Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses
Abstract
:1. Introduction
2. Materials and Methods
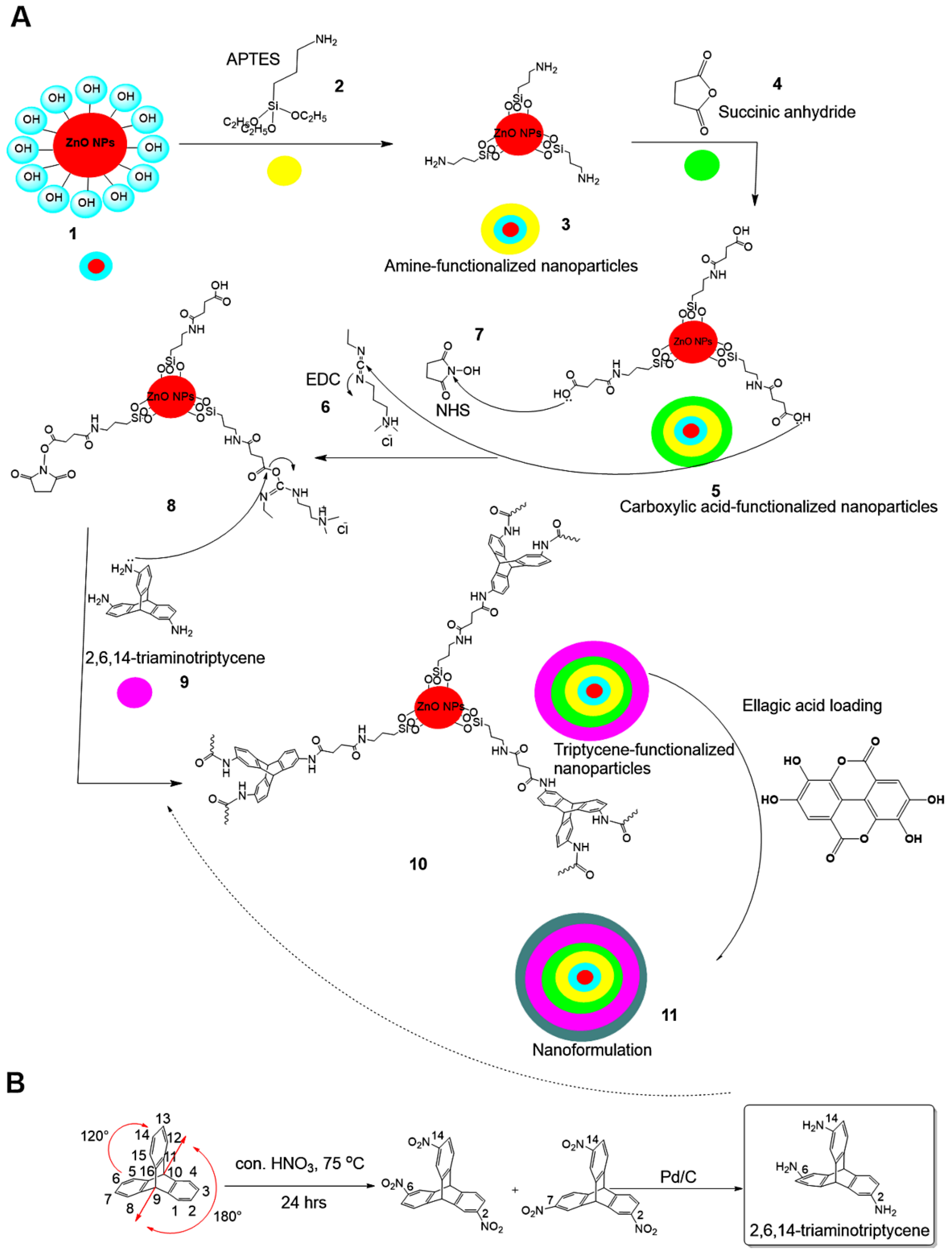
2.1. Triptycene Functionalization
- In the first step, ZnO NPs were modified with amino groups (–NH2) by adding 1.250 g ZnO NPs to 150 mL anhydrous toluene (POCH, Gliwice, Poland) and reacting it with 1.5 mL 3-aminopropyltriethoxysilane (APTES, Sigma-Aldrich, St. Louis, MO, USA) for 24 h under medium stirring speed (250 rpm) at room temperature (RT). The surface-modified NPs were collected using a Sigma 3-30KS cooling centrifuge (Sigma Laborzentrifugen GmbH, Osterode am Harz, Germany) and washed with water (18.2 MW, Milli-Q® system, Millipore, Darmstadt, Germany) to remove unreacted APTES. The collected surface-modified NPs were finally oven-dried at 50 °C to complete dryness, and the product was designated as ZnO NPs–NH2.
- In the second step, we functionalized the ZnO NPs–NH2 with carboxylic groups (–COOH) through a modified synthesis protocol, according to Hakeem et al. [35]. ZnO NPs–NH2 were suspended in acetone (50 mL, Fisher Scientific, Loughborough, UK) under water-bath sonication (5 min, Elma GmbH, Singen, Germany) and stirred at RT for 4 h (DAIHAN Scientific, Seoul, Korea). Next, succinic acid anhydride (0.7 M, 40 mL, Acros Organics, Geel, Belgium), dissolved in acetone, was added dropwise under stirring (350 rpm) and allowed to react for 24 h at RT. Subsequently, the functionalized particles were collected by centrifugation and washed with both deionized water and methanol (Fisher Scientific, Loughborough, UK). Finally, the obtained particles were oven-dried for 12 h at 60 °C and designated as ZnO NPs–NH2–COOH.
- In the final step, TRP-functionalized NPs were obtained in the following sequence: (1) ZnO NPs–NH2–COOH were activated through 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) coupling in 0.5% acetone (25 mL EDC and NHS, Acros Organics, Geel, Belgium) under stirring (400 rpm) for 6 h at 40 °C, resulting in solution A. (2) In separate screw cap bottles, TRP (300 mg) was activated in acetone containing EDC/NHS (0.5%, 50 mL), under stirring (400 rpm) for 6 h at 40 °C, resulting in solution B. (3) Solution B (25 mL) was slowly dropped into solution A and stirred (250 rpm) at RT for 24 h. (4) We separated the obtained NPs by centrifugation and washed them with deionized water and acetone to remove unreacted TRP, before drying at 60 °C. The obtained material was designated ZnO NPs–NH2–COOH–TRP.
