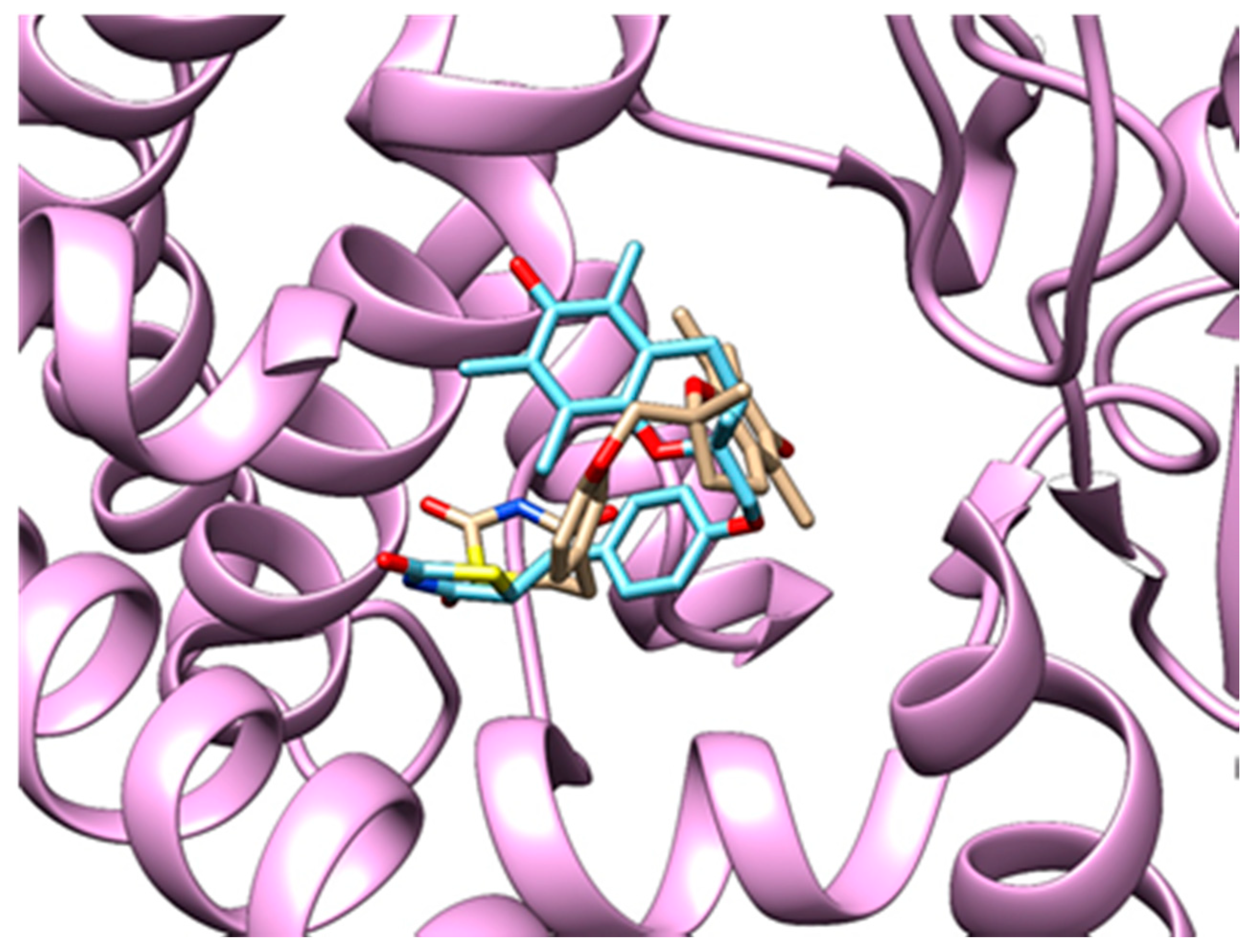
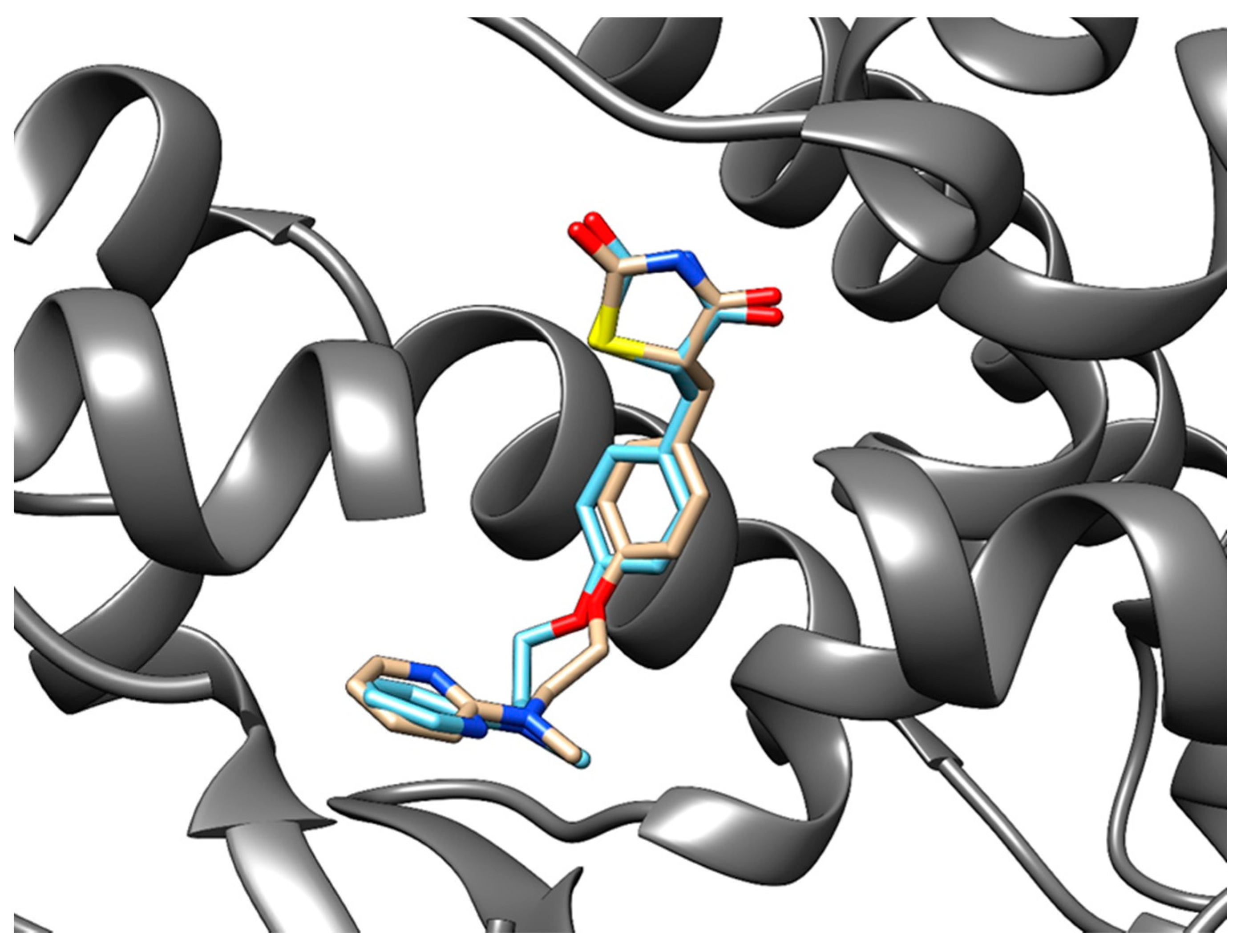
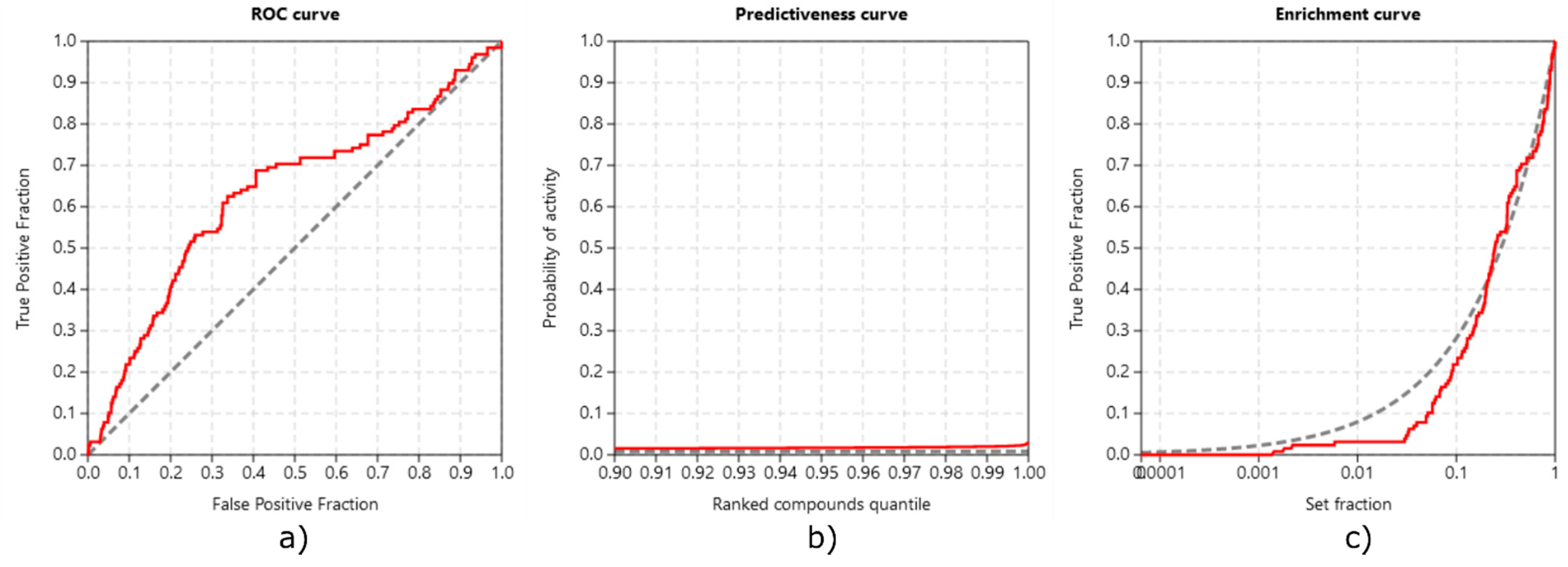
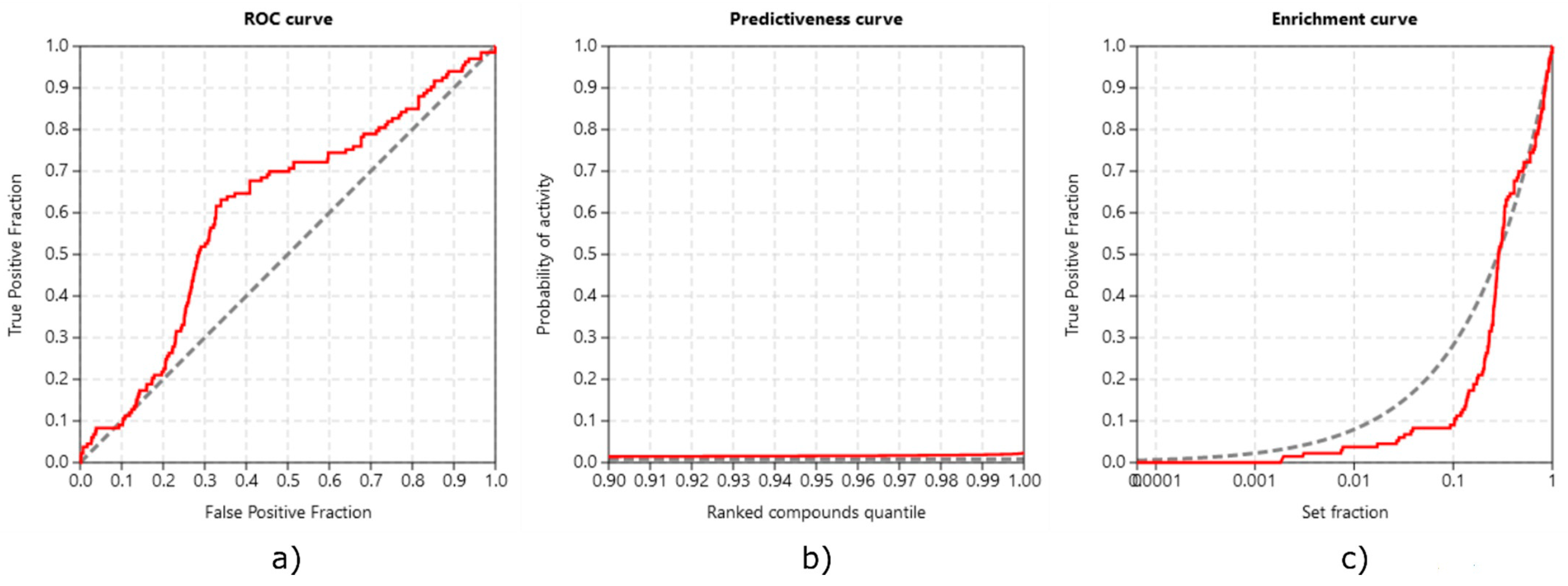
3.3.3. Method Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lebovitz, H.E. Thiazolidinediones: The Forgotten Diabetes Medications. Curr. Diab. Rep. 2019, 19, 151. [Google Scholar] [CrossRef] [PubMed]
- Schoonjans, K.; Auwerx, J. Thiazolidinediones: An Update. Lancet 2000, 355, 1008–1010. [Google Scholar] [CrossRef]
- DeFronzo, R.A. Pharmacologic Therapy for Type 2 Diabetes Mellitus. Ann. Intern. Med. 1999, 131, 281–303. [Google Scholar] [CrossRef] [PubMed]
- Martín-Timón, I.; Sevillano-Collantes, C.; Segura-Galindo, A.; del Cañizo-Gómez, F.J. Type 2 Diabetes and Cardiovascular Disease: Have All Risk Factors the Same Strength? World J. Diabetes 2014, 5, 444–470. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.M.; Moore, L.B.; Smith-Oliver, T.A.; Wilkison, W.O.; Willson, T.M.; Kliewer, S.A. An Antidiabetic Thiazolidinedione Is a High Affinity Ligand for Peroxisome Proliferator-Activated Receptor γ (PPARγ). J. Biol. Chem. 1995, 270, 12953–12956. [Google Scholar] [CrossRef] [PubMed]
- Fanny, L. Staels Bart Fibrates, Glitazones, and Peroxisome Proliferator—Activated Receptors. Arter. Thromb. Vasc. Biol. 2010, 30, 894–899. [Google Scholar] [CrossRef]
- Nanjan, M.J.; Mohammed, M.; Prashantha Kumar, B.R.; Chandrasekar, M.J.N. Thiazolidinediones as Antidiabetic Agents: A Critical Review. Bioorganic Chem. 2018, 77, 548–567. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.T. Mechanisms of Troglitazone Hepatotoxicity. Chem. Res. Toxicol. 2003, 16, 679–687. [Google Scholar] [CrossRef]
- Yokoi, T. Troglitazone. In Adverse Drug Reactions; Uetrecht, J., Ed.; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 419–435. ISBN 978-3-642-00663-0. [Google Scholar]
- Ogimura, E.; Nakagawa, T.; Deguchi, J.; Sekine, S.; Ito, K.; Bando, K. Troglitazone Inhibits Bile Acid Amidation: A Possible Risk Factor for Liver Injury. Toxicol. Sci. 2017, 158, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, H.; Zhang, C.; Zhang, Q.; Guo, J.; Li, J.; Yuan, H.; Li, L.; Carmichael, P.; Peng, S. Integrating in Vitro Testing and Physiologically-Based Pharmacokinetic (PBPK) Modelling for Chemical Liver Toxicity Assessment—A Case Study of Troglitazone. Environ. Toxicol. Pharm. 2020, 74, 103296. [Google Scholar] [CrossRef]
- Przybelski, S.; Ulloa, M.E.; Rudie, M.; Cornett, M.; Strom, D. Characterization of the Troglitazone Induced Apoptosis in a Human Colon Cancer Cell Line. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Jung, K.-H.; Lee, J.H.; Park, J.-W.; Moon, S.-H.; Cho, Y.S.; Lee, K.-H. Troglitazone Exerts Metabolic and Antitumor Effects on T47D Breast Cancer Cells by Suppressing Mitochondrial Pyruvate Availability. Oncol. Rep. 2020, 43, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Nissen, S.E. The Rise and Fall of Rosiglitazone. Eur. Heart J. 2010, 31, 773–776. [Google Scholar] [CrossRef]
- Home, P.D.; Pocock, S.J.; Beck-Nielsen, H.; Gomis, R.; Hanefeld, M.; Jones, N.P.; Komajda, M.; McMurray, J.J.V. Rosiglitazone Evaluated for Cardiovascular Outcomes—An Interim Analysis. N. Engl. J. Med. 2007, 357, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Nissen, S.E.; Wolski, K. Effect of Rosiglitazone on the Risk of Myocardial Infarction and Death from Cardiovascular Causes. N. Engl. J. Med. 2007, 356, 2457–2471. [Google Scholar] [CrossRef]
- Research, C.D.E.; FDA. Drug Safety Communication: Updated Risk Evaluation and Mitigation Strategy (REMS) to Restrict Access to Rosiglitazone-Containing Medicines Including Avandia, Avandamet, and Avandaryl; FDA: Silver Spring, MD, USA, 2019.
