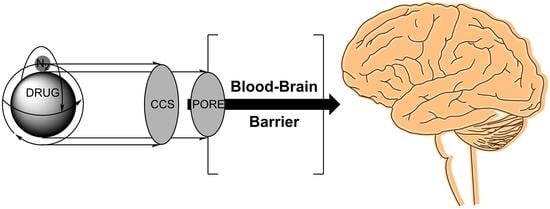
Large-Scale Evaluation of Collision Cross Sections to Investigate Blood-Brain Barrier Permeation of Drugs
Abstract
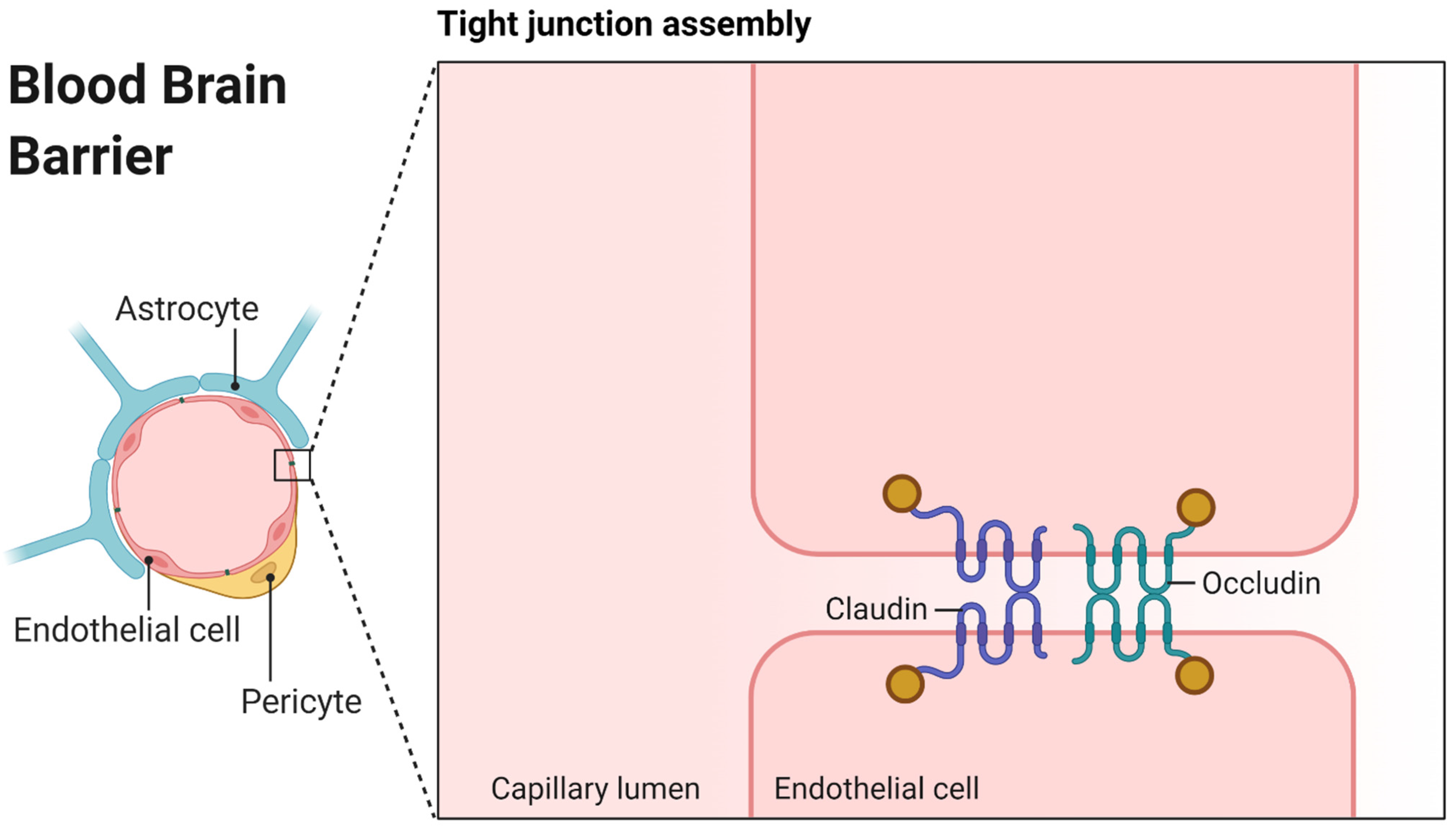
:1. Introduction
2. Materials and Methods
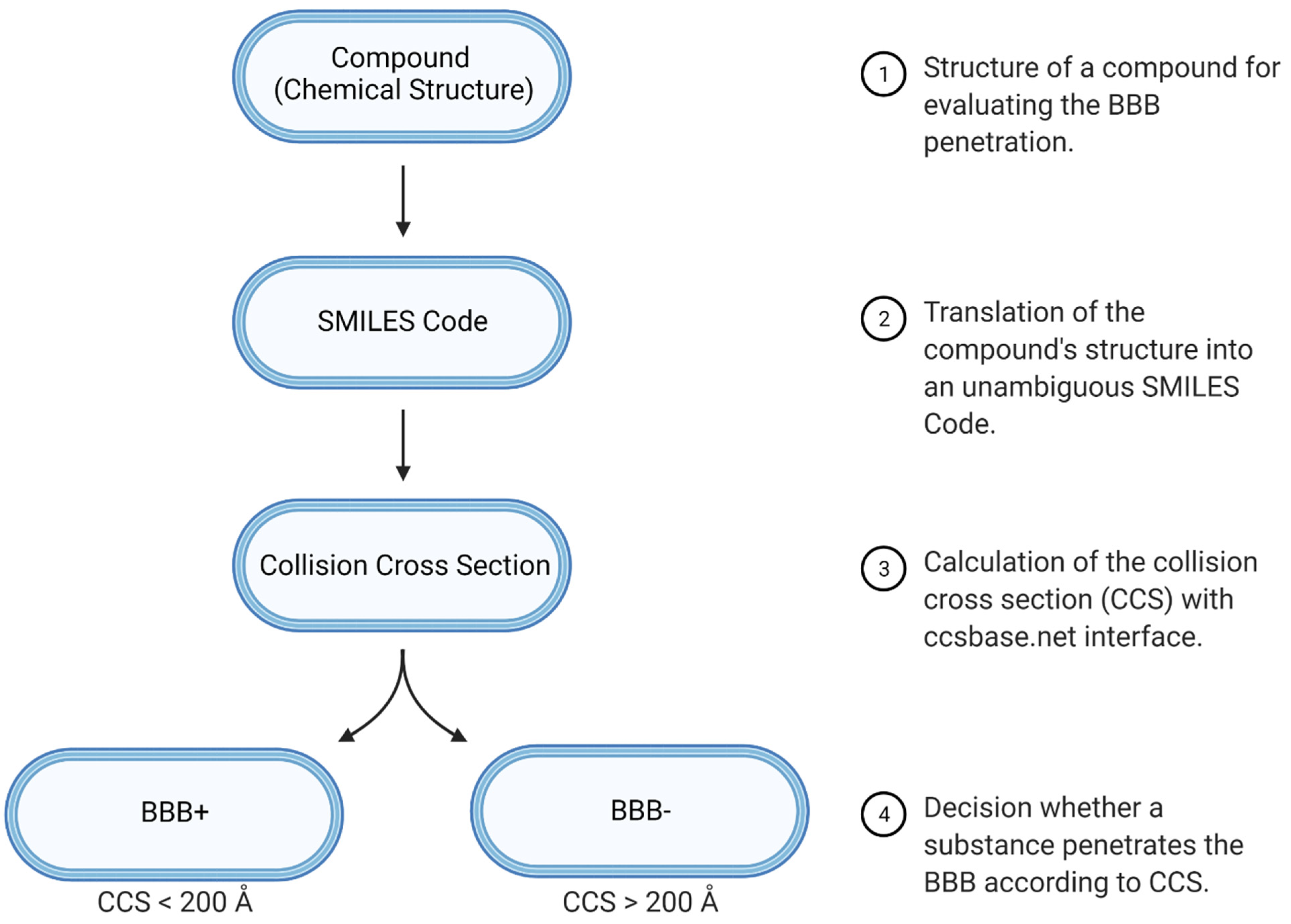
2.1. Statistical Evaluation of CCS as a Molecular Descriptor of BBB Penetration
2.2. Performance Evaluation of CCS Next to the BOILED-Egg Approach
2.3. Performance Evaluation of CCS as a Single Molecular Descriptor
3. Results and Discussion
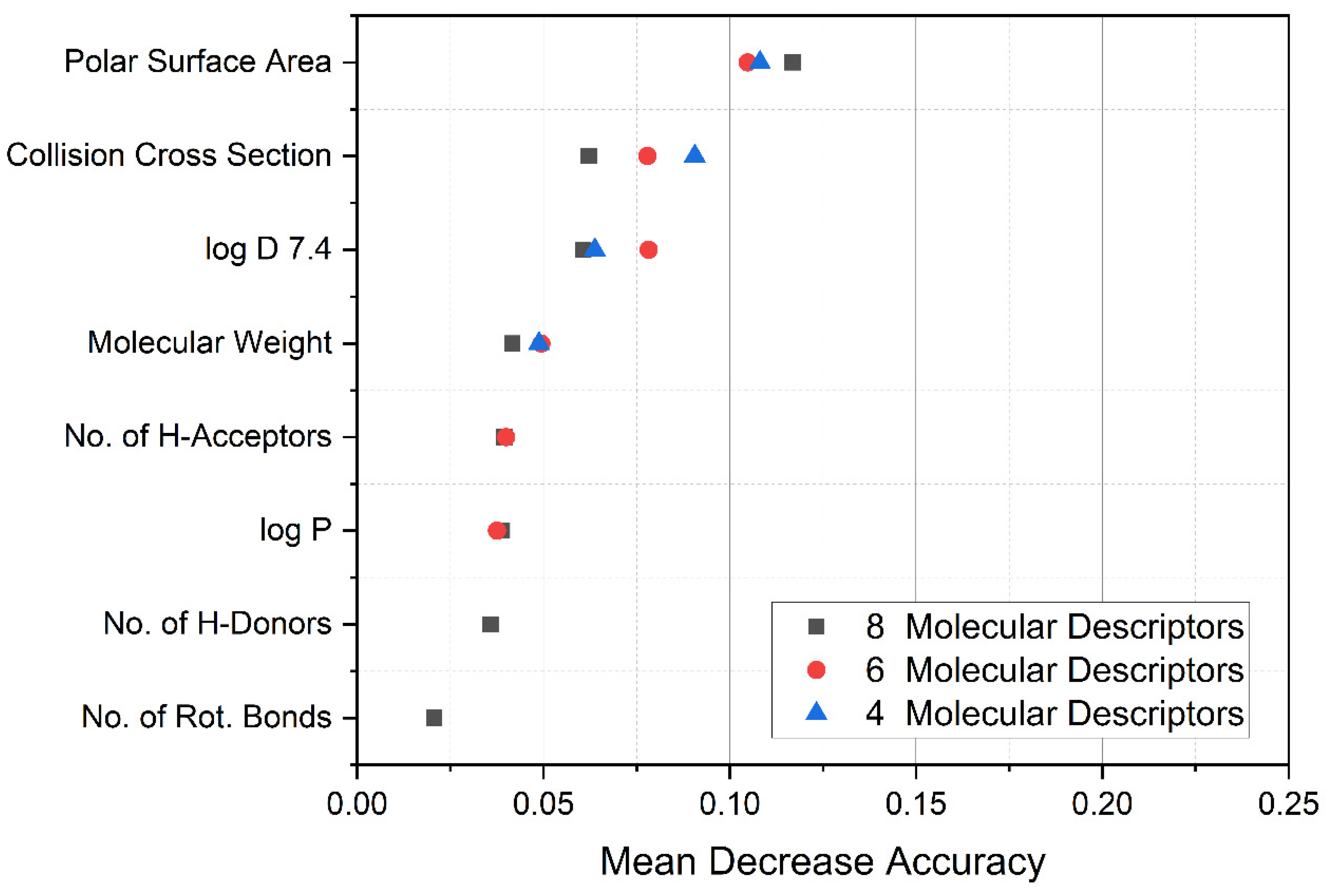
3.1. Random Forest Data Evaluation
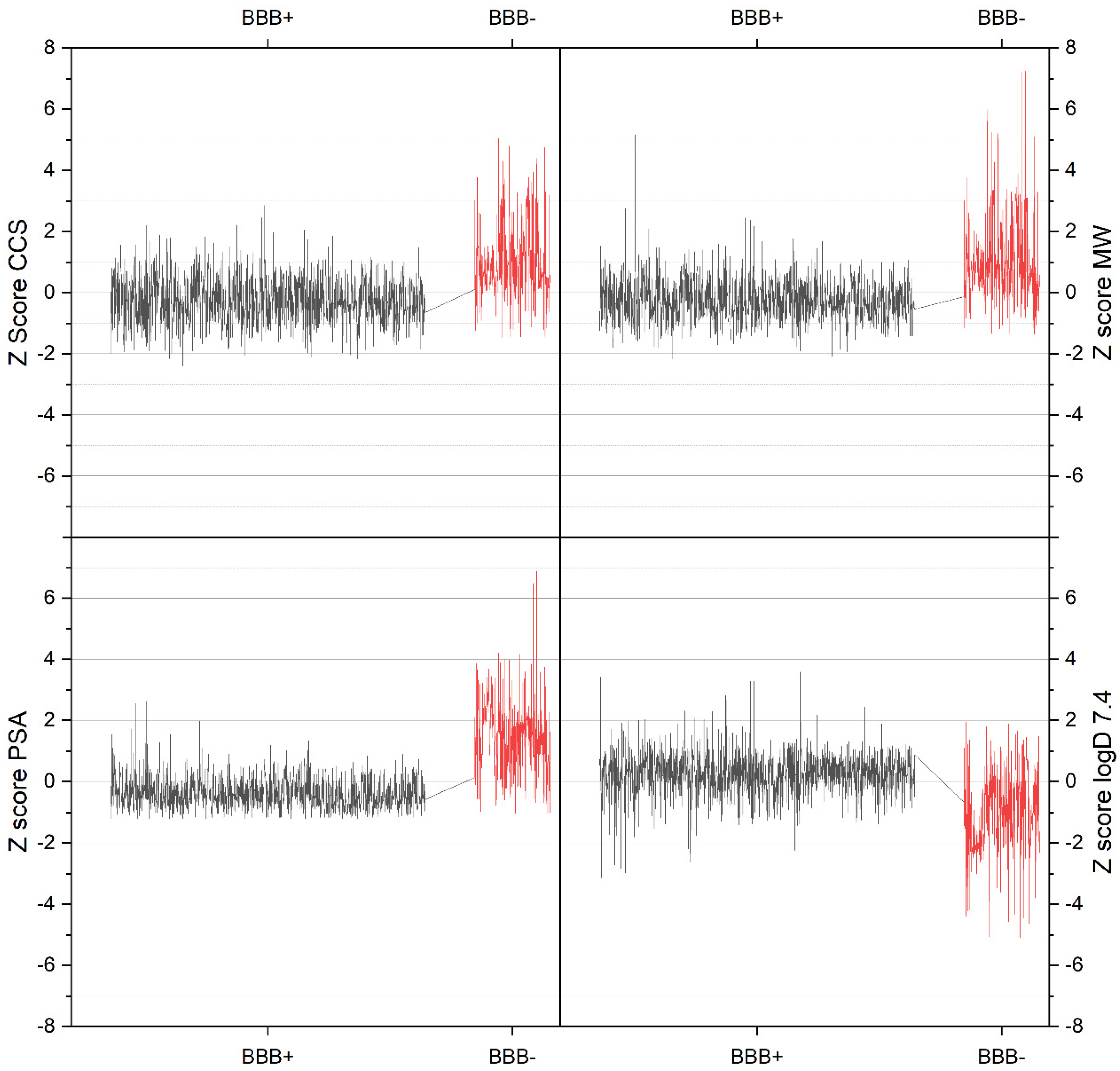
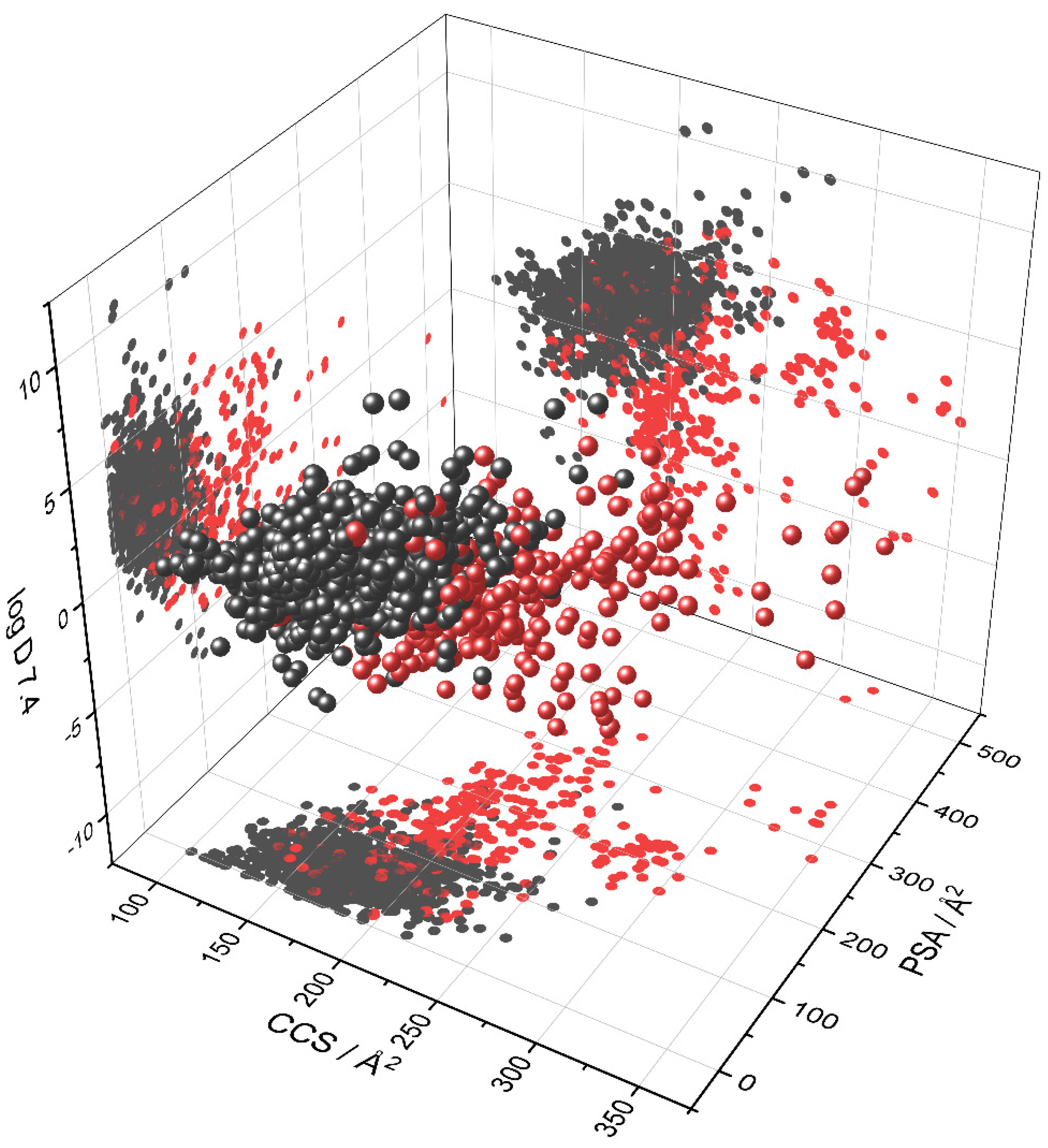
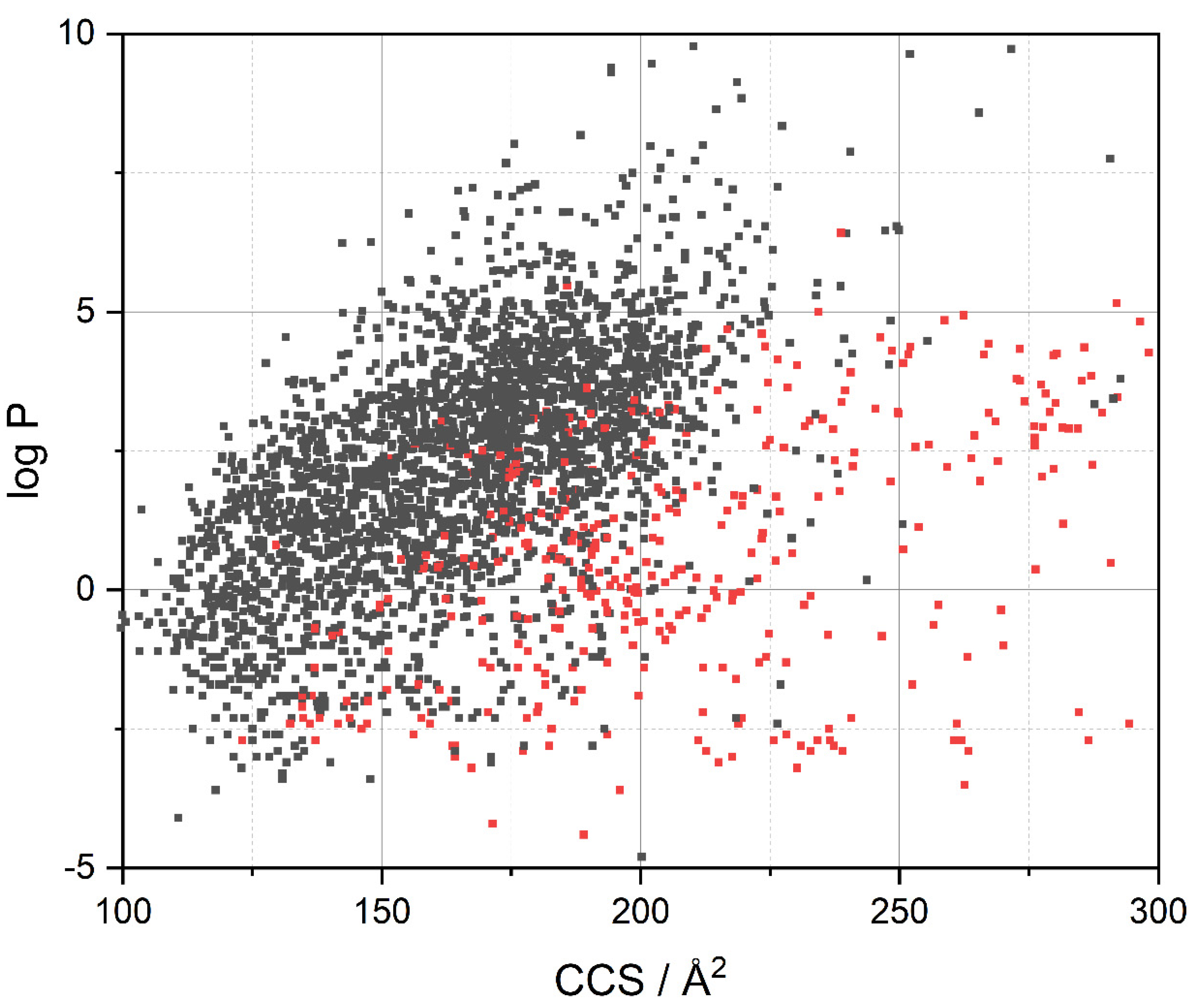
3.2. Visualization of Compound Properties for BBB Penetration
3.3. Drugbank Evaluation Regarding the BBB
3.4. Prediction Performance of CCS for Evaluating BBB Penetration Properties
3.5. Correlation of CCS with Pore Dimensions in the Brain
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reichel, A. Addressing Central Nervous System (CNS) Penetration in Drug Discovery: Basics and Implications of the Evolving New Concept. Chem. Biodivers. 2009, 6, 2030–2049. [Google Scholar] [CrossRef] [PubMed]
