Inhibitory Effects of Cinnamaldehyde Derivatives on Biofilm Formation and Virulence Factors in Vibrio Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain, Chemicals, and Culture Materials
2.2. Planktonic Cell Growth and Minimum Inhibitory Concentrations (MICs)
2.3. Crystal Violet Biofilm Inhibition Assay
2.4. Biofilm Dispersal Assay
2.5. Confocal Laser Scanning Microscopy (CLSM) and COMSTAT Analysis
2.6. Scanning Electron Microscopy (SEM)
2.7. Analysis of Swimming and Swarming Motility
2.8. Yeast Agglutination Assay
2.9. Bacterial Surface Hydrophobicity Assay
2.10. Exoprotease Assay
2.11. Indole Production Assay
2.12. Biotic Surface Assay
2.13. RNA Isolation and Quantitative Reverse Transcriptase PCR (qRT-PCR)
2.14. Statistical Analysis
3. Results
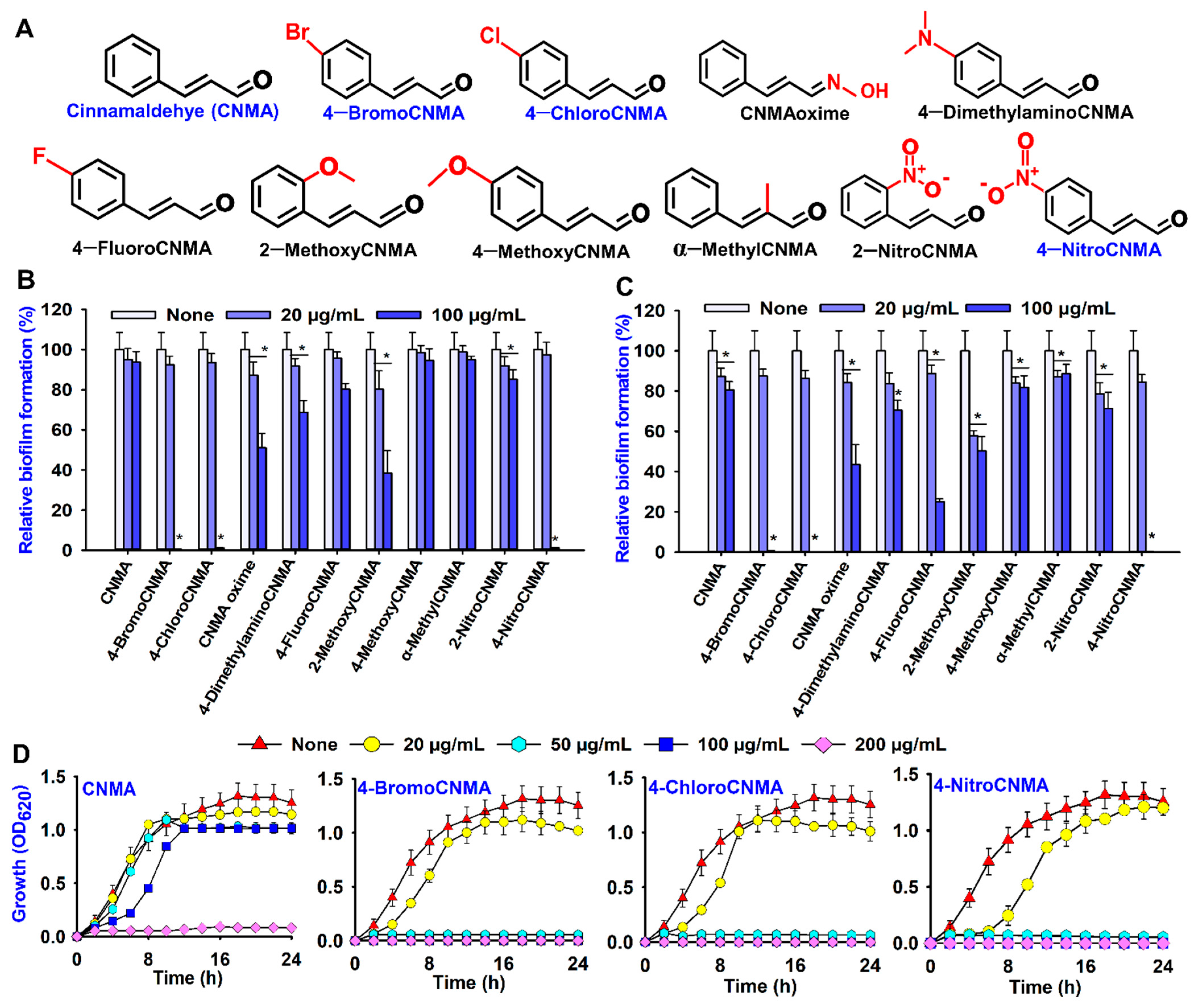
3.1. Biofilm Inhibitory and Antimicrobial Activities of Cinnamaldehyde (CNMA) and Its Derivatives against V. parahaemolyticus and V. harveyi
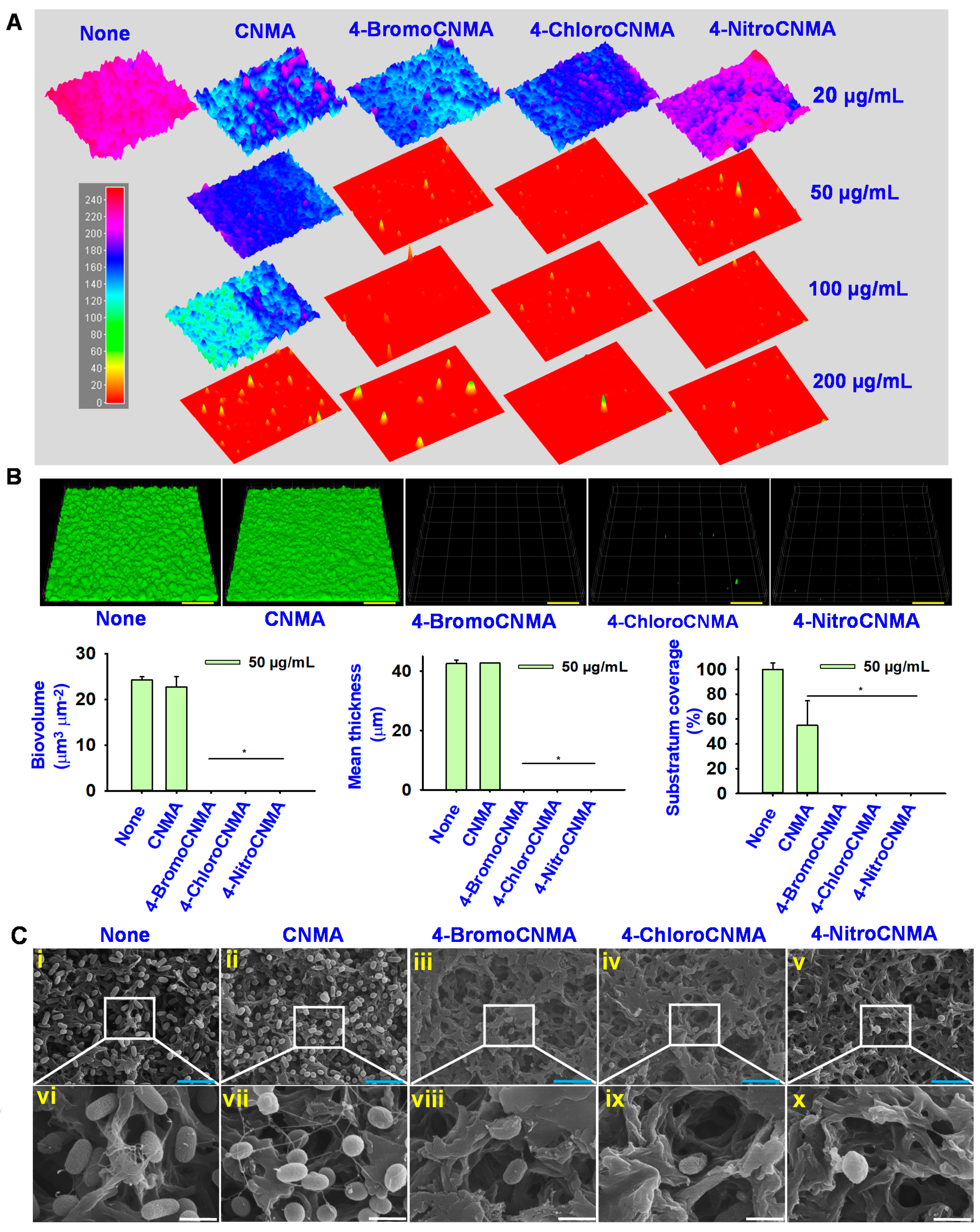
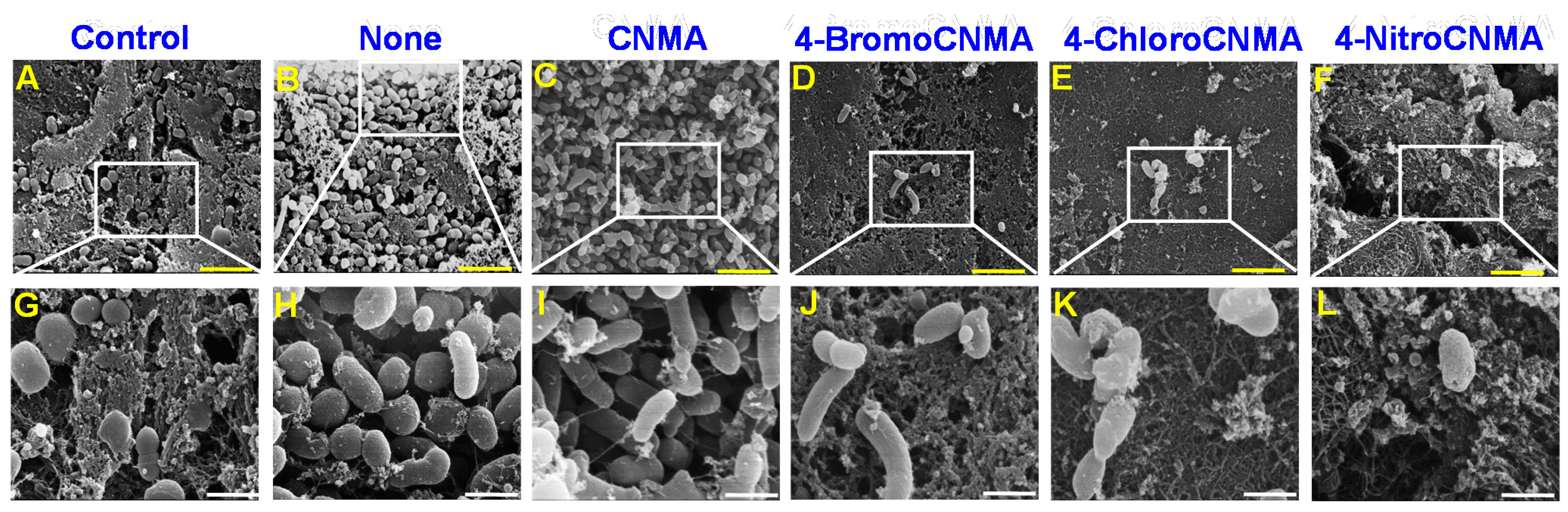
3.2. Light, Confocal and Electron Microscopy Observation of Biofilm Inhibition by CNMA Derivatives
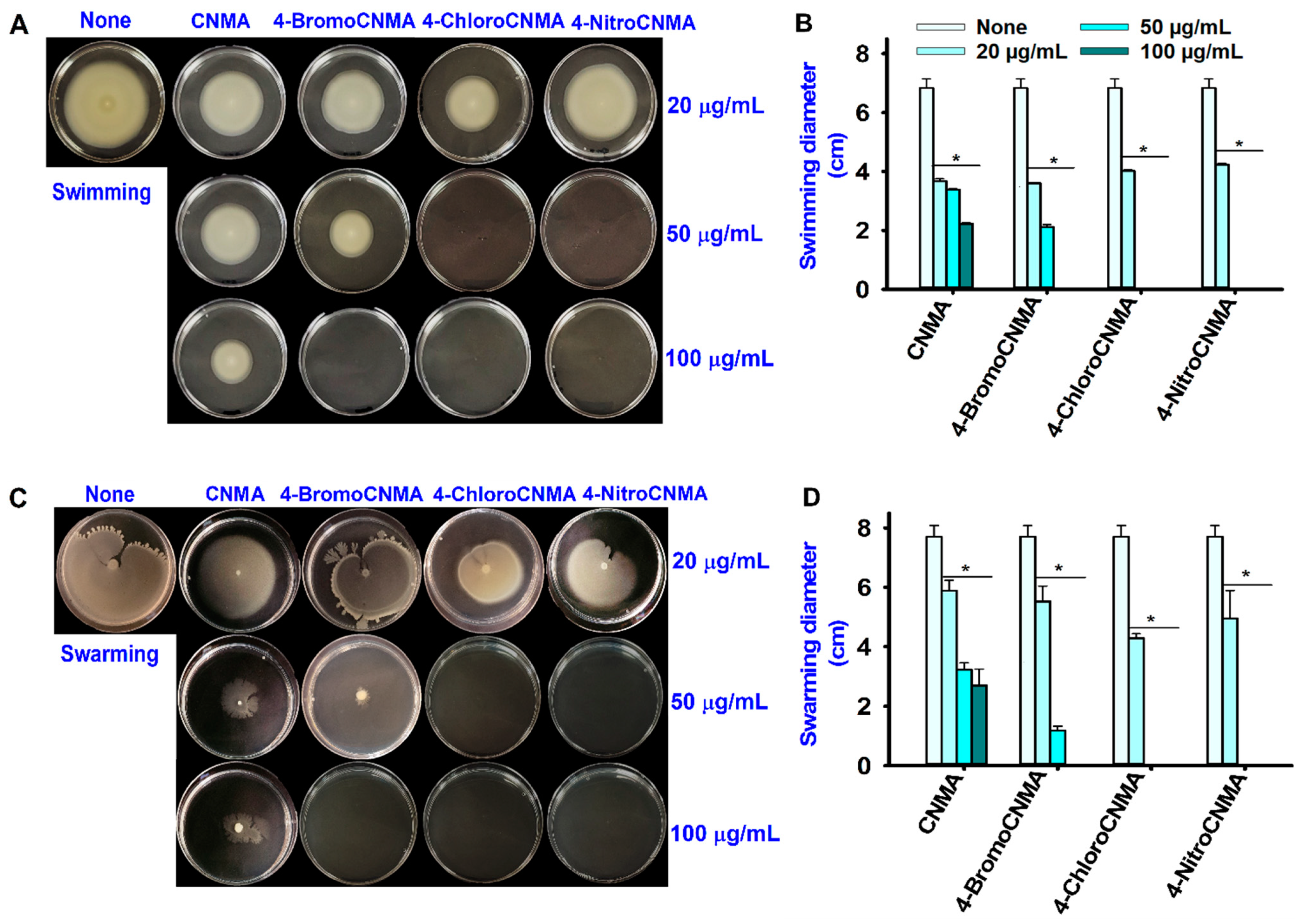
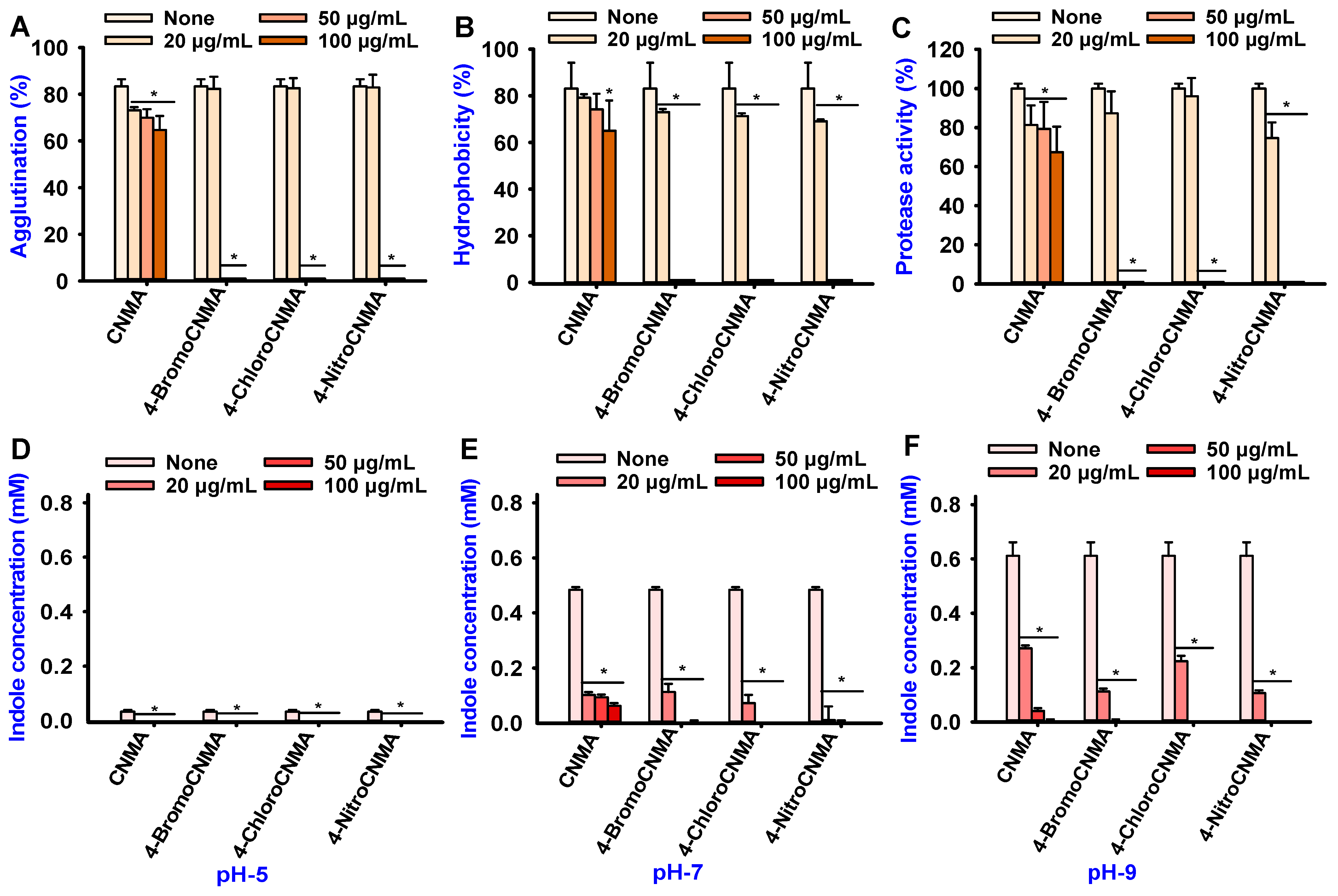
3.3. CNMA Derivatives Reduced Surface Motility, Fimbriae, Hydrophobicity, and Protease Production
3.4. CNMA and Its Derivatives Suppressed Indole Production by V. parahaemolyticus
3.5. CNMA Derivatives Eradicated V. parahaemolyticus on Squid Surface
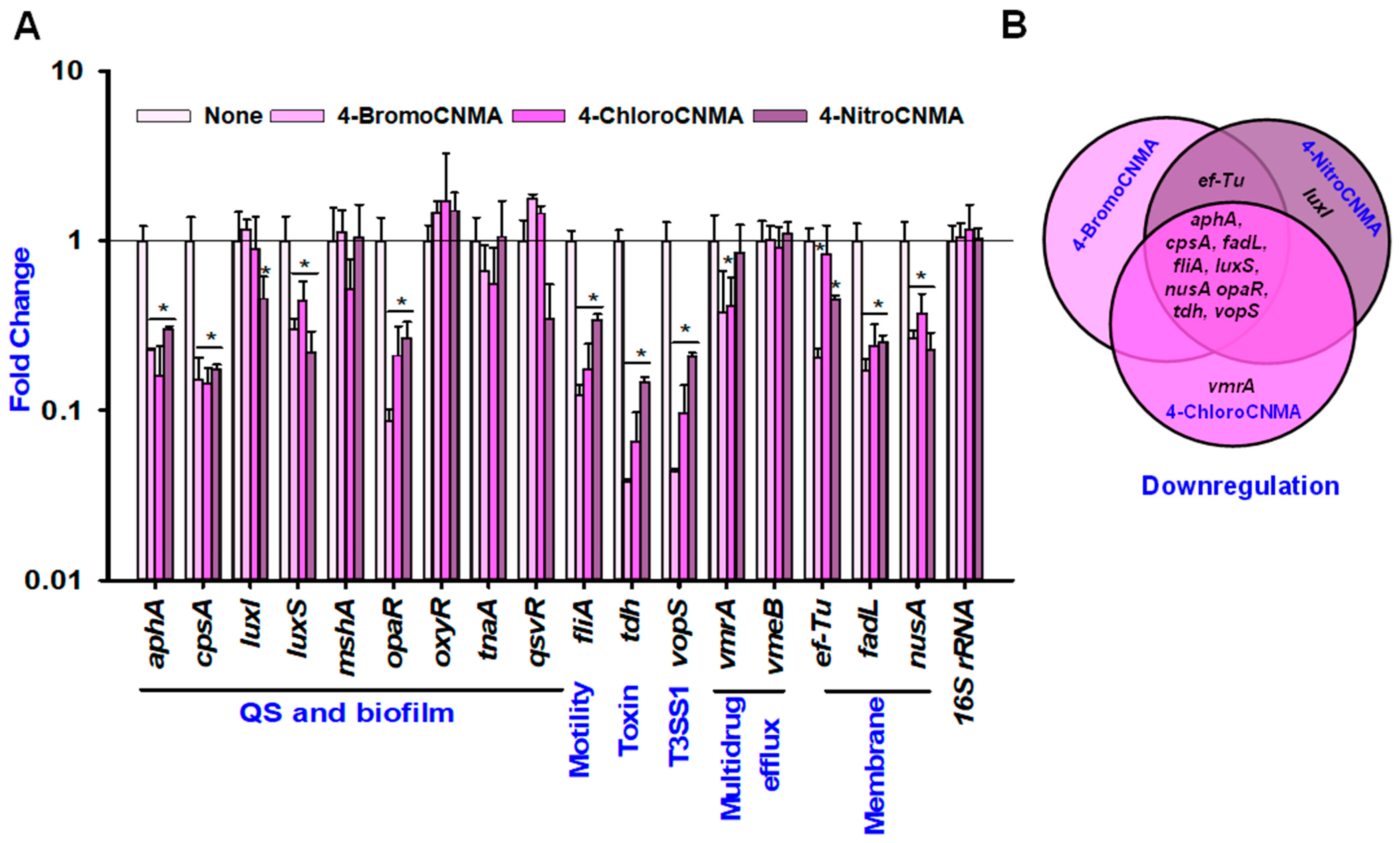
3.6. CNMA Derivatives Repressed the Expressions of Biofilm, Quorum Sensing, and Other Virulence-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drake, S.L.; DePaola, A.; Jaykus, L.A. An overview of Vibrio vulnificus and Vibrio parahaemolyticus. Compr. Rev. Food Sci. Food Saf. 2007, 6, 120–144. [Google Scholar] [CrossRef]
- Huang, J.Y.; Henao, O.L.; Griffin, P.M.; Vugia, D.J.; Cronquist, A.B.; Hurd, S.; Tobin-D’Angelo, M.; Ryan, P.; Smith, K.; Lathrop, S.; et al. Infection with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance—Foodborne diseases active surveillance network, 10 US sites, 2012–2015. Morb. Mortal. Wkly. Rep. 2016, 65, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Defoirdt, T.; Miyamoto, C.; Bossier, P.; Van Calenbergh, S.; Nelis, H.; Coenye, T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008, 8, 149. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Prim. 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Khouadja, S.; Lamari, F.; Bakhrouf, A. Characterization of Vibrio parahaemolyticus isolated from farmed sea bass (Dicentrarchus labrax) during disease outbreaks. Int. Aquat. Res. 2013, 5, 1–11. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Ashrafudoulla, M.; Mizan, M.; Rahaman, F.; Park, H.; Byun, K.-H.; Lee, N.; Park, S.H.; Ha, S.-D. Genetic relationship, virulence factors, drug resistance profile and biofilm formation ability of Vibrio parahaemolyticus isolated from mussel. Front. Microbiol. 2019, 10, 513. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, Y.; Tan, Y.; Guo, Z.; Yang, R.; Zhou, D. Transcriptional regulation of opaR, qrr2–4 and aphA by the master quorum-sensing regulator OpaR in Vibrio parahaemolyticus. PLoS ONE 2012, 7, e34622. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Osei-Adjei, G.; Huang, X.; Zhang, Y. Role and regulation of the orphan AphA protein of quorum sensing in pathogenic Vibrios. Future Microbiol. 2018, 13, 383–391. [Google Scholar] [CrossRef]
- Lu, R.; Tang, H.; Qiu, Y.; Yang, W.; Yang, H.; Zhou, D.; Huang, X.; Hu, L.; Zhang, Y. Quorum sensing regulates the transcription of lateral flagellar genes in Vibrio parahaemolyticus. Future Microbiol. 2019, 14, 1043–1053. [Google Scholar] [CrossRef]
- Yuan, L.; Hansen, M.F.; Røder, H.L.; Wang, N.; Burmølle, M.; He, G. Mixed-species biofilms in the food industry: Current knowledge and novel control strategies. Crit. Rev. Food Sci. Nutr. 2020, 60, 2277–2293. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Ashrafudoulla, M.; Sadekuzzaman, M.; Kang, I.; Ha, S.-D. Effects of NaCl, glucose, and their combinations on biofilm formation on black tiger shrimp (Penaeus monodon) surfaces by Vibrio parahaemolyticus. Food Control 2018, 89, 203–209. [Google Scholar] [CrossRef]
- Han, N.; Mizan, M.F.R.; Jahid, I.K.; Ha, S.-D. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 2016, 70, 161–166. [Google Scholar] [CrossRef]
- Concha-Meyer, A.; Schöbitz, R.; Brito, C.; Fuentes, R. Lactic acid bacteria in an alginate film inhibit Listeria monocytogenes growth on smoked salmon. Food Control 2011, 22, 485–489. [Google Scholar] [CrossRef]
- Elexson, N.; Yaya, R.; Nor, A.M.; Kantilal, H.K.; Ubong, A.; Nishibuchi, M.; Yoshitsugu, N.; Son, R. Biofilm assessment of Vibrio parahaemolyticus from seafood using random amplified polymorphism DNA-PCR. Int. Food Res. J. 2014, 21, 59–65. [Google Scholar]
- Xu, X.; Cheng, J.; Wu, Q.; Zhang, J.; Xie, T. Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol. 2016, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.A.; El Bayomi, R.M.; Hussein, M.A.; Khedr, M.H.E.; Remela, E.M.A.; El-Ashram, A.M.M. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int. J. Food Microbiol. 2018, 274, 31–37. [Google Scholar] [CrossRef]
- Cao, J.; Liu, H.; Wang, Y.; He, X.; Jiang, H.; Yao, J.; Xia, F.; Zhao, Y.; Chen, X. Antimicrobial and antivirulence efficacies of citral against foodborne pathogen Vibrio parahaemolyticus RIMD2210633. Food Control 2021, 120, 107507. [Google Scholar] [CrossRef]