2.2. Nanoformulation Preparation
2.3. Characterization Techniques
2.4. Antiviral Evaluations
2.4.1. Viruses and Cell Lines
2.4.2. Cytotoxicity Assessment
2.5. Antiviral Assessment
2.5.1. Cytopathic Assay
2.5.2. Virucidal Mechanism
2.6. Statistical Analysis
3. Results and Discussion
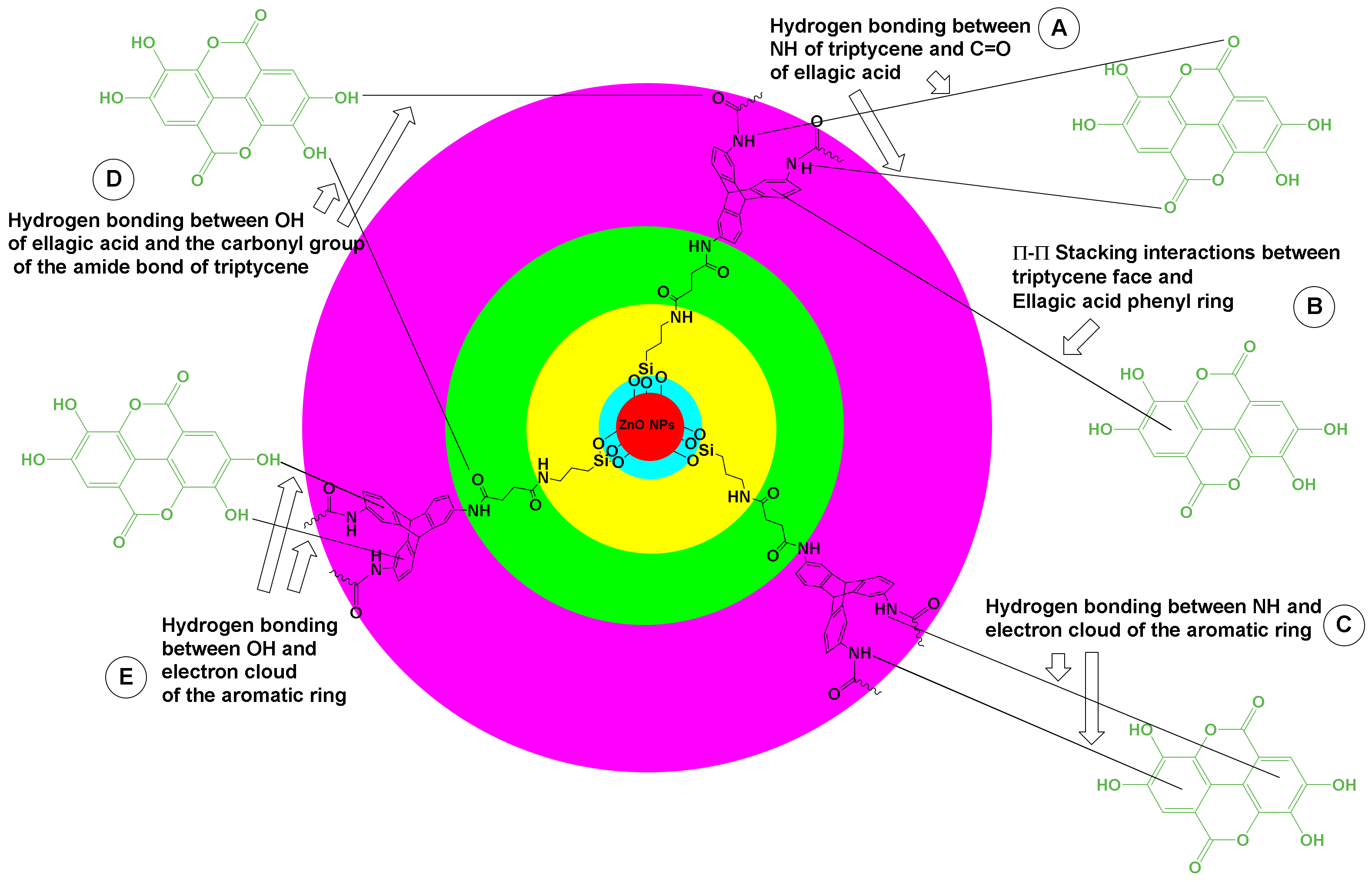
3.1. Hybrid Nanoformulation Preparation
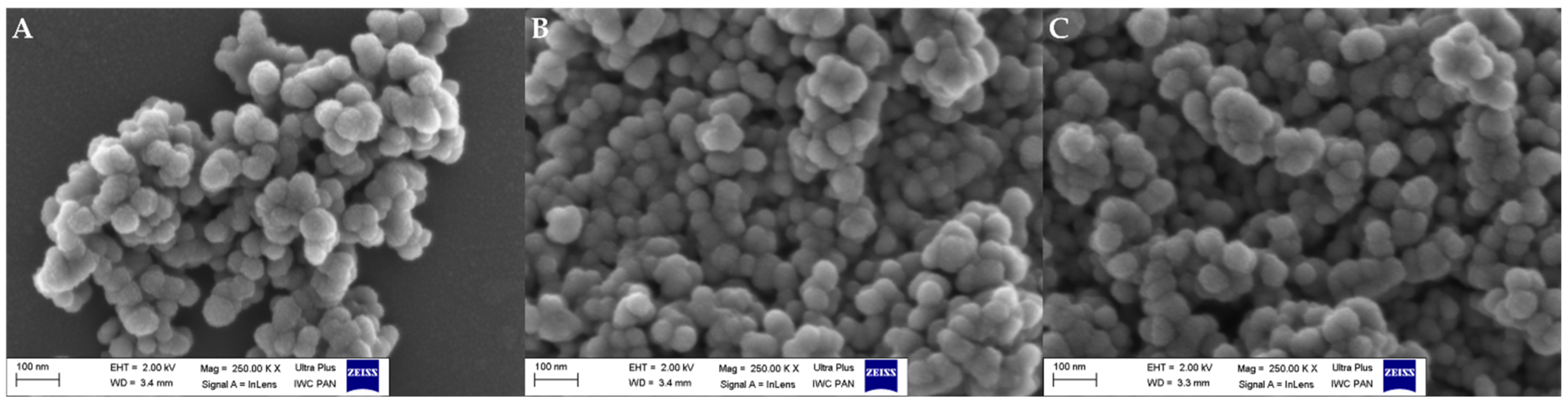
3.2. Microscopic Observations
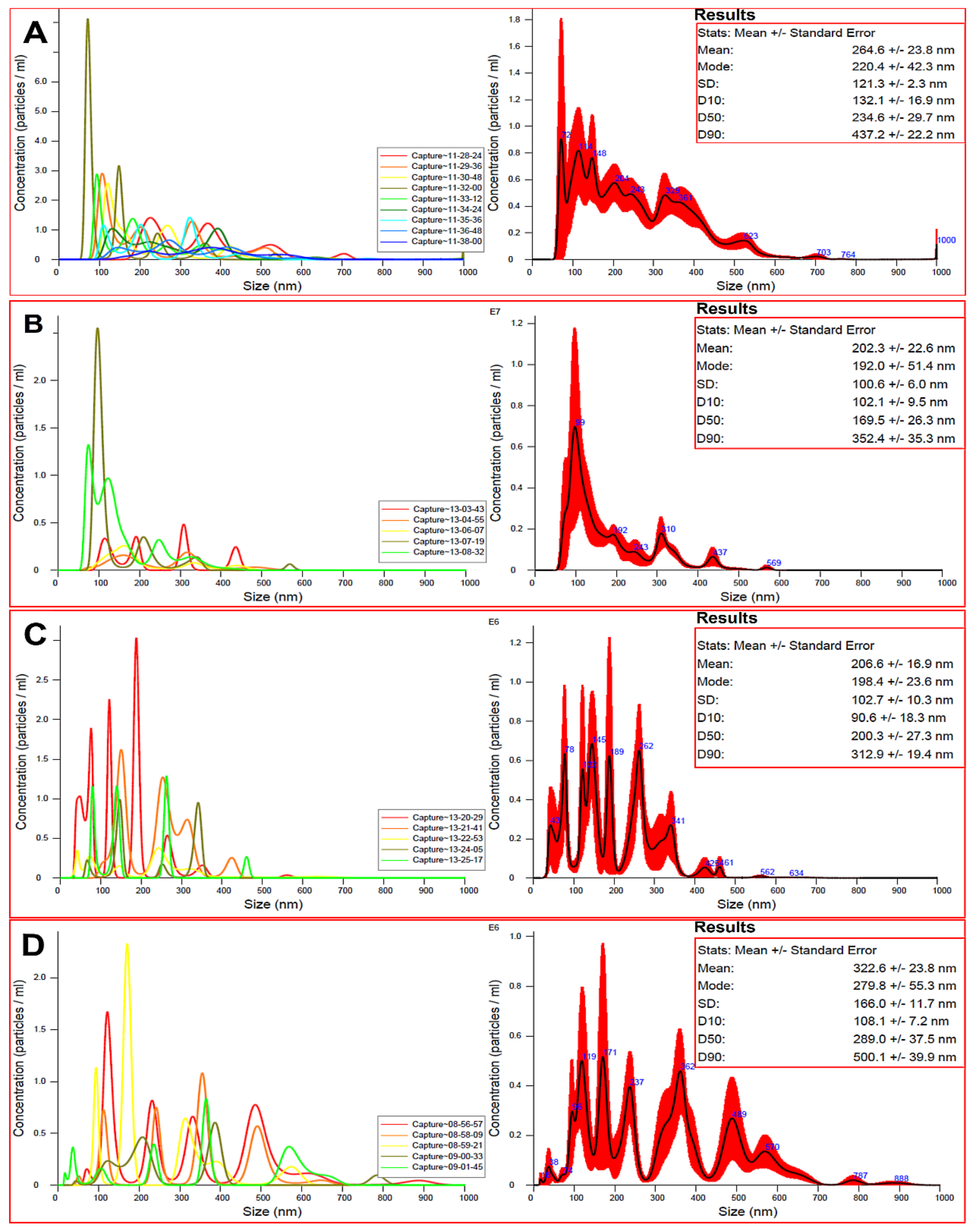
3.3. Particle Size Measurements
3.4. Specific Surface Area
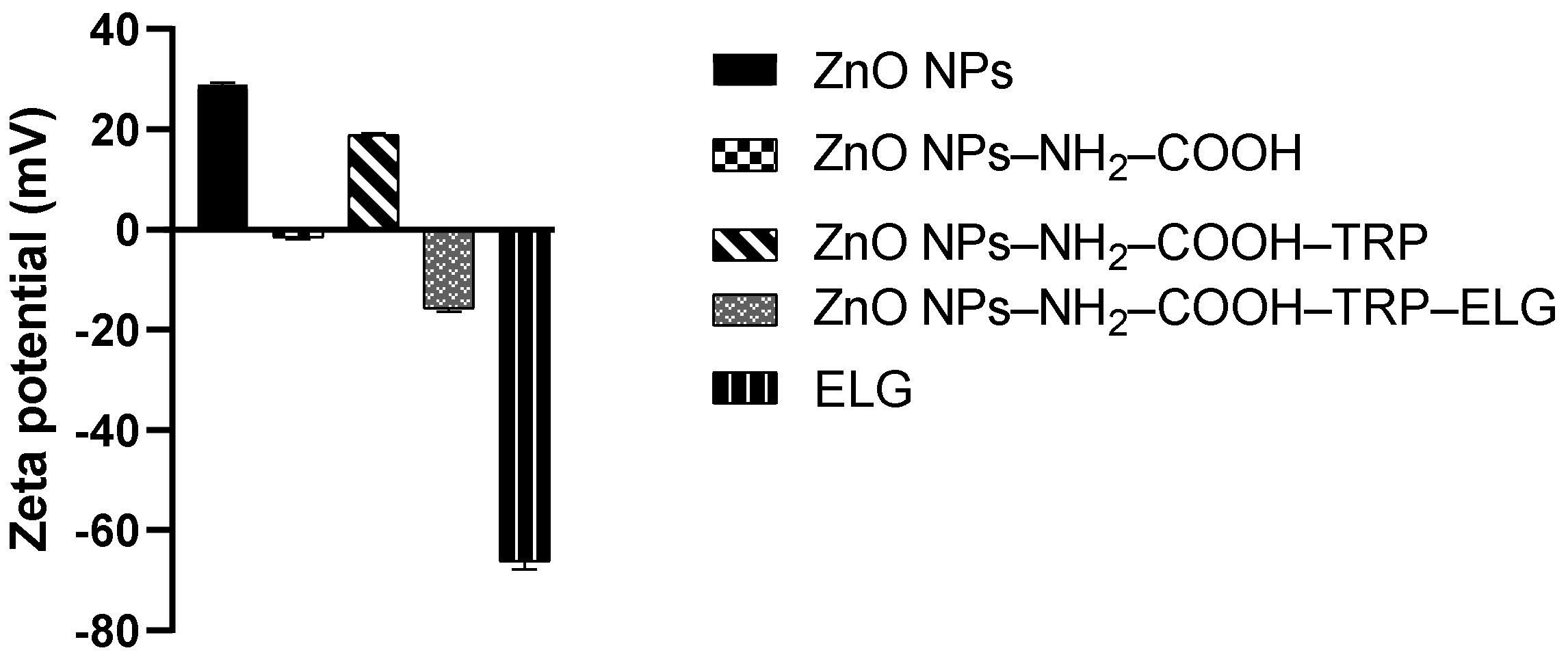
3.5. Zeta Potential Measurements
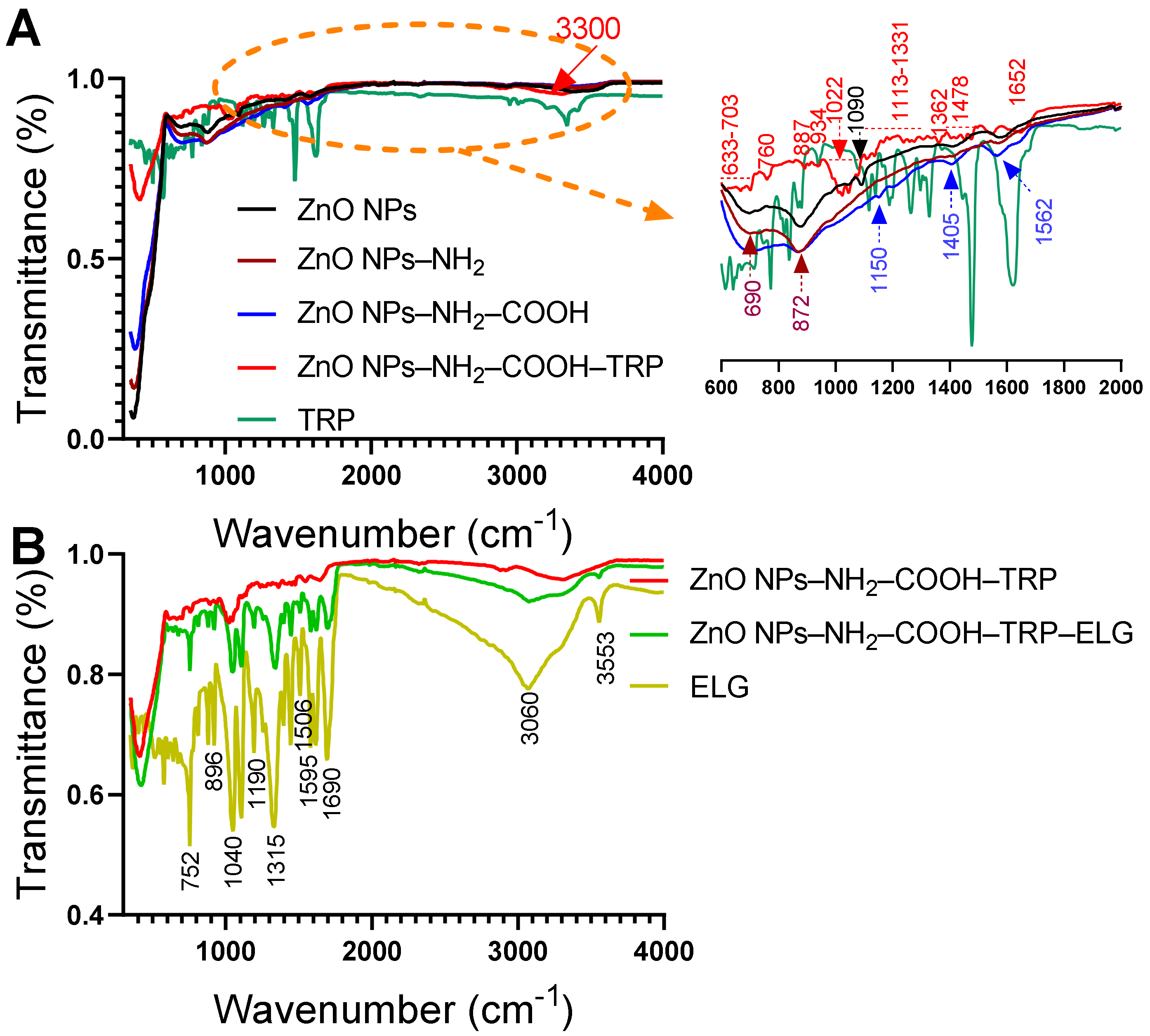
3.6. FTIR–ATR Analysis
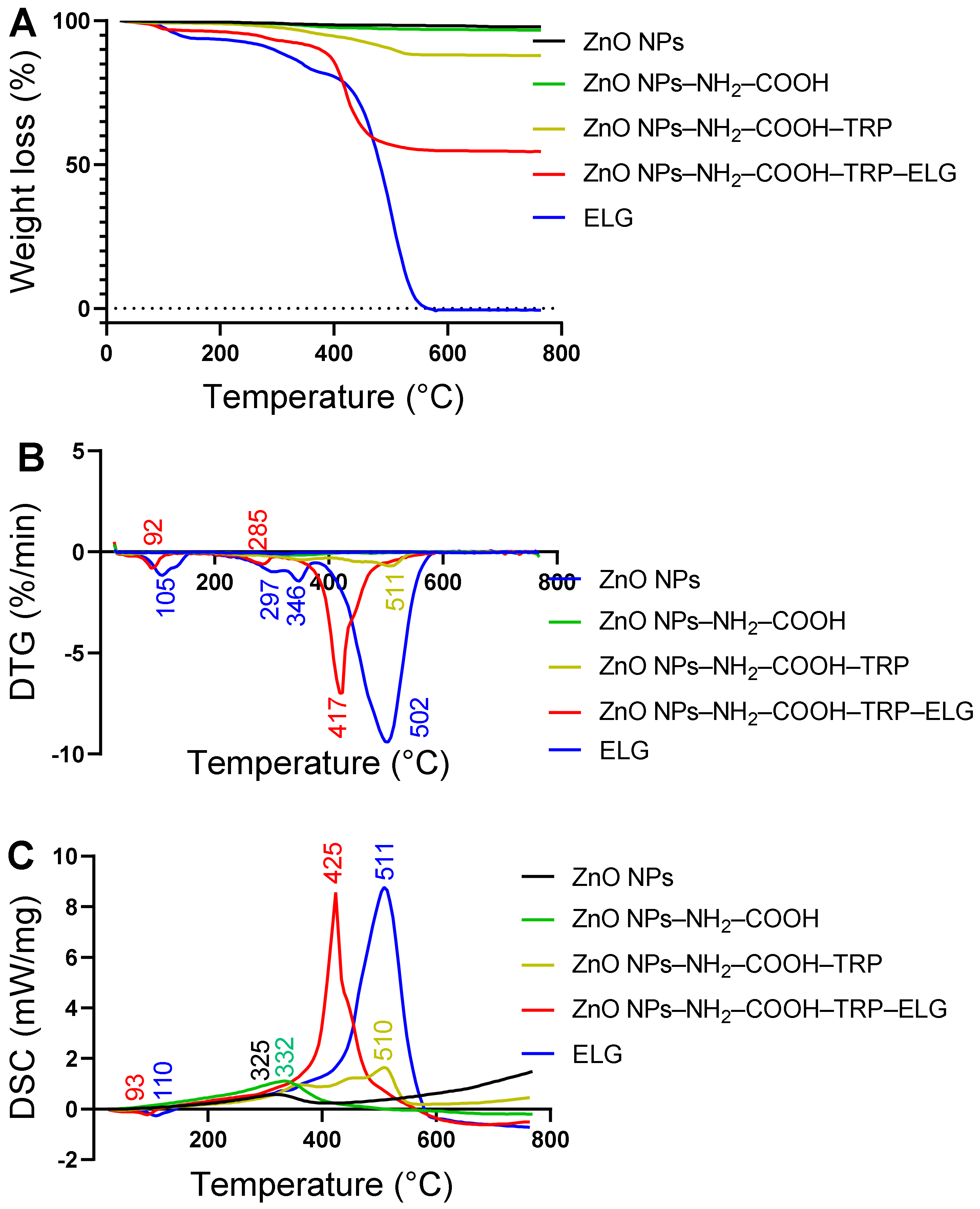
3.7. Thermal and DSC Analysis
3.8. Cytotoxicity Evaluations
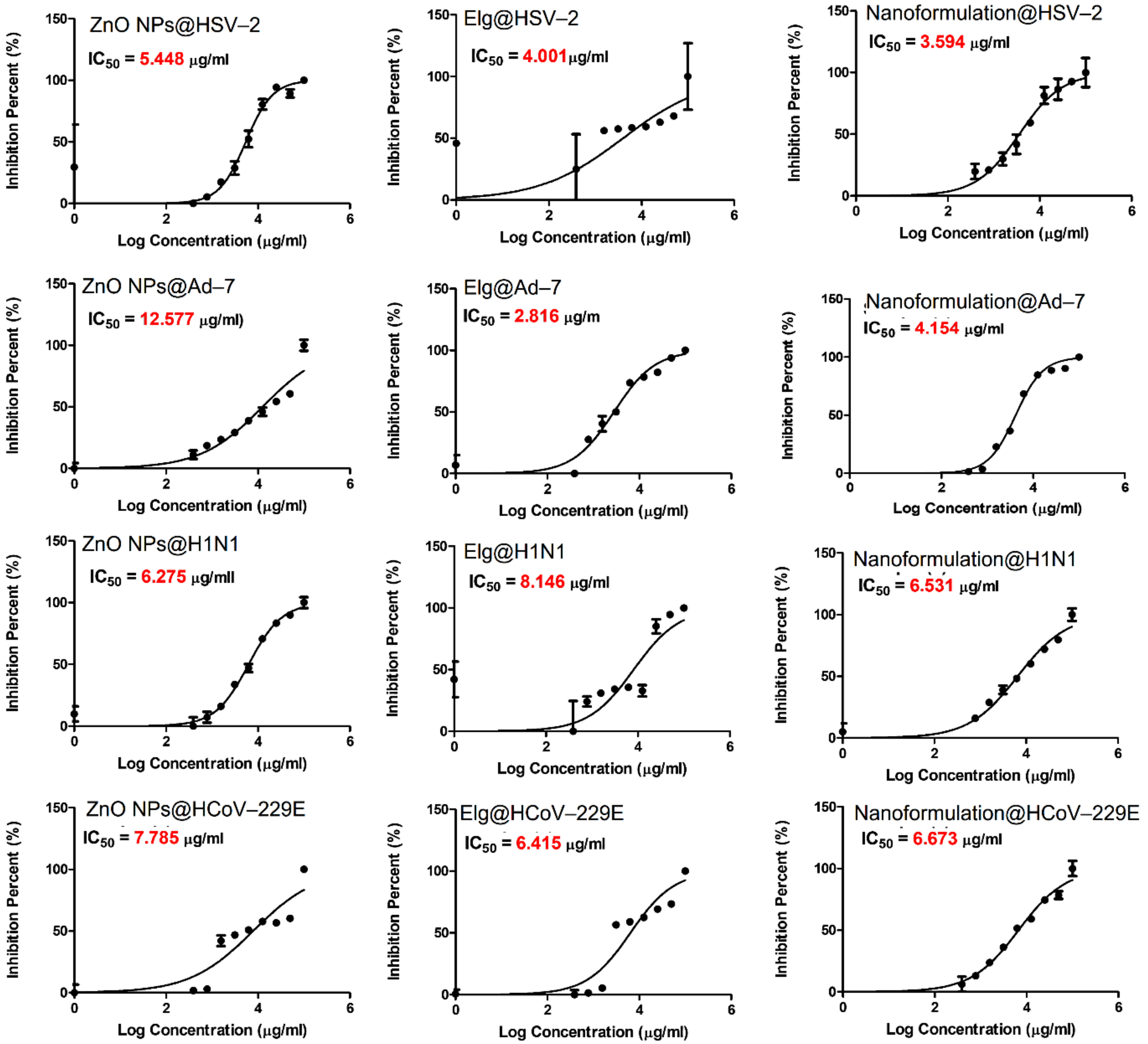
3.9. Antiviral Evaluation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vecitis, C.D. Antiviral-nanoparticle interactions and reactions. Environ. Sci. Nano 2021, 8, 11–19. [Google Scholar] [CrossRef]
- Tharayil, A.; Rajakumari, R.; Kumar, A.; Choudhary, M.D.; Palit, P.; Thomas, S. New insights into application of nanoparticles in the diagnosis and screening of novel coronavirus (SARS-CoV-2). Emergent. Mater. 2021, 4, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Tan, X. Current Strategies of Antiviral Drug Discovery for COVID-19. Front. Mol. Biosci. 2021, 8, 310. [Google Scholar] [CrossRef]
- Yang, K.C.; Lin, J.C.; Tsai, H.H.; Hsu, C.Y.; Shih, V.; Hu, C.J. Nanotechnology advances in pathogen- and host-targeted antiviral delivery: Multipronged therapeutic intervention for pandemic control. Drug Deliv. Transl. Res. 2021, 11, 1420–1437. [Google Scholar] [CrossRef]
- Shah, S.; Chougule, M.B.; Kotha, A.K.; Kashikar, R.; Godugu, C.; Raghuvanshi, R.S.; Singh, S.B.; Srivastava, S. Nanomedicine based approaches for combating viral infections. J. Control. Release 2021, 338, 80–104. [Google Scholar] [CrossRef] [PubMed]
- Jamalipour Soufi, G.; Iravani, S. Nanomaterials against pathogenic viruses: Greener and sustainable approaches. Inorg. Nano-Met. Chem. 2021, 51, 1598–1614. [Google Scholar] [CrossRef]