- Pardridge, W.M. Blood–brain barrier delivery. Drug Discov. Today 2007, 12, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Reichel, A. The Role of Blood-Brain Barrier Studies in the Pharmaceutical Industry. Curr. Drug Metab. 2006, 7, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund-Udenaes, M.; Fridén, M.; Syvänen, S.; Gupta, A. On the Rate and Extent of Drug Delivery to the Brain. Pharm. Res. 2008, 25, 1737–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, H.-C.; Krizbai, I.A.; Bauer, H.; Traweger, A. “You Shall Not Pass”—Tight junctions of the blood brain barrier. Front. Neurosci. 2014, 8, 392. [Google Scholar] [CrossRef]
- Abbott, N.J. Blood–brain barrier structure and function and the challenges for CNS drug delivery. J. Inherit. Metab. Dis. 2013, 36, 437–449. [Google Scholar] [CrossRef]
- Engelhardt, B.; Sorokin, L. The blood–brain and the blood–cerebrospinal fluid barriers: Function and dysfunction. Semin. Immunopathol. 2009, 31, 497–511. [Google Scholar] [CrossRef] [Green Version]
- Pajouhesh, H.; Lenz, G.R. Medicinal chemical properties of successful central nervous system drugs. NeuroRX 2005, 2, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Fong, C.W. Permeability of the Blood–Brain Barrier: Molecular Mechanism of Transport of Drugs and Physiologically Important Compounds. J. Membr. Biol. 2015, 248, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Gerebtzoff, G.; Seelig, A. In Silico Prediction of Blood−Brain Barrier Permeation Using the Calculated Molecular Cross-Sectional Area as Main Parameter. J. Chem. Inf. Model. 2006, 46, 2638–2650. [Google Scholar] [CrossRef]
- Fischer, H.; Gottschlich, R.; Seelig, A. Blood-Brain Barrier Permeation: Molecular Parameters Governing Passive Diffusion. J. Membr. Biol. 1998, 165, 201–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Bree, J.B.; De Boer, A.G.; Danhof, A.; Ginsel, L.A.; Breimer, D.D. Characterization of an “in vitro” blood-brain barrier: Effects of molecular size and lipophilicity on cerebrovascular endothelial transport rates of drugs. J. Pharmacol. Exp. Ther. 1988, 247, 1233–1239. [Google Scholar]