- Research, C.D.E.; FDA. Drug Safety Communication: FDA Requires Removal of Some Prescribing and Dispensing Restrictions for Rosiglitazone-Containing Diabetes Medicines; FDA: Silver Spring, MD, USA, 2019.
- Research, C.D.E.; FDA. Drug Safety Communication: FDA Eliminates the Risk Evaluation and Mitigation Strategy (REMS) for Rosiglitazone-Containing Diabetes Medicines; FDA: Silver Spring, MD, USA, 2019.
- Wagstaff, A.J.; Goa, K.L. Rosiglitazone. Drugs 2002, 62, 1805–1837. [Google Scholar] [CrossRef]
- Wang, C.-H.; Weisel, R.D.; Liu, P.P.; Fedak, P.W.M. Verma Subodh Glitazones and Heart Failure. Circulation 2003, 107, 1350–1354. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Ung, C.Y. Prediction of Potential Toxicity and Side Effect Protein Targets of a Small Molecule by a Ligand–Protein Inverse Docking Approach. J. Mol. Graph. Model. 2001, 20, 199–218. [Google Scholar] [CrossRef]
- Ma, C.; Kang, H.; Liu, Q.; Zhu, R.; Cao, Z. Insight into Potential Toxicity Mechanisms of Melamine: An in Silico Study. Toxicology 2011, 283, 96–100. [Google Scholar] [CrossRef]
- Vallone, A.; D’Alessandro, S.; Brogi, S.; Brindisi, M.; Chemi, G.; Alfano, G.; Lamponi, S.; Lee, S.G.; Jez, J.M.; Koolen, K.J.M.; et al. Antimalarial Agents against Both Sexual and Asexual Parasites Stages: Structure-Activity Relationships and Biological Studies of the Malaria Box Compound 1-[5-(4-Bromo-2-Chlorophenyl)Furan-2-Yl]-N-[(Piperidin-4-Yl)Methyl]Methanamine (MMV019918) and Analogues. Eur. J. Med. Chem. 2018, 150, 698–718. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Huang, M.; Zou, X. Docking-Based Inverse Virtual Screening: Methods, Applications, and Challenges. Biophys. Rep. 2018, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.; Konc, J.; Samudrala, R.; Chopra, G. CANDOCK: Chemical Atomic Network-Based Hierarchical Flexible Docking Algorithm Using Generalized Statistical Potentials. J. Chem. Inf. Model. 2020, 60, 1509–1527. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Konc, J.; Janežič, D. ProBiS Algorithm for Detection of Structurally Similar Protein Binding Sites by Local Structural Alignment. Bioinformatics 2010, 26, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Konc, J.; Janežič, D. ProBiS-2012: Web Server and Web Services for Detection of Structurally Similar Binding Sites in Proteins. Nucleic Acids Res. 2012, 40, W214–W221. [Google Scholar] [CrossRef] [PubMed]
- Konc, J.; Janežič, D. ProBiS-Ligands: A Web Server for Prediction of Ligands by Examination of Protein Binding Sites. Nucleic Acids Res. 2014, 42, W215–W220. [Google Scholar] [CrossRef]
- Štular, T.; Lešnik, S.; Rožman, K.; Schink, J.; Zdouc, M.; Ghysels, A.; Liu, F.; Aldrich, C.C.; Haupt, V.J.; Salentin, S.; et al. Discovery of Mycobacterium Tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction. J. Med. Chem. 2016, 59, 11069–11078. [Google Scholar] [CrossRef] [PubMed]
- Furlan, V.; Konc, J.; Bren, U. Inverse Molecular Docking as a Novel Approach to Study Anticarcinogenic and Anti-Neuroinflammatory Effects of Curcumin. Molecules 2018, 23, 3351. [Google Scholar] [CrossRef] [PubMed]
- Konc, J.; Janežič, D. A Branch and Bound Algorithm for Matching Protein Structures. In Proceedings of the Adaptive and Natural Computing Algorithms; Beliczynski, B., Dzielinski, A., Iwanowski, M., Ribeiro, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 399–406. [Google Scholar]
- Kores, K.; Lešnik, S.; Bren, U.; Janežič, D.; Konc, J. Discovery of Novel Potential Human Targets of Resveratrol by Inverse Molecular Docking. J. Chem. Inf. Model. 2019, 59, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. A.03; Gaussian, Inc.: Wallingford, UK, 2016. [Google Scholar]