- Sadique, M.A.; Yadav, S.; Ranjan, P.; Verma, S.; Salammal, S.T.; Khan, M.A.; Kaushik, A.; Khan, R. High-performance antiviral nano-systems as a shield to inhibit viral infections: SARS-CoV-2 as a model case study. J. Mater. Chem. B 2021, 9, 4620–4642. [Google Scholar] [CrossRef] [PubMed]
- Innocenzi, P.; Stagi, L. Carbon-based antiviral nanomaterials: Graphene, C-dots, and fullerenes. A perspective. Chem. Sci. 2020, 11, 6606–6622. [Google Scholar] [CrossRef]
- AbouAitah, K.; Swiderska-Sroda, A.; Kandeil, A.; Salman, A.M.M.; Wojnarowicz, J.; Ali, M.A.; Opalinska, A.; Gierlotka, S.; Ciach, T.; Lojkowski, W. Virucidal Action against Avian Influenza H5N1 Virus and Immunomodulatory Effects of Nanoformulations Consisting of Mesoporous Silica Nanoparticles Loaded with Natural Prodrugs. Int. J. Nanomed. 2020, 15, 5181–5202. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, H.; Tavakoli, A.; Moradi, A.; Tabarraei, A.; Bokharaei-Salim, F.; Zahmatkeshan, M.; Farahmand, M.; Javanmard, D.; Kiani, S.J.; Esghaei, M.; et al. Inhibition of H1N1 influenza virus infection by zinc oxide nanoparticles: Another emerging application of nanomedicine. J. Biomed. Sci. 2019, 26, 70. [Google Scholar] [CrossRef]
- Jaros, S.W.; Król, J.; Bażanów, B.; Poradowski, D.; Chrószcz, A.; Nesterov, D.S.; Kirillov, A.M.; Smoleński, P. Antiviral, Antibacterial, Antifungal, and Cytotoxic Silver(I) BioMOF Assembled from 1,3,5-Triaza-7-Phoshaadamantane and Pyromellitic Acid. Molecules 2020, 25, 2119. [Google Scholar] [CrossRef]
- Galdiero, S.; Falanga, A.; Vitiello, M.; Cantisani, M.; Marra, V.; Galdiero, M. Silver nanoparticles as potential antiviral agents. Molecules 2011, 16, 8894–8918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbouAitah, K.; Piotrowska, U.; Wojnarowicz, J.; Swiderska-Sroda, A.; El-Desoky, A.H.H.; Lojkowski, W. Enhanced Activity and Sustained Release of Protocatechuic Acid, a Natural Antibacterial Agent, from Hybrid Nanoformulations with Zinc Oxide Nanoparticles. Int. J. Mol. Sci. 2021, 22, 5287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Luo, S.; Weidman, J.; Guo, R. Surface modification of ZIF-90 with triptycene for enhanced interfacial interaction in mixed-matrix membranes for gas separation. J. Polym. Sci. 2020, 58, 2675–2687. [Google Scholar] [CrossRef]