- Firmino, D.F.; Cavalcante, T.T.A.; Gomes, G.A.; Firmino, N.; Rosa, L.D.; de Carvalho, M.G.; Catunda, F.E.A., Jr. Antibacterial and antibiofilm activities of Cinnamomum sp. essential oil and cinnamaldehyde: Antimicrobial activities. Sci. World J. 2018, 2018, 9. [Google Scholar] [CrossRef]
- Adams, T.B.; Cohen, S.M.; Doull, J.; Feron, V.J.; Goodman, J.I.; Marnett, L.J.; Munro, I.C.; Portoghese, P.S.; Smith, R.L.; Waddell, W.J. The FEMA GRAS assessment of cinnamyl derivatives used as flavor ingredients. Food Chem. Toxicol. 2004, 42, 157–185. [Google Scholar] [CrossRef]
- Zhu, L.; Olsen, C.; McHugh, T.; Friedman, M.; Jaroni, D.; Ravishankar, S. Apple, Carrot, and Hibiscus edible films containing the plant antimicrobials carvacrol and cinnamaldehyde inactivate Salmonella Newport on organic leafy greens in sealed plastic bags. J. Food Sci. 2014, 79, M61–M66. [Google Scholar] [CrossRef]
- Han, C.; Wang, J.; Li, Y.; Cui, Y. In vitro antimicrobial activity and effect on E. coli integrity of cinnamon essential oil and rhubarb ethanol extract. Food Sci. Technol. Res. 2013, 19, 1155–1163. [Google Scholar] [CrossRef][Green Version]
- Shen, Q.; Zhou, W.; Hu, L.; Qi, Y.; Ning, H.; Chen, J.; Mo, H. Bactericidal activity of alpha-bromocinnamaldehyde against persisters in Escherichia coli. PLoS ONE 2017, 12, e0182122. [Google Scholar] [CrossRef]
- Kot, B.; Wicha, J.; Piechota, M.; Wolska, K.; Gruzewska, A. Antibiofilm activity of trans-cinnamaldehyde, p-coumaric, and ferulic acids on uropathogenic Escherichia coli. Turk. J. Med. Sci. 2015, 45, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Lee, J.-H.; Kim, S.-I.; Baek, K.-H.; Lee, J. Cinnamon bark oil and its components inhibit biofilm formation and toxin production. Int. J. Food Microbiol. 2015, 195, 30–39. [Google Scholar] [CrossRef]
- Malheiro, J.F.; Maillard, J.Y.; Borges, F.; Simões, M. Evaluation of cinnamaldehyde and cinnamic acid derivatives in microbial growth control. Int. Biodeterior. Biodegrad. 2019, 141, 71–78. [Google Scholar] [CrossRef]
- Kavanaugh, N.L.; Ribbeck, K. Selected antimicrobial essential oils eradicate Pseudomonas spp. and Staphylococcus aureus biofilms. Appl. Environ. Microbiol. 2012, 78, 4057–4061. [Google Scholar] [CrossRef] [PubMed]
- Beema Shafreen, R.M.; Selvaraj, C.; Singh, S.K.; Karutha Pandian, S. In silico and in vitro studies of cinnamaldehyde and their derivatives against LuxS in Streptococcus pyogenes: Effects on biofilm and virulence genes. J. Mol. Recognit. 2014, 27, 106–116. [Google Scholar] [CrossRef]