- Triballeau, N.; Acher, F.; Brabet, I.; Pin, J.-P.; Bertrand, H.-O. Virtual Screening Workflow Development Guided by the “Receiver Operating Characteristic” Curve Approach. Application to High-Throughput Docking on Metabotropic Glutamate Receptor Subtype 4. J. Med. Chem. 2005, 48, 2534–2547. [Google Scholar] [CrossRef]
- Truchon, J.-F.; Bayly, C.I. Evaluating Virtual Screening Methods: Good and Bad Metrics for the “Early Recognition” Problem. J. Chem. Inf. Model. 2007, 47, 488–508. [Google Scholar] [CrossRef] [PubMed]
- Empereur-mot, C.; Guillemain, H.; Latouche, A.; Zagury, J.-F.; Viallon, V.; Montes, M. Predictiveness Curves in Virtual Screening. J. Cheminformatics 2015, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, R.P.; Singh, S.B.; Fluder, E.M.; Kearsley, S.K. Protocols for Bridging the Peptide to Nonpeptide Gap in Topological Similarity Searches. J. Chem. Inf. Comput. Sci. 2001, 41, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Empereur-Mot, C.; Zagury, J.-F.; Montes, M. Screening Explorer–An Interactive Tool for the Analysis of Screening Results. J. Chem. Inf. Model. 2016, 56, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.H.; Kai, M.H.; Setoguchi, Y.; Eggertsen, G.; Sjöblom, P.; Setoguchi, T.; Okuda, K.I.; Björkhem, I. Cloning and Expression of CDNA of Human Delta 4-3-Oxosteroid 5 Beta-Reductase and Substrate Specificity of the Expressed Enzyme. Eur. J. Biochem. 1994, 219, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Tumber, A.; Nuzzi, A.; Hookway, E.S.; Hatch, S.B.; Velupillai, S.; Johansson, C.; Kawamura, A.; Savitsky, P.; Yapp, C.; Szykowska, A.; et al. Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells. Cell Chem. Biol. 2017, 24, 371–380. [Google Scholar] [CrossRef]
- Roesch, A.; Mueller, A.M.; Stempfl, T.; Moehle, C.; Landthaler, M.; Vogt, T. RBP2-H1/JARID1B Is a Transcriptional Regulator with a Tumor Suppressive Potential in Melanoma Cells. Int. J. Cancer 2008, 122, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Dodson, C.A.; Kosmopoulou, M.; Richards, M.W.; Atrash, B.; Bavetsias, V.; Blagg, J.; Bayliss, R. Crystal Structure of an Aurora-A Mutant That Mimics Aurora-B Bound to MLN8054: Insights into Selectivity and Drug Design. Biochem. J. 2010, 427, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Carvalhal, S.; Ribeiro, S.A.; Arocena, M.; Kasciukovic, T.; Temme, A.; Koehler, K.; Huebner, A.; Griffis, E.R. The Nucleoporin ALADIN Regulates Aurora A Localization to Ensure Robust Mitotic Spindle Formation. Mol. Biol. Cell 2015, 26, 3424–3438. [Google Scholar] [CrossRef] [PubMed]
- Friedmann, D.R.; Aguilar, A.; Fan, J.; Nachury, M.V.; Marmorstein, R. Structure of the α-Tubulin Acetyltransferase, ATAT1, and Implications for Tubulin-Specific Acetylation. Proc. Natl. Acad. Sci. USA 2012, 109, 19655–19660. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.-J. Tubulin Acetylation: Responsible Enzymes, Biological Functions and Human Diseases. Cell. Mol. Life Sci. 2015, 72, 4237–4255. [Google Scholar] [CrossRef] [PubMed]
- Stams, T.; Spurlino, J.C.; Smith, D.L.; Wahl, R.C.; Ho, T.F.; Qoronfleh, M.W.; Banks, T.M.; Rubin, B. Structure of Human Neutrophil Collagenase Reveals Large S1’ Specificity Pocket. Nat. Struct. Biol. 1994, 1, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Matter, H.; Schwab, W.; Barbier, D.; Billen, G.; Haase, B.; Neises, B.; Schudok, M.; Thorwart, W.; Schreuder, H.; Brachvogel, V.; et al. Quantitative Structure−Activity Relationship of Human Neutrophil Collagenase (MMP-8) Inhibitors Using Comparative Molecular Field Analysis and X-Ray Structure Analysis. J. Med. Chem. 1999, 42, 1908–1920. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Liu, L.; Cao, C.-L.; Li, M.-J.; Tan, K.; Yang, X.; Yun, C.-H. Structure and Function of Human Naa60 (NatF), a Golgi-Localized Bi-Functional Acetyltransferase. Sci. Rep. 2016, 6, 31425. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.C.; Heffernan, M.L.R.; Saraswat, L.D.; Bowen, C.A.; Melnick, L.; Hardy, L.W.; Orsini, M.A.; Allen, M.S.; Koch, P.; Spear, K.L.; et al. Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules. J. Med. Chem. 2013, 56, 3710–3724. [Google Scholar] [CrossRef]