- Moylan, C.; Rogers, L.; Shaker, Y.M.; Davis, M.; Eckhardt, H.-G.; Eckert, R.; Ryan, A.A.; Senge, M.O. Preparation of Tri- and Hexasubstituted Triptycene Synthons by Transition Metal Catalyzed Cross-Coupling Reactions for Post-Modifications. Eur. J. Org. Chem. 2016, 2016, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.H.; Ardakani, S.J.; Smith, K.J.; MacLachlan, M.J. Triptycene-Based Metal Salphens—Exploiting Intrinsic Molecular Porosity for Gas Storage. Chem.—Eur. J. 2009, 15, 11824–11828. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, K.; Müller-Bunz, H.; Ortin, Y.; Muldoon, J.; McGlinchey, M.J. Molecular Dials: Hindered Rotations in Mono- and Diferrocenyl Anthracenes and Triptycenes. J. Am. Chem. Soc. 2010, 132, 17617–17622. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, E.M.; Rein, R.; Solladié, N.; Dahms, K.; Götz, D.C.G.; Bringmann, G.; Senge, M.O. Synthesis and ligand binding properties of triptycene-linked porphyrin arrays. Tetrahedron 2011, 67, 1126–1134. [Google Scholar] [CrossRef] [Green Version]
- Jacquot de Rouville, H.-P.; Garbage, R.; Cook, R.E.; Pujol, A.R.; Sirven, A.M.; Rapenne, G. Synthesis of Polycyclic Aromatic Hydrocarbon-Based Nanovehicles Equipped with Triptycene Wheels. Chem.—Eur. J. 2012, 18, 3023–3031. [Google Scholar] [CrossRef]
- Rogerio, A.P.; Fontanari, C.; Borducchi, E.; Keller, A.C.; Russo, M.; Soares, E.G.; Albuquerque, D.A.; Faccioli, L.H. Anti-inflammatory effects of Lafoensia pacari and ellagic acid in a murine model of asthma. Eur. J. Pharm. 2008, 580, 262–270. [Google Scholar] [CrossRef]
- Bell, C.; Hawthorne, S. Ellagic acid, pomegranate and prostate cancer—A mini review. J. Pharm. Pharm. 2008, 60, 139–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, E.H.; Kown, T.Y.; Oh, G.T.; Park, W.F.; Park, S.I.; Park, S.K.; Lee, Y.I. The flavonoid ellagic acid from a medicinal herb inhibits host immune tolerance induced by the hepatitis B viruse antigen. Antivir. Res. 2006, 72, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, E.L.; Zografov, N.N.; Simeonova, L.S. Comparative study on the antioxidant capacities of synthetic influenza inhibitors and ellagic acid in model systems. Biomed. Pharm. 2016, 83, 755–762. [Google Scholar] [CrossRef]