- Da Nóbrega Alves, D.; Monteiro, A.F.M.; Andrade, P.N.; Lazarini, J.G.; Abílio, G.M.F.; Guerra, F.Q.S.; Scotti, M.T.; Scotti, L.; Rosalen, P.L.; Castro, R.D. Docking prediction, antifungal activity, anti-biofilm effects on Candida spp., and toxicity against human cells of cinnamaldehyde. Molecules 2020, 25, 5969. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Shang, B.; Wang, L.; Lu, Z.; Liu, Y. Cinnamaldehyde inhibits fungal growth and aflatoxin B1 biosynthesis by modulating the oxidative stress response of Aspergillus flavus. Appl. Microbiol. Biotechnol. 2016, 100, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Banu, S.F.; Rubini, D.; Murugan, R.; Vadivel, V.; Gowrishankar, S.; Pandian, S.K.; Nithyanand, P. Exploring the antivirulent and sea food preservation efficacy of essential oil combined with DNase on Vibrio parahaemolyticus. LWT 2018, 95, 107–115. [Google Scholar] [CrossRef]
- Lu, C.; Liu, H.; Shangguan, W.; Chen, S.; Zhong, Q. Antibiofilm activities of the cinnamon extract against Vibrio parahaemolyticus and Escherichia coli. Arch. Microbiol. 2021, 203, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Feyaerts, A.F.; Van Dijck, P.; Bossier, P. Inhibitory activity of essential oils against Vibrio campbellii and Vibrio parahaemolyticus. Microorganisms 2020, 8, 1946. [Google Scholar] [CrossRef]
- Doyle, A.A.; Stephens, J.C. A review of cinnamaldehyde and its derivatives as antibacterial agents. Fitoterapia 2019, 139, 104405. [Google Scholar] [CrossRef]
- Khadke, S.K.; Lee, J.-H.; Woo, J.-T.; Lee, J. Inhibitory effects of honokiol and magnolol on biofilm formation by Acinetobacter baumannii. Biotechnol. Bioprocess Eng. 2019, 24, 359–365. [Google Scholar] [CrossRef]
- Seo, S.; Jung, J.; Kim, C.Y.; Kang, H.; Lee, I.H. Antimicrobial peptides encounter resistance of aureolysin during their action on Staphylococcus aureus biofilm. Biotechnol. Bioprocess Eng. 2021, 26, 216–222. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Raorane, C.J.; Ryu, S.Y.; Shim, J.-J.; Lee, J. The anti-biofilm and anti-virulence activities of trans-resveratrol and oxyresveratrol against uropathogenic Escherichia coli. Biofouling 2019, 35, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Lee, J.-H.; Lee, S.; Lee, Y.-K.; Hwang, B.S.; Lee, J. Antibiofilm activity of phorbaketals from the marine sponge Phorbas sp. against Staphylococcus aureus. Mar. Drugs 2021, 19, 301. [Google Scholar] [CrossRef]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Raorane, C.J.; Lee, J.-H.; Lee, J. Rapid killing and biofilm inhibition of multidrug-resistant Acinetobacter baumannii strains and other microbes by iodoindoles. Biomolecules 2020, 10, 1186. [Google Scholar] [CrossRef] [PubMed]