- Pollegioni, L.; Piubelli, L.; Sacchi, S.; Pilone, M.S.; Molla, G. Physiological Functions of D-Amino Acid Oxidases: From Yeast to Humans. Cell. Mol. Life Sci. 2007, 64, 1373–1394. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Uslaner, J.M.; Hutson, P.H. The Therapeutic Potential of D-Amino Acid Oxidase (DAAO) Inhibitors. Open Med. Chem. J. 2010, 4, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Steele, D.L.; El-Kabbani, O.; Dunten, P.; Windsor, L.J.; Kammlott, R.U.; Crowther, R.L.; Michoud, C.; Engler, J.A.; Birktoft, J.J. Expression, Characterization and Structure Determination of an Active Site Mutant (Glu202-Gln) of Mini-Stromelysin-1. Protein Eng. 2000, 13, 397–405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sternlicht, M.D.; Lochter, A.; Sympson, C.J.; Huey, B.; Rougier, J.-P.; Gray, J.W.; Pinkel, D.; Bissell, M.J.; Werb, Z. The Stromal Proteinase MMP3/Stromelysin-1 Promotes Mammary Carcinogenesis. Cell 1999, 98, 137–146. [Google Scholar] [CrossRef]
- McAndrew, R.P.; Wang, Y.; Mohsen, A.-W.; He, M.; Vockley, J.; Kim, J.-J.P. Structural Basis for Substrate Fatty Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase. J. Biol. Chem. 2008, 283, 9435–9443. [Google Scholar] [CrossRef]
- Merinero, B.; Pérez-Cerdá, C.; Garcia, M.J.; Gangoiti, J.; Font, L.M.; Garcia Silva, M.T.; Vianey-Saban, C.; Duran, M.; Ugarte, M. Mitochondrial Very Long-Chain Acyl-CoA Dehydrogenase Deficiency with a Mild Clinical Course. J. Inherit. Metab. Dis. 1996, 19, 173–176. [Google Scholar] [CrossRef]
- Chaudhry, A.S.; Thirumaran, R.K.; Yasuda, K.; Yang, X.; Fan, Y.; Strom, S.C.; Schuetz, E.G. Genetic Variation in Aldo-Keto Reductase 1D1 (AKR1D1) Affects the Expression and Activity of Multiple Cytochrome P450s. Drug Metab. Dispos. 2013, 41, 1538–1547. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmthera 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tamayo, R. Pathology of Collagen Degradation. A Review. Am. J. Pathol. 1978, 92, 508–566. [Google Scholar] [PubMed]
- Van Damme, P.; Hole, K.; Pimenta-Marques, A.; Helsens, K.; Vandekerckhove, J.; Martinho, R.G.; Gevaert, K.; Arnesen, T. NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef]
- Clark-Taylor, T.; Clark-Taylor, B.E. Is Autism a Disorder of Fatty Acid Metabolism? Possible Dysfunction of Mitochondrial β-Oxidation by Long Chain Acyl-CoA Dehydrogenase. Med. Hypotheses 2004, 62, 970–975. [Google Scholar] [CrossRef]
- Tranchant, I.; Vera, L.; Czarny, B.; Amoura, M.; Cassar, E.; Beau, F.; Stura, E.A.; Dive, V. Halogen Bonding Controls Selectivity of FRET Substrate Probes for MMP-9. Chem. Biol. 2014, 21, 408–413. [Google Scholar] [CrossRef]
- Wang, J.; Tsirka, S.E. Neuroprotection by Inhibition of Matrix Metalloproteinases in a Mouse Model of Intracerebral Haemorrhage. Brain 2005, 128, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Groblewska, M.; Siewko, M.; Mroczko, B.; Szmitkowski, M. The Role of Matrix Metalloproteinases (MMPs) and Their Inhibitors (TIMPs) in the Development of Esophageal Cancer. Folia Histochem. Cytobiol. 2012, 50, 12–19. [Google Scholar] [CrossRef]