- Soh, P.N.; Witkowski, B.; Olagnier, D.; Nicolau, M.L.; Garcia-Alvarez, M.C.; Berry, A.; Benoit-Vical, F. In vitro and in vivo properties of ellagic acid in malaria treatment. Antimicrob. Agents Chemother. 2009, 53, 1100–1106. [Google Scholar] [CrossRef] [Green Version]
- Park, S.W.; Kwon, M.J.; Yoo, J.Y.; Choi, H.-J.; Ahn, Y.-J. Antiviral activity and possible mode of action of ellagic acid identified in Lagerstroemia speciosa leaves toward human rhinoviruses. BMC Complement. Altern. Med. 2014, 14, 171. [Google Scholar] [CrossRef] [Green Version]
- Promsong, A.; Chuenchitra, T.; Saipin, K.; Tewtrakul, S.; Panichayupakaranant, P.; Satthakarn, S.; Nittayananta, W. Ellagic acid inhibits HIV-1 infection in vitro: Potential role as a novel microbicide. Oral Dis. 2018, 24, 249–252. [Google Scholar] [CrossRef]
- Vilhelmova-Ilieva, N.; Jacquet, R.; Deffieux, D.; Pouységu, L.; Sylla, T.; Chassaing, S.; Nikolova, I.; Quideau, S.; Galabov, A.S. Anti-Herpes Simplex Virus Type 1 Activity of Specially Selected Groups of Tannins. Drug Res. 2019, 69, 374. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Du, R.; Anantpadma, M.; Schafer, A.; Hou, L.; Tian, J.; Davey, R.A.; Cheng, H.; Rong, L. Identification of Ellagic Acid from Plant Rhodiola rosea L. as an Anti-Ebola Virus Entry Inhibitor. Viruses 2018, 10, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundararajan, A.; Ganapathy, R.; Huan, L.; Dunlap, J.R.; Webby, R.J.; Kotwal, G.J.; Sangster, M.Y. Influenza virus variation in susceptibility to inactivation by pomegranate polyphenols is determined by envelope glycoproteins. Antivir. Res. 2010, 88, 1–9. [Google Scholar] [CrossRef]
- Karimi, A.; Moradi, M.-T.; Rabiei, M.; Alidadi, S. In vitro anti-adenoviral activities of ethanol extract, fractions, and main phenolic compounds of pomegranate (Punica granatum L.) peel. Antivir. Chem. Chemother. 2020, 28, 2040206620916571. [Google Scholar] [CrossRef]