- Heering, J.; Alvarado, A.; Ringgaard, S. Induction of cellular differentiation and single cell imaging of Vibrio parahaemolyticus swimmer and swarmer cells. J. Vis. Exp. 2017, 123, e55842. [Google Scholar] [CrossRef]
- Sathiyamoorthi, E.; Faleye, O.S.; Lee, J.-H.; Raj, V.; Lee, J. Antibacterial and antibiofilm activities of chloroindoles against Vibrio parahaemolyticus. Front. Microbiol. 2021, 12, 714371. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, S.; Sathiyamoorthi, E.; Kim, Y.-G.; Lee, J.-H.; Lee, J. Antibiofilm and antivirulence properties of indoles against Serratia marcescens. Front. Microbiol. 2020, 11, 584812. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Jahid, I.K.; Kim, M.; Lee, K.-H.; Kim, T.J.; Ha, S.-D. Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling 2016, 32, 497–509. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Huang, Z.; Jiang, W.; Zhou, W. Antimicrobial activity of cinnamaldehyde on Streptococcus mutans biofilms. Front. Microbiol. 2019, 10, 2241. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, P.; Fursy, R.; Bret, L.; Sundararaju, B.; Phillips, R.S. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 2003, 49, 443–449. [Google Scholar] [CrossRef]
- Mueller, R.S.; Beyhan, S.; Saini, S.G.; Yildiz, F.H.; Bartlett, D.H. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 2009, 191, 3504–3516. [Google Scholar] [CrossRef] [PubMed]
- Han, T.H.; Lee, J.-H.; Cho, M.H.; Wood, T.K.; Lee, J. Environmental factors affecting indole production in Escherichia coli. Res. Microbiol. 2011, 162, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Toushik, S.H.; Kim, K.; Ashrafudoulla, M.; Mizan, M.F.R.; Roy, P.K.; Nahar, S.; Kim, Y.; Ha, S.-D. Korean kimchi-derived lactic acid bacteria inhibit foodborne pathogenic biofilm growth on seafood and food processing surface materials. Food Control 2021, 129, 108276. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Lee, J.-H.; Gwon, G.; Kim, S.-I.; Park, J.G.; Lee, J. Essential oils and eugenols inhibit biofilm formation and the virulence of Escherichia coli O157: H7. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Li, X.-H.; Lee, J.-H. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bajpai, V.K.; Baek, K.H. Determination of antibacterial mode of action of Allium sativum essential oil against foodborne pathogens using membrane permeability and surface characteristic parameters. J. Food Saf. 2013, 33, 197–208. [Google Scholar] [CrossRef]
- Denis, K.; Le Bris, M.; Le Guennec, L.; Barnier, J.-P.; Faure, C.; Gouge, A.; Bouzinba-Ségard, H.; Jamet, A.; Euphrasie, D.; Durel, B.; et al. Targeting Type IV pili as an antivirulence strategy against invasive meningococcal disease. Nat. Microbiol. 2019, 4, 972–984. [Google Scholar] [CrossRef]