- Dollery, C.M.; McEwan, J.R.; Henney, A.M. Matrix Metalloproteinases and Cardiovascular Disease. Circ. Res. 1995, 77, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.M.; Polizzi, S.J.; Kadirvelraj, R.; Howard, W.W.; Wood, Z.A. Man o’ War Mutation in UDP-α-d-Xylose Synthase Favors the Abortive Catalytic Cycle and Uncovers a Latent Potential for Hexamer Formation. Biochemistry 2015, 54, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Moriarity, J.L.; Hurt, K.J.; Resnick, A.C.; Storm, P.B.; Laroy, W.; Schnaar, R.L.; Snyder, S.H. UDP-Glucuronate Decarboxylase, a Key Enzyme in Proteoglycan Synthesis Cloning, Characterization, and Localization. J. Biol. Chem. 2002, 277, 16968–16975. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.-F.; Dirk, L.M.A.; Brunzelle, J.S.; Houtz, R.L.; Trievel, R.C. Structural Origins for the Product Specificity of SET Domain Protein Methyltransferases. Proc. Natl. Acad. Sci. USA 2008, 105, 20659–20664. [Google Scholar] [CrossRef] [PubMed]
- Milite, C.; Feoli, A.; Viviano, M.; Rescigno, D.; Cianciulli, A.; Balzano, A.L.; Mai, A.; Castellano, S.; Sbardella, G. The Emerging Role of Lysine Methyltransferase SETD8 in Human Diseases. Clin. Epigenetics 2016, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, L.; Li, J.; Ding, Z.; Xiao, J.; Yin, X.; He, S.; Shi, P.; Dong, L.; Li, G.; et al. Structural Insight into Autoinhibition and Histone H3-Induced Activation of DNMT3A. Nature 2015, 517, 640–644. [Google Scholar] [CrossRef]
- Kim, G.-D.; Ni, J.; Kelesoglu, N.; Roberts, R.J.; Pradhan, S. Co-Operation and Communication between the Human Maintenance and de Novo DNA (Cytosine-5) Methyltransferases. Embo J. 2002, 21, 4183–4195. [Google Scholar] [CrossRef]
- Yang, L.; Rau, R.; Goodell, M.A. DNMT3A in Haematological Malignancies. Nat. Rev. Cancer 2015, 15, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Rajavelu, A.; Tulyasheva, Z.; Jaiswal, R.; Jeltsch, A.; Kuhnert, N. The Inhibition of the Mammalian DNA Methyltransferase 3a (Dnmt3a) by Dietary Black Tea and Coffee Polyphenols. BMC Biochem. 2011, 12, 16. [Google Scholar] [CrossRef]
- Stura, E.A.; Visse, R.; Cuniasse, P.; Dive, V.; Nagase, H. Crystal Structure of Full-Length Human Collagenase 3 (MMP-13) with Peptides in the Active Site Defines Exosites in the Catalytic Domain. FASEB J. 2013, 27, 4395–4405. [Google Scholar] [CrossRef] [PubMed]
- Woessner, J.F. Matrix Metalloproteinases and Their Inhibitors in Connective Tissue Remodeling. FASEB J. 1991, 5, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Knäuper, V.; López-Otin, C.; Smith, B.; Knight, G.; Murphy, G. Biochemical Characterization of Human Collagenase-3. J. Biol. Chem. 1996, 271, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Mazza, C.; Breton, R.; Housset, D.; Fontecilla-Camps, J.C. Unusual Charge Stabilization of NADP+ in 17β-Hydroxysteroid Dehydrogenase. J. Biol. Chem. 1998, 273, 8145–8152. [Google Scholar] [CrossRef] [PubMed]
- Mindnich, R.; Möller, G.; Adamski, J. The Role of 17 Beta-Hydroxysteroid Dehydrogenases. Mol. Cell. Endocrinol. 2004, 218, 7–20. [Google Scholar] [CrossRef]
- Liu, P.; Jia, M.; Zhou, X.E.; De Waal, P.W.; Dickson, B.M.; Liu, B.; Hou, L.; Yin, Y.; Kang, Y.; Shi, Y.; et al. The Structural Basis of the Dominant Negative Phenotype of the Gαi1β1γ2 G203A/A326S Heterotrimer. Acta Pharm. Sin. 2016, 37, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.E.; Sprang, S.R. Crystal Structures of the G Protein Giα1 Complexed with GDP and Mg2+: A Crystallographic Titration Experiment. Biochemistry 1998, 37, 14376–14385. [Google Scholar] [CrossRef] [PubMed]
- Milligan, G.; Kostenis, E. Heterotrimeric G-Proteins: A Short History. Br. J. Pharm. 2006, 147, S46–S55. [Google Scholar] [CrossRef]