- Acquadro, S.; Civra, A.; Cagliero, C.; Marengo, A.; Rittà, M.; Francese, R.; Sanna, C.; Bertea, C.; Sgorbini, B.; Lembo, D.; et al. Punica granatum Leaf Ethanolic Extract and Ellagic Acid as Inhibitors of Zika Virus Infection. Planta Med. 2020, 86, 1363–1374. [Google Scholar] [CrossRef]
- Pavlova, E.L.; Simeonova, L.S.; Gegova, G.A. Combined efficacy of oseltamivir, isoprinosine and ellagic acid in influenza A(H3N2)-infected mice. Biomed Pharm. 2018, 98, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Wojnarowicz, J.; Chudoba, T.; Gierlotka, S.; Lojkowski, W. Effect of Microwave Radiation Power on the Size of Aggregates of ZnO NPs Prepared Using Microwave Solvothermal Synthesis. Nanomaterials 2018, 8, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojnarowicz, J.; Chudoba, T.; Koltsov, I.; Gierlotka, S.; Dworakowska, S.; Lojkowski, W. Size control mechanism of ZnO nanoparticles obtained in microwave solvothermal synthesis. Nanotechnology 2018, 29, 065601. [Google Scholar] [CrossRef] [PubMed]
- Hakeem, A.; Duan, R.; Zahid, F.; Dong, C.; Wang, B.; Hong, F.; Ou, X.; Jia, Y.; Lou, X.; Xia, F. Dual stimuli-responsive nano-vehicles for controlled drug delivery: Mesoporous silica nanoparticles end-capped with natural chitosan. Chem. Commun. 2014, 50, 13268–13271. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Song, J.H.; Choi, H.J.; Song, H.H.; Hong, E.H.; Lee, B.R.; Oh, S.R.; Choi, K.; Yeo, S.G.; Lee, Y.P.; Cho, S.; et al. Antiviral activity of ginsenosides against coxsackievirus B3, enterovirus 71, and human rhinovirus 3. J. Ginseng Res. 2014, 38, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Schuhmacher, A.; Reichling, J.; Schnitzler, P. Virucidal effect of peppermint oil on the enveloped viruses herpes simplex virus type 1 and type 2 in vitro. Phytomedicine 2003, 10, 504–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malloy, A. Count, size and visualize nanoparticles. Mater. Today 2011, 14, 170–173. [Google Scholar] [CrossRef]
- Tang, E.; Cheng, G.; Ma, X.; Pang, X.; Zhao, Q. Surface modification of zinc oxide nanoparticle by PMAA and its dispersion in aqueous system. Appl. Surf. Sci. 2006, 252, 5227–5232. [Google Scholar] [CrossRef]