- Guntner, A.S.; Thalhamer, B.; Klampfl, C.; Buchberger, W. Collision cross sections obtained with ion mobility mass spectrometry as new descriptor to predict blood-brain barrier permeation by drugs. Sci. Rep. 2019, 9, 19182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Atri, V.; Causon, T.; Hernandez-Alba, O.; Mutabazi, A.; Veuthey, J.L.; Cianferani, S.; Guillarme, D. Adding a new separation dimension to MS and LC-MS: What is the utility of ion mobility spectrometry? J. Sep. Sci. 2018, 41, 20–67. [Google Scholar] [CrossRef] [PubMed]
- Stow, S.M.; Causon, T.J.; Zheng, X.; Kurulugama, R.T.; Mairinger, T.; May, J.C.; Rennie, E.E.; Baker, E.S.; Smith, R.D.; McLean, J.A.; et al. An Interlaboratory Evaluation of Drift Tube Ion Mobility–Mass Spectrometry Collision Cross Section Measurements. Anal. Chem. 2017, 89, 9048–9055. [Google Scholar] [CrossRef] [Green Version]
- Siems, W.F.; Viehland, L.A.; Hill, H.H. Improved Momentum-Transfer Theory for Ion Mobility. 1. Derivation of the Fundamental Equation. Anal. Chem. 2012, 84, 9782–9791. [Google Scholar] [CrossRef]
- Mason, E.A.; McDaniel, E.W. Transport Properties of Ions in Gases; Wiley: Hoboken, NJ, USA, 1988; ISBN 9780471883852. [Google Scholar]