- Thoden, J.B.; Wohlers, T.M.; Fridovich-Keil, J.L.; Holden, H.M. Molecular Basis for Severe Epimerase Deficiency Galactosemia X-RAY Structure of The Human V94m-Substituted Udp-Galactose 4-Epimerase. J. Biol. Chem. 2001, 276, 20617–20623. [Google Scholar] [CrossRef]
- Thoden, J.B.; Wohlers, T.M.; Fridovich-Keil, J.L.; Holden, H.M. Crystallographic Evidence for Tyr 157 Functioning as the Active Site Base in Human UDP−Galactose 4-Epimerase. Biochemistry 2000, 39, 5691–5701. [Google Scholar] [CrossRef]
- Daenzer, J.M.I.; Sanders, R.D.; Hang, D.; Fridovich-Keil, J.L. UDP-Galactose 4’-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of Drosophila Melanogaster. PLoS Genet. 2012, 8, e1002721. [Google Scholar] [CrossRef] [PubMed]
- Tempel, W.; Wu, H.; Dombrovsky, L.; Zeng, H.; Loppnau, P.; Zhu, H.; Plotnikov, A.N.; Bochkarev, A. An Intact SAM-Dependent Methyltransferase Fold Is Encoded by the Human Endothelin-Converting Enzyme-2 Gene. Proteins Struct. Funct. Bioinforma. 2009, 74, 789–793. [Google Scholar] [CrossRef]
- Liu, H.; Zang, C.; Fenner, M.H.; Possinger, K.; Elstner, E. PPARγ Ligands and ATRA Inhibit the Invasion of Human Breast Cancer Cells in Vitro. Breast Cancer Res. Treat. 2003, 79, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Egger, S.; Chaikuad, A.; Kavanagh, K.L.; Oppermann, U.; Nidetzky, B. Structure and Mechanism of Human UDP-Glucose 6-Dehydrogenase. J. Biol. Chem. 2011, 286, 23877–23887. [Google Scholar] [CrossRef] [PubMed]
- Wise, P.M.; Suzuki, S.; Brown, C.M. Estradiol: A Hormone with Diverse and Contradictory Neuroprotective Actions. Dialogues Clin. Neurosci. 2009, 11, 297–303. [Google Scholar] [PubMed]
- Hickie, R.A.; Walker, C.M.; Croll, G.A. Decreased Basal Cyclic Adenosine 3′,5′-Monophosphate Levels in Morris Hepatoma 5123 t.c.(h). Biochem. Biophys. Res. Commun. 1974, 59, 167–173. [Google Scholar] [CrossRef]
- Robinson, E.A.; Kalckar, H.M.; Troedsson, H.; Sanford, K. Metabolic Inhibition of Mammalian Uridine Diphosphate Galactose 4-Epimerase in Cell Cultures and in Tumor Cells. J. Biol. Chem. 1966, 241, 2737–2745. [Google Scholar] [CrossRef]
- Aubert, G.; Vega, R.B.; Kelly, D.P. Perturbations in the Gene Regulatory Pathways Controlling Mitochondrial Energy Production in the Failing Heart. Biochim. Biophys. Acta Bba Mol. Cell Res. 2013, 1833, 840–847. [Google Scholar] [CrossRef]
- Warren, G.L.; Andrews, C.W.; Capelli, A.-M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Lindvall, M.; Nevins, N.; Semus, S.F.; Senger, S.; et al. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Koshizuka, K.; Williamson, E.A.; Asou, H.; Said, J.W.; Holden, S.; Miyoshi, I.; Koeffler, H.P. Ligand for Peroxisome Proliferator-Activated Receptor γ (Troglitazone) Has Potent Antitumor Effect against Human Prostate Cancer Both in Vitro and in Vivo. Cancer Res. 1998, 58, 3344–3352. [Google Scholar] [PubMed]
- Hosokawa, M.; Kudo, M.; Maeda, H.; Kohno, H.; Tanaka, T.; Miyashita, K. Fucoxanthin Induces Apoptosis and Enhances the Antiproliferative Effect of the PPARγ Ligand, Troglitazone, on Colon Cancer Cells. Biochim. Biophys. Acta BBA Gen. Subj. 2004, 1675, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P450 and Chemical Toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Takada, I.; Makishima, M. Peroxisome Proliferator-Activated Receptor Agonists and Antagonists: A Patent Review (2014-Present). Expert Opin. Pat. 2020, 30, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Somody, J.C.; MacKinnon, S.S.; Windemuth, A. Structural Coverage of the Proteome for Pharmaceutical Applications. Drug Discov. Today 2017, 22, 1792–1799. [Google Scholar] [CrossRef]
- RCSB PDB: Homepage. Available online: http://www.rcsb.org/ (accessed on 5 February 2018).