- Akbar, N.; Aslam, Z.; Siddiqui, R.; Shah, M.R.; Khan, N.A. Zinc oxide nanoparticles conjugated with clinically-approved medicines as potential antibacterial molecules. AMB Express 2021, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Shen, C.; Feltis, B.N.; Martin, L.L.; Hughes, A.E.; Wright, P.F.A.; Turney, T.W. Reducing ZnO nanoparticle cytotoxicity by surface modification. Nanoscale 2014, 6, 5791–5798. [Google Scholar] [CrossRef] [PubMed]
- Halbus, A.F.; Horozov, T.S.; Paunov, V.N. Surface-Modified Zinc Oxide Nanoparticles for Antialgal and Antiyeast Applications. ACS Appl. Nano Mater. 2020, 3, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Hu, Y.; Zhang, Y.; Wei, W.; Hou, X.; Guo, Y.; Hu, X.; Jiang, D. Enhancing the mechanical performance of poly(ether ether ketone)/zinc oxide nanocomposites to provide promising biomaterials for trauma and orthopedic implants. RSC Adv. 2018, 8, 27304–27317. [Google Scholar] [CrossRef] [Green Version]
- Baghdadi, Y.N.; Youssef, L.; Bouhadir, K.; Harb, M.; Mustapha, S.; Patra, D.; Tehrani-Bagha, A.R. The effects of modified zinc oxide nanoparticles on the mechanical/thermal properties of epoxy resin. J. Appl. Polym. Sci. 2020, 137, 49330. [Google Scholar] [CrossRef]
- Feifel, S.C.; Lisdat, F. Silica nanoparticles for the layer-by-layer assembly of fully electro-active cytochrome c multilayers. J. Nanobiotechnol. 2011, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, S.; Raj, C.J.; Robert, R.; Ramanand, A.; Das, S.J. Growth and characterization of succinic acid single crystals. Cryst. Res. Technol. 2007, 42, 1087–1090. [Google Scholar] [CrossRef]
- Barahuie, F.; Hussein, M.Z.; Gani, S.A.; Fakurazi, S.; Zainal, Z. Synthesis of protocatechuic acid–zinc/aluminium–layered double hydroxide nanocomposite as an anticancer nanodelivery system. J. Solid State Chem. 2015, 221, 21–31. [Google Scholar] [CrossRef]
- Usman, M.S.; Hussein, M.Z.; Kura, A.U.; Fakurazi, S.; Masarudin, M.J.; Ahmad Saad, F.F. Graphene Oxide as a Nanocarrier for a Theranostics Delivery System of Protocatechuic Acid and Gadolinium/Gold Nanoparticles. Molecules 2018, 23, 500. [Google Scholar] [CrossRef] [Green Version]
- Parang, K.; El-Sayed, N.S.; Kazeminy, A.J.; Tiwari, R.K. Comparative Antiviral Activity of Remdesivir and Anti-HIV Nucleoside Analogs against Human Coronavirus 229E (HCoV-229E). Molecules 2020, 25, 2343. [Google Scholar] [CrossRef]