- McCarter, L.L. Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 2004, 7, 18–29. [Google Scholar] [CrossRef]
- Van Houdt, R.; Michiels, C.W. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef]
- Paranjpye, R.N.; Johnson, A.B.; Baxter, A.E.; Strom, M.S. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl. Environ. Microbiol. 2007, 73, 5041–5044. [Google Scholar] [CrossRef] [PubMed]
- Letchumanan, V.; Chan, K.-G.; Lee, L.-H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef]
- Ottaviani, D.; Santarelli, S.; Bacchiocchi, S.; Masini, L.; Ghittino, C.; Bacchiocchi, I. Presence of pathogenic Vibrio parahaemolyticus strains in mussels from the Adriatic Sea, Italy. Food Microbiol. 2005, 22, 585–590. [Google Scholar] [CrossRef]
- Lee, J.-H.; Wood, T.K.; Lee, J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef]
- Wyeth, F.J.S. The effects of acids, alkalies, and sugars on the growth and indole formation of Bacillus coli: A report to the medical research committee. Biochem. J. 1919, 13, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Yohannes, E.; Barnhart, D.M.; Slonczewski, J.L. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J. Bacteriol. 2004, 186, 192–199. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Kim, C.-J.; Lee, J.-C.; Cho, M.H.; Lee, J. Indole-3-acetaldehyde from Rhodococcus sp. BFI 332 inhibits Escherichia coli O157: H7 biofilm formation. Appl. Microbiol. Biotechnol. 2012, 96, 1071–1078. [Google Scholar] [CrossRef]
- Malheiro, J.; Gomes, I.; Borges, A.; Bastos, M.; Maillard, J.Y.; Borges, F.; Simões, M. Phytochemical profiling as a solution to palliate disinfectant limitations. Biofouling 2016, 32, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Gilbert, E.S. Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 2004, 70, 6951–6956. [Google Scholar] [CrossRef]
- Alkawash, M.A.; Soothill, J.S.; Schiller, N.L. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 2006, 114, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Darouiche, R.O.; Mansouri, M.D.; Gawande, P.V.; Madhyastha, S. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB® combination. J. Antimicrob. Chemother. 2009, 64, 88–93. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry, antimicrobial mechanisms, and antibiotic activities of cinnamaldehyde against pathogenic bacteria in animal feeds and human foods. J. Agric. Food Chem. 2017, 65, 10406–10423. [Google Scholar] [CrossRef] [PubMed]