| PDB ID with Chain | Docking Score [Arbitrary Units 1] | Protein Name | Protein Function and Connection with Diseases | References |
|---|---|---|---|---|
| 3uzxA 2 | −68.2 | 3-oxo-5-beta-steroid 4-dehydrogenase | Catalyzes the reduction of the Δ4 double bond of bile acid intermediates and steroid hormones carrying the Δ4-3-one structure in the A/B cis configuration. | [44] |
| 5fyyA 2 | −68.0 | Lysine-specific demethylase 5B | A member of the KDM5 sub-family, acting as a histone 3 lysine 4 trimethyl demethylase, regulating proliferation, stem cell self-renewal, and differentiation. Connected with various forms of cancer. | [45,46] |
| 2wtvA | −66.1 | Aurora kinase A | Involved in the regulation of centrosomes and segregation of chromosomes. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in several mitotic events. Known connections to various forms of cancer. | [47,48] |
| 4gs4A 3 | −64.7 | Alpha-tubulin N-acetyltransferase 1 | Acetylates K40 on α-tubulin at the lumenal side of microtubules. Connections with neurological disorders, cancer, heart diseases. | [49,50] |
| 1mncA | −64.0 | Neutrophil collagenase | A member of the matrix metalloproteinase family. Degrades fibrillar type I, II, and III collagens. Implicated in several degenerative diseases with a slow matrix degradation rate, such as osteoporosis and Alzheimer’s disease. | [51,52] |
| 5hh1A | −63.5 | N-alpha-acetyl transferase 60 | Specifically catalyzes acetylation of the N-terminal α-amine in a majority of transmembrane proteins, mediates lysine acetylation of free histone H4. Possible connections with different forms of cancer. | [53] |
| 3znnA | −62.7 | d-amino-acid oxidase | Catalyzes the oxidative deamination of d-isomers of neutral and polar amino acids. The physiological role in the kidney and liver represents the detoxification of accumulated d-amino acids. Known connection with schizophrenia. | [54,55,56] |
| 1c8tA | −62.3 | Stromelysin-1 | A member of the matrix metalloproteinase family. It degrades numerous ECM substrates, including collagens III, IV, V, IX, X, and XI, laminins, and elastin. Known connections with fibrosis, neovascularization, and potentially cancer progression. | [57,58] |
| 3b96A | −60.3 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial (VLCAD) | Catalyzes the initial, rate-limiting step of the mitochondrial fatty acid β-oxidation, exerts optimal chain length specificity for fatty acyl-CoAs having 16–24 carbons in length. Connected with hypoketotic hypoglycemia, liver damage, and hypertrophic myocardiopathy. | [59,60] |
| PDB ID with Chain | Docking score [Arbitrary Units 1] | Protein Name | Protein Function and Connection with Diseases | References |
|---|---|---|---|---|
| 4jijA | −60.5 | Matrix metalloproteinase 9 | Degrades components of the extracellular matrix, mostly collagens. Known connections with cancer, neurological disorders, and cardiovascular diseases. | [66,67,68,69] |
| 4lk3A | −59.9 | UDP-glucuronic acid decarboxylase 1 | Catalyzes the formation of UDP-xylose from UDP-glucuronate. No known connections with human diseases. | [70,71] |
| 3f9zA | −59.8 | N-lysine methyltransferase KMT5A | Monomethylates lysine 20 of histone H4 (H4K20), proliferating cell nuclear antigen (PCNA) and p53. Potential connections with cancer tumorigenesis and neurodegenerative diseases. | [72,73] |
| 4u7pA | −58.8 | DNA (cytosine-5)-methyltransferase 3A | Responsible for the de novo methylation of 5-methylcytosine during embryogenesis for the establishment of the somatic methylation pattern, which is crucial for embryonic development. Known connections with hematological malignancies, that is tumors of the hematopoietic and lymphoid tissues, as well as with defects in brain development or neurological defects. | [74,75,76,77] |
| 4fvlA | −58.6 | Collagenase 3 | One of the matrix metalloproteinases. Plays a key role in tissue remodeling and repair. Known connections with cancer, aneurysm, fibrosis, and other diseases. | [78,79,80] |
| 1fduA | −58.4 | Estradiol 17-beta-dehydrogenase 1 | Catalyzes the reversible transformation of estrone into biologically active estradiol. Connected with cancer, the biology of reproduction, and neuronal diseases. | [81,82] |
| 5tdhA | −58.0 | Guanine nucleotide-binding protein G(i) subunit alpha-1 | Transduces intracellular signals from membrane-bound receptors to downstream effector molecules. Cooperates in different biological activities, such as vision or synaptic nerve signal transmissions. Connected with neurological problems, cardiovascular defects, and certain forms of cancer. | [83,84,85] |