- Ross, D.H.; Xu, L. Determination of drugs and drug metabolites by ion mobility-mass spectrometry: A review. Anal. Chim. Acta 2021, 1154, 338270. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, V.; Nahin, M.; Hogan, C.J.; Larriba-Andaluz, C. Benchmark Comparison for a Multi-Processing Ion Mobility Calculator in the Free Molecular Regime. J. Am. Soc. Mass Spectrom. 2017, 28, 1540–1551. [Google Scholar] [CrossRef]
- Zhou, Z.; Luo, M.; Chen, X.; Yin, Y.; Xiong, X.; Wang, R.; Zhu, Z.-J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 2020, 11, 4334. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.H.; Cho, J.H.; Xu, L. Breaking Down Structural Diversity for Comprehensive Prediction of Ion-Neutral Collision Cross Sections. Anal. Chem. 2020, 92, 4548–4557. [Google Scholar] [CrossRef]
- Adenot, M.; Lahana, R. Blood-Brain Barrier Permeation Models: Discriminating between Potential CNS and Non-CNS Drugs Including P-Glycoprotein Substrates. J. Chem. Inf. Comput. Sci. 2004, 44, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Abraham, M.H.; Ibrahim, A.; Fish, P.V.; Cole, S.; Lewis, M.L.; de Groot, M.J.; Reynolds, D.P. Predicting Penetration Across the Blood-Brain Barrier from Simple Descriptors and Fragmentation Schemes. J. Chem. Inf. Model. 