- Wolfram, J.; Zhu, M.; Yang, Y.; Shen, J.; Gentile, E.; Paolino, D.; Fresta, M.; Nie, G.; Chen, C.; Shen, H. Safety of nanoparticles in medicine. Curr. Drug Targets 2015, 16, 1671–1681. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, C.; Cui, H.; Lin, L. Encapsulation strategies to enhance the antibacterial properties of essential oils in food system. Food Control 2021, 123, 107856. [Google Scholar] [CrossRef]
- Zhan, Q.; Xu, Y.; Zhan, L.; Wang, B.; Guo, Y.; Wu, X.; Ai, W.; Song, Z.; Yu, F. Chromone derivatives CM3a potently eradicate Staphylococcus aureus biofilms by inhibiting cell adherence. Infect. Drug Resist. 2021, 14, 979–986. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Cheng, M.-F.; Yu, M.-S.; Pan, M.-J. Purification and characterization of a putative virulence factor, serine protease, from Vibrio parahaemolyticus. FEMS Microbiol. Lett. 2002, 209, 31–37. [Google Scholar] [CrossRef]
- Valiente, E.; Lee, C.T.; Hor, L.I.; Fouz, B.; Amaro, C. Role of the metalloprotease Vvp and the virulence plasmid pR99 of Vibrio vulnificus serovar E in surface colonization and fish virulence. Environ. Microbiol. 2008, 10, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Denkin, S.M.; Nelson, D.R. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 2004, 70, 4193–4204. [Google Scholar] [CrossRef]
- Miyoshi, S. Vibrio vulnificus infection and metalloprotease. J. Dermatol. 2006, 33, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Maurer, L.M.; Yohannes, E.; Bondurant, S.S.; Radmacher, M.; Slonczewski, J.L. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. Res. 2005, 187, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Blankenhorn, D.; Phillips, J.; Slonczewski, J.L. Acid-and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. Res. 1999, 181, 2209–2216. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jayaraman, A.; Wood, T.K. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007, 7, 42. [Google Scholar] [CrossRef]
- Wang, L.; Ling, Y.; Jiang, H.; Qiu, Y.; Qiu, J.; Chen, H.; Yang, R.; Zhou, D. AphA is required for biofilm formation, motility, and virulence in pandemic Vibrio parahaemolyticus. Int. J. Food Microbiol. 2013, 160, 245–251. [Google Scholar] [CrossRef]
- Qian, H.; Li, W.; Guo, L.; Tan, L.; Liu, H.; Wang, J.; Pan, Y.; Zhao, Y. Stress response of Vibrio parahaemolyticus and Listeria monocytogenes biofilms to different modified atmospheres. Front. Microbiol. 2020, 11, 23. [Google Scholar] [CrossRef]
- Zhou, D.; Yan, X.; Qu, F.; Wang, L.; Zhang, Y.; Hou, J.; Hu, Y.; Li, J.; Xin, S.; Qiu, J. Quorum sensing modulates transcription of cpsQ-mfpABC and mfpABC in Vibrio parahaemolyticus. Int. J. Food Microbiol. 2013, 166, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Bansal, T.; Jayaraman, A.; Bentley, W.E.; Wood, T.K. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl. Environ. Microbiol. 2007, 73, 4100–4109. [Google Scholar] [CrossRef]