| 1i3kA | −57.6 | UDP-glucose 4-epimerase | Catalyzes the interconversion of UDP-galactose and UDP-glucose during normal galactose metabolism, as well as interconversion of UDP-N-acetyl galactosamine (UDP-galNAc) and UDP-N-acetylglucosamine (UDP-glcNAc). Loss of this protein results in the spectrum disorder epimerase deficiency galactosemia. | [86,87,88] |
| 2pxxA | −57.6 | EEF1A lysine methyltransferase 4 | Catalyzes methylations on Lys36 in eukaryotic translation elongation factor 1 alpha. No known connections with human diseases. | [89] |
| Query Protein | Proteins with Similar Binding Sites | ||
|---|---|---|---|
| PDB ID with Chain | PDB ID with Chain | Protein | Z-Score |
| 2vn0A | 1r9oA | Cytochrome P450 2C9 | 3.83 |
| 4gqsA | Cytochrome P450 2C19 | 3.82 | |
| 3tbgA | Cytochrome P450 2D6 | 3.21 | |
| 3ibdA | Cytochrome P450 2B6 | 3.13 | |
| 3czhA | Cytochrome P450 2R1 | 3.11 | |
| 4nkyA | Steroid 17-Alpha-Hydroxylase/17,20 Lyase | 3.00 | |
| 4y8wA | Cytochrome P450 21-Hydroxylase | 2.93 | |
| 3e6iA | Cytochrome P450 2E1 | 2.54 | |
| 3ld6A | Lanosterol 14-Alpha Demethylase | 2.06 | |
| Query Proteins | Proteins with Similar Binding Sites | ||
|---|---|---|---|
| PDB ID with Chain | PDB ID with Chain | Protein | Z-Score |
| 1fm6DX | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.84 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.77 | |
| 3ipqA | Oxysterols Receptor LXR-Alpha | 2.04 | |
| 1zgyA | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.78 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.66 | |
| 2prgAB | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.60 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.59 | |
| 3cs8A | 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.30 |
| 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 2.98 | |
| 3dzyD | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.25 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.05 | |
| 4emaA | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.73 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.14 | |
| 4o8fAB | 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.63 |
| 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.45 | |
| 4xldA | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.85 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.44 | |
| 5ji0D | 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.94 |
| 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.86 | |
| 5ycpA | 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.15 |
| 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 2.73 | |
| 4a5xA | MIT Domain-Containing Protein 1 | 2.04 | |
| 6md4A | 3tkmA | Peroxisome Proliferator-Activated Receptor Delta | 3.76 |
| 3vi8A | Peroxisome Proliferator-Activated Receptor Alpha | 3.36 | |
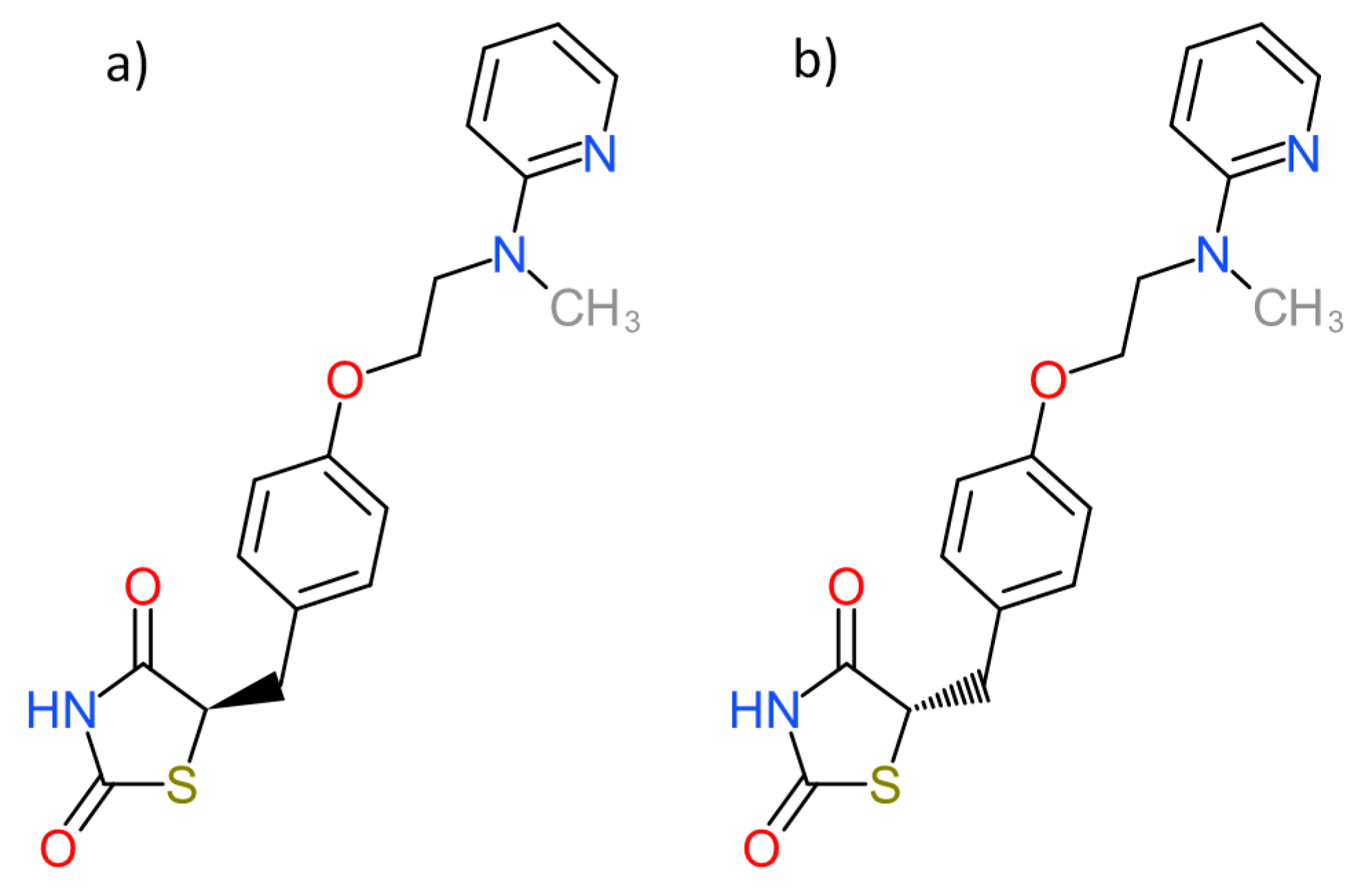
| PDB ID with Chain (Crystal Structure) | Stereoisomer (Figure 1) | Docking Score [Arbitrary Units] | RMSD [Å] |
|---|---|---|---|
| 2vn0A | a | −24.77 | 4.51 |
| b | −14.1 | 5.45 | |
| c | −26.5 | 5.84 | |
| d | −25.1 | 6.12 |
| PDB ID with Chain (Crystal Structure) | Stereoisomer (Figure 2) | Docking Score [Arbitrary Units] | RMSD [Å] |
|---|---|---|---|
| 1fm6D | a | −30.6 | 2.15 |
| b | −32.1 | 2.25 | |
| 1zgyA | a | −32.9 | 2.26 |
| b | −38.2 | 2.47 | |
| 2prgA | a | −37.1 | 0.85 |
| b | −46.2 | 2.70 | |
| 3cs8A | a | −29.0 | 2.89 |
| b | −28.2 | 8.07 | |
| 3dzyD | a | −41.4 | 2.22 |
| b | −32.0 | 1.09 | |
| 4emaA | a | −43.6 | 1.96 |
| b | −21.0 | 3.20 | |
| 4o8fA | a | −32.2 | 1.12 |
| b | −23.8 | 2.03 | |
| 4xldA | a | −39.3 | 1.78 |
| b | −35.8 | 2.62 | |
| 5ji0D | a | −37.2 | 1.44 |
| b | −39.7 | 2.14 | |
| 5ycpA | a | −34.3 | 1.76 |
| b | −44.9 | 1.90 | |
| 6md4A | a | −31.8 | 2.06 |
| b | −37.6 | 2.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kores, K.; Konc, J.; Bren, U. Mechanistic Insights into Side Effects of Troglitazone and Rosiglitazone Using a Novel Inverse Molecular Docking Protocol. Pharmaceutics 2021, 13, 315. https://doi.org/10.3390/pharmaceutics13030315
Kores K, Konc J, Bren U. Mechanistic Insights into Side Effects of Troglitazone and Rosiglitazone Using a Novel Inverse Molecular Docking Protocol. Pharmaceutics. 2021; 13(3):315. https://doi.org/10.3390/pharmaceutics13030315
Chicago/Turabian StyleKores, Katarina, Janez Konc, and Urban Bren. 2021. "Mechanistic Insights into Side Effects of Troglitazone and Rosiglitazone Using a Novel Inverse Molecular Docking Protocol" Pharmaceutics 13, no. 3: 315. https://doi.org/10.3390/pharmaceutics13030315
APA StyleKores, K., Konc, J., & Bren, U. (2021). Mechanistic Insights into Side Effects of Troglitazone and Rosiglitazone Using a Novel Inverse Molecular Docking Protocol. Pharmaceutics, 13(3), 315. https://doi.org/10.3390/pharmaceutics13030315