2007, 47, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yap, C.W.; Ung, C.Y.; Xue, Y.; Cao, Z.W.; Chen, Y.Z. Effect of Selection of Molecular Descriptors on the Prediction of Blood−Brain Barrier Penetrating and Nonpenetrating Agents by Statistical Learning Methods. J. Chem. Inf. Model. 2005, 45, 1376–1384. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Muehlbacher, M.; Spitzer, G.M.; Liedl, K.R.; Kornhuber, J. Qualitative prediction of blood–brain barrier permeability on a large and refined dataset. J. Comput. Aided Mol. Des. 2011, 25, 1095–1106. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, S.; Kumar, S.; Parkesh, R. Chemical Space Exploration of DprE1 Inhibitors Using Chemoinformatics and Artificial Intelligence. ACS Omega 2021, 6, 14430–14441. [Google Scholar] [CrossRef]
- Langdon, S.R.; Brown, N.; Blagg, J. Scaffold Diversity of Exemplified Medicinal Chemistry Space. J. Chem. Inf. Model. 2011, 51, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Minikel, E.V. Properties of CNS Drugs vs. All FDA-Approved Drugs. Available online: https://www.cureffi.org/2013/10/04/properties-of-cns-drugs-vs-all-fda-approved-drugs/ (accessed on 21 May 2021).
- Roy, D.; Hinge, V.K.; Kovalenko, A. To Pass or not to Pass: Predicting the Blood–Brain Barrier Permeability with the 3D-RISM-KH Molecular Solvation Theory. ACS Omega 2019, 4, 16774–16780. [Google Scholar] [CrossRef]
- Gabelica, V.; Shvartsburg, A.A.; Afonso, C.; Barran, P.; Benesch, J.L.P.; Bleiholder, C.; Bowers, M.T.; Bilbao, A.; Bush, M.F.; Campbell, J.L.; et al. Recommendations for reporting ion mobility Mass Spectrometry measurements. Mass Spectrom. Rev. 2019, 38, 291–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyttenbach, T.; Bleiholder, C.; Bowers, M.T. Factors Contributing to the Collision Cross Section of Polyatomic Ions in the Kilodalton to Gigadalton Range: Application to Ion Mobility Measurements. Anal. Chem. 2013, 85, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Dunaway, K. Collisional Cross Section. Available online: https://chem.libretexts.org/@go/page/1403 (accessed on 8 December 2021).