- Cheng, P.-W.; Ng, L.-T.; Chiang, L.-C.; Lin, C.-C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006, 33, 612–616. [Google Scholar] [CrossRef] [PubMed]



| Materials and Formulation | Weight Loss, wt.% a | SBET (m2/g) b | Total Drug Content c | Entrapment Efficiency, % d |
|---|---|---|---|---|
| ZnO NPs | 2.2 | 51.2 | ||
| ZnO NPs–NH2–COOH | 3.3 | 21.6 | ||
| ZnO NPs–NH2–COOH–TRP | 12.1 | 13.1 | ||
| ZnO NPs–NH2–COOH–TRP–ELG | 45.5 | 11.7 | 33.3 | 83.2 |
| Treatment/Virus | CC50, µg/mL | IC50, µg/mL | Therapeutic Index |
|---|---|---|---|
| ZnO NPs | |||
| HSV-2 | 222.7 | 5.4 | 40.8 |
| Ad-7 | 404.5 | 12.5 | 32.1 |
| H1N1 | 329.1 | 6.2 | 52.4 |
| HCoV-229E | 329.1 | 7.7 | 42.3 |
| ELG | |||
| HSV-2 | 112.5 | 4.0 | 28.1 |
| Ad-7 | 127.4 | 2.8 | 45.2 |
| H1N1 | 230.9 | 8.1 | 28.3 |
| HCoV-229E | 230.9 | 6.4 | 36.0 |
| ZnO NPs–NH2–COOH–TRP–ELG | |||
| HSV-2 | 206.9 | 3.6 | 57.5 |
| Ad-7 | 215.0 | 4.1 | 51.7 |
| H1N1 | 505.2 | 6.5 | 77.3 |
| HCoV-229E | 505.2 | 6.6 | 75.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AbouAitah, K.; Allayh, A.K.; Wojnarowicz, J.; Shaker, Y.M.; Swiderska-Sroda, A.; Lojkowski, W. Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses. Pharmaceutics 2021, 13, 2174. https://doi.org/10.3390/pharmaceutics13122174
AbouAitah K, Allayh AK, Wojnarowicz J, Shaker YM, Swiderska-Sroda A, Lojkowski W. Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses. Pharmaceutics. 2021; 13(12):2174. https://doi.org/10.3390/pharmaceutics13122174
Chicago/Turabian StyleAbouAitah, Khaled, Abdou K. Allayh, Jacek Wojnarowicz, Yasser M. Shaker, Anna Swiderska-Sroda, and Witold Lojkowski. 2021. "Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses" Pharmaceutics 13, no. 12: 2174. https://doi.org/10.3390/pharmaceutics13122174
APA StyleAbouAitah, K., Allayh, A. K., Wojnarowicz, J., Shaker, Y. M., Swiderska-Sroda, A., & Lojkowski, W. (2021). Nanoformulation Composed of Ellagic Acid and Functionalized Zinc Oxide Nanoparticles Inactivates DNA and RNA Viruses. Pharmaceutics, 13(12), 2174. https://doi.org/10.3390/pharmaceutics13122174