- Sun, X.-H.; Hao, L.-R.; Xie, Q.-C.; Lan, W.-Q.; Zhao, Y.; Pan, Y.-J.; Wu, V.C.H. Antimicrobial effects and membrane damage mechanism of blueberry (Vaccinium corymbosum L.) extract against Vibrio parahaemolyticus. Food Control 2020, 111, 107020. [Google Scholar] [CrossRef]
- Lowery, C.A.; Dickerson, T.J.; Janda, K.D. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 2008, 37, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Pompeani, A.J.; Irgon, J.J.; Berger, M.F.; Bulyk, M.L.; Wingreen, N.S.; Bassler, B.L. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 2008, 70, 76–88. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, M.; Quan, C.S.; Fan, S.D. Mechanisms of quorum sensing and strategies for quorum sensing disruption in aquaculture pathogens. J. Fish Dis. 2015, 38, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Afre, S.; Gilbert, E.S. Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett. Appl. Microbiol. 2006, 43, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, M.H.; Subhedar, D.D.; Shingate, B.B.; Khan, F.A.K.; Sangshetti, J.N.; Khedkar, V.M.; Nawale, L.; Sarkar, D.; Navale, G.R.; Shinde, S.S. Synthesis, biological evaluation and molecular docking of novel coumarin incorporated triazoles as antitubercular, antioxidant and antimicrobial agents. Med. Chem. Res. 2016, 25, 790–804. [Google Scholar] [CrossRef]
- Doyle, A.A.; Krämer, T.; Kavanagh, K.; Stephens, J.C. Cinnamaldehydes: Synthesis, antibacterial evaluation, and the effect of molecular structure on antibacterial activity. Results Chem. 2019, 25, 790–804. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faleye, O.S.; Sathiyamoorthi, E.; Lee, J.-H.; Lee, J. Inhibitory Effects of Cinnamaldehyde Derivatives on Biofilm Formation and Virulence Factors in Vibrio Species. Pharmaceutics 2021, 13, 2176. https://doi.org/10.3390/pharmaceutics13122176
Faleye OS, Sathiyamoorthi E, Lee J-H, Lee J. Inhibitory Effects of Cinnamaldehyde Derivatives on Biofilm Formation and Virulence Factors in Vibrio Species. Pharmaceutics. 2021; 13(12):2176. https://doi.org/10.3390/pharmaceutics13122176
Chicago/Turabian StyleFaleye, Olajide Sunday, Ezhaveni Sathiyamoorthi, Jin-Hyung Lee, and Jintae Lee. 2021. "Inhibitory Effects of Cinnamaldehyde Derivatives on Biofilm Formation and Virulence Factors in Vibrio Species" Pharmaceutics 13, no. 12: 2176. https://doi.org/10.3390/pharmaceutics13122176
APA StyleFaleye, O. S., Sathiyamoorthi, E., Lee, J.-H., & Lee, J. (2021). Inhibitory Effects of Cinnamaldehyde Derivatives on Biofilm Formation and Virulence Factors in Vibrio Species. Pharmaceutics, 13(12), 2176. https://doi.org/10.3390/pharmaceutics13122176