- Hinnenkamp, V.; Klein, J.; Meckelmann, S.W.; Balsaa, P.; Schmidt, T.C.; Schmitz, O.J. Comparison of CCS Values Determined by Traveling Wave Ion Mobility Mass Spectrometry and Drift Tube Ion Mobility Mass Spectrometry. Anal. Chem. 2018, 90, 12042–12050. [Google Scholar] [CrossRef]
- Aguilar-Armenta, G.; Patiño-Iglesias, M.E.; Leyva-Ramos, R. Adsorption Kinetic Behaviour of Pure CO2, N2 and CH4 in Natural Clinoptilolite at Different Temperatures. Adsorpt. Sci. Technol. 2003, 21, 81–91. [Google Scholar] [CrossRef]
- Guntner, A.S. Into the Depth: The Key Role of Modern Analytical Chemistry in Pharmaceutical Development and Medicinal Research. Ph.D. Thesis, Johannes Kepler University Linz, Linz, Austria, 2020. [Google Scholar]
- Marrink, S.J.; Jähnig, F.; Berendsen, H.J. Proton transport across transient single-file water pores in a lipid membrane studied by molecular dynamics simulations. Biophys. J. 1996, 71, 632–647. [Google Scholar] [CrossRef] [Green Version]
- Träuble, H. The movement of molecules across lipid membranes: A molecular theory. J. Membr. Biol. 1971, 4, 193–208. [Google Scholar] [CrossRef]
- Kadry, H.; Noorani, B.; Cucullo, L. A blood–brain barrier overview on structure, function, impairment, and biomarkers of integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Irudayanathan, F.J.; Wang, N.; Wang, X.; Nangia, S. Architecture of the paracellular channels formed by claudins of the blood–brain barrier tight junctions. Ann. N. Y. Acad. Sci. 2017, 1405, 131–146. [Google Scholar] [CrossRef] [PubMed]

| Data Set | Compounds (N) | Ring System with Atomic-No Subst. Pattern Scaffolds (Nrs) | Murcko Scaffolds (Nms) | Most Central Ring Scaffolds (Ncr) | Skeleton Scaffolds (Nsc) | Nrs/N | Nms/N | Ncr/N | Nsc/N |
|---|---|---|---|---|---|---|---|---|---|
| Adenot | 1592 | 961 | 842 | 320 | 581 | 0.60 | 0.53 | 0.20 | 0.36 |
| Adenot+ | 1282 | 683 | 672 | 276 | 447 | 0.53 | 0.52 | 0.22 | 0.35 |
| Adenot− | 310 | 215 | 168 | 68 | 150 | 0.69 | 0.54 | 0.22 | 0.48 |
| Drugbank | 3261 | 2101 | 1729 | 572 | 1107 | 0.64 | 0.53 | 0.18 | 0.34 |
| Drugbank+ | 2841 | 1754 | 1462 | 478 | 903 | 0.62 | 0.51 | 0.17 | 0.32 |
| Drugbank− | 420 | 432 | 282 | 123 | 235 | 1.03 | 0.67 | 0.29 | 0.56 |
| Li | 400 | 324 | 222 | 118 | 164 | 0.81 | 0.56 | 0.30 | 0.41 |
| Li+ | 267 | 222 | 144 | 87 | 105 | 0.83 | 0.54 | 0.33 | 0.39 |
| Li- | 133 | 149 | 88 | 54 | 78 | 1.12 | 0.66 | 0.41 | 0.59 |
| Muehlbacher | 362 | 233 | 148 | 86 | 117 | 0.64 | 0.41 | 0.24 | 0.32 |
| Muehlbacher+ | 327 | 203 | 132 | 78 | 104 | 0.62 | 0.40 | 0.24 | 0.32 |
| Muehlbacher− | 35 | 46 | 25 | 21 | 22 | 1.31 | 0.71 | 0.60 | 0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guntner, A.S.; Bögl, T.; Mlynek, F.; Buchberger, W. Large-Scale Evaluation of Collision Cross Sections to Investigate Blood-Brain Barrier Permeation of Drugs. Pharmaceutics 2021, 13, 2141. https://doi.org/10.3390/pharmaceutics13122141
Guntner AS, Bögl T, Mlynek F, Buchberger W. Large-Scale Evaluation of Collision Cross Sections to Investigate Blood-Brain Barrier Permeation of Drugs. Pharmaceutics. 2021; 13(12):2141. https://doi.org/10.3390/pharmaceutics13122141
Chicago/Turabian StyleGuntner, Armin Sebastian, Thomas Bögl, Franz Mlynek, and Wolfgang Buchberger. 2021. "Large-Scale Evaluation of Collision Cross Sections to Investigate Blood-Brain Barrier Permeation of Drugs" Pharmaceutics 13, no. 12: 2141. https://doi.org/10.3390/pharmaceutics13122141
APA StyleGuntner, A. S., Bögl, T., Mlynek, F., & Buchberger, W. (2021). Large-Scale Evaluation of Collision Cross Sections to Investigate Blood-Brain Barrier Permeation of Drugs. Pharmaceutics, 13(12), 2141. https://doi.org/10.3390/pharmaceutics13122141