Hepatocyte-Specific Co-Delivery of Zinc Ions and Plasmid DNA by Lactosylated Poly(1-vinylimidazole) for Suppression of Insulin Receptor Internalization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of PVIm-Lac with Various Lactosylated Degrees
2.3. Gel Filtration Chromatography (GFC)
2.4. 1H NMR Spectroscopy
2.5. Agarose Gel Retardation Assay
2.6. Particle Size and ζ-Potential Measurement
2.7. Cell Viability Assay
2.8. Cellular Uptake of Zn2+ Ions
2.9. Transfection Procedure
2.10. Insulin Receptor Cell-Based Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of PVIm-Lac with Various Lactosylated Degrees
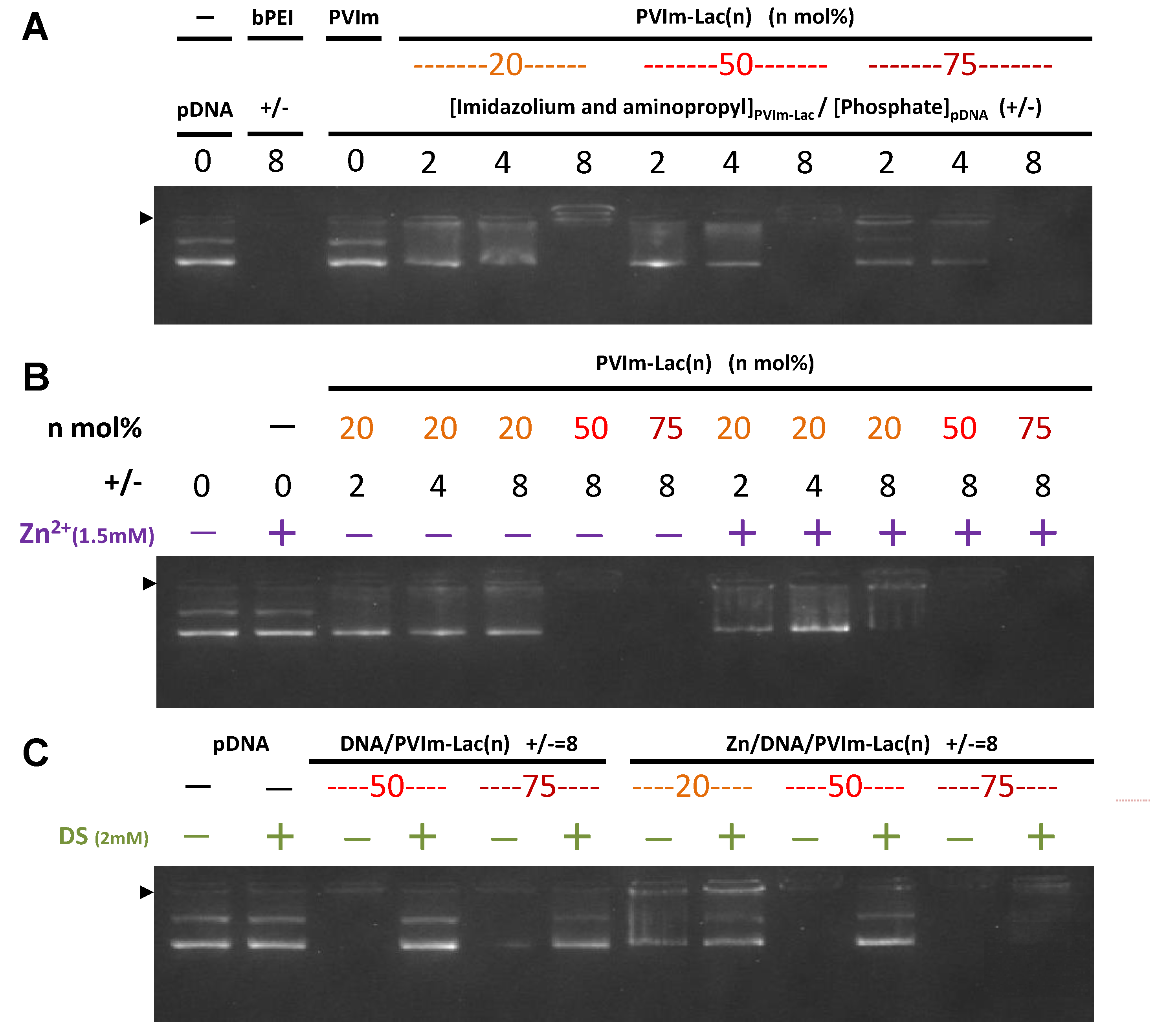
3.2. Formation of Zn/DNA/PVIm-Lac Complexes
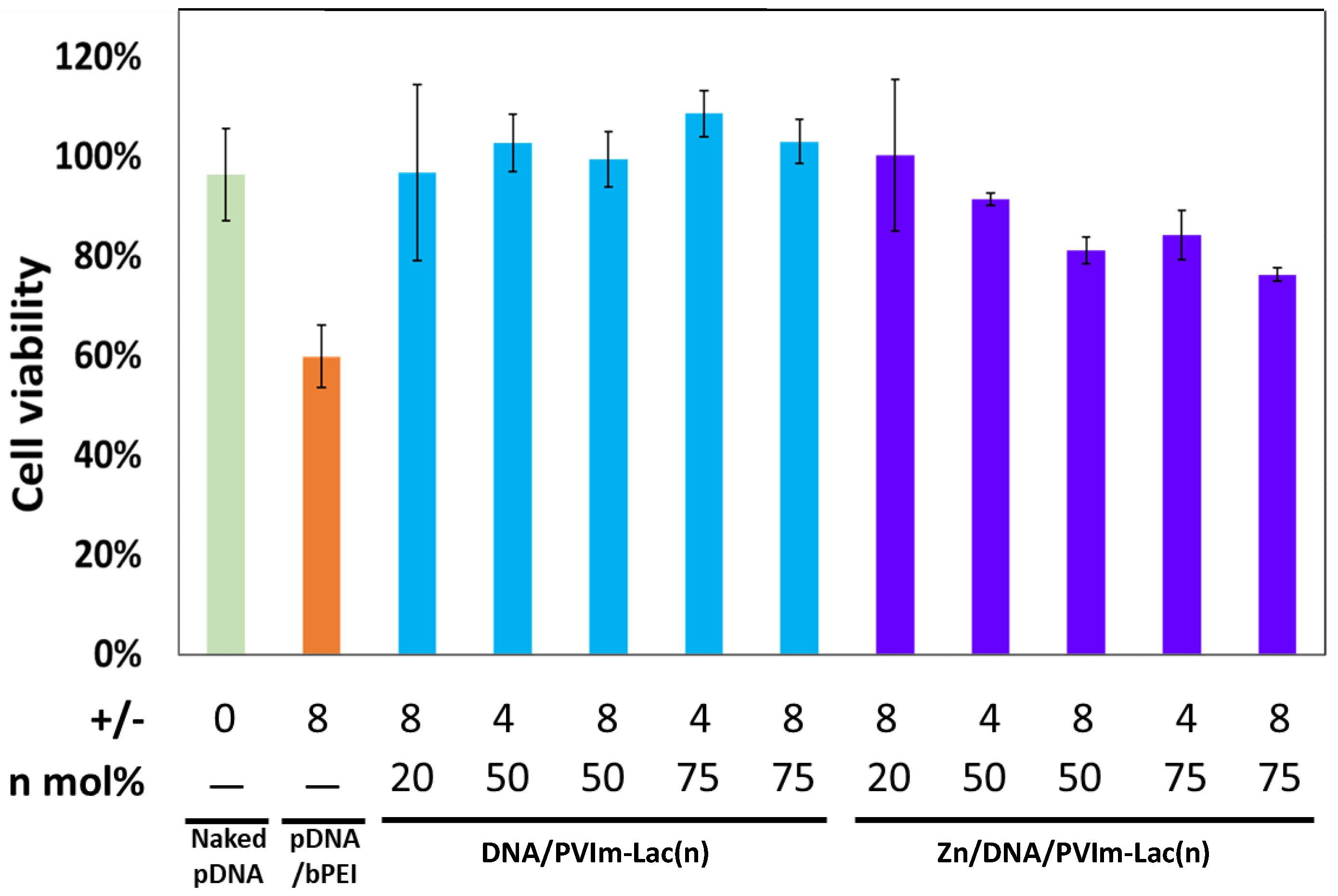
3.3. Cell Viability in the Presence of Zn/DNA/PVIm-Lac Complexes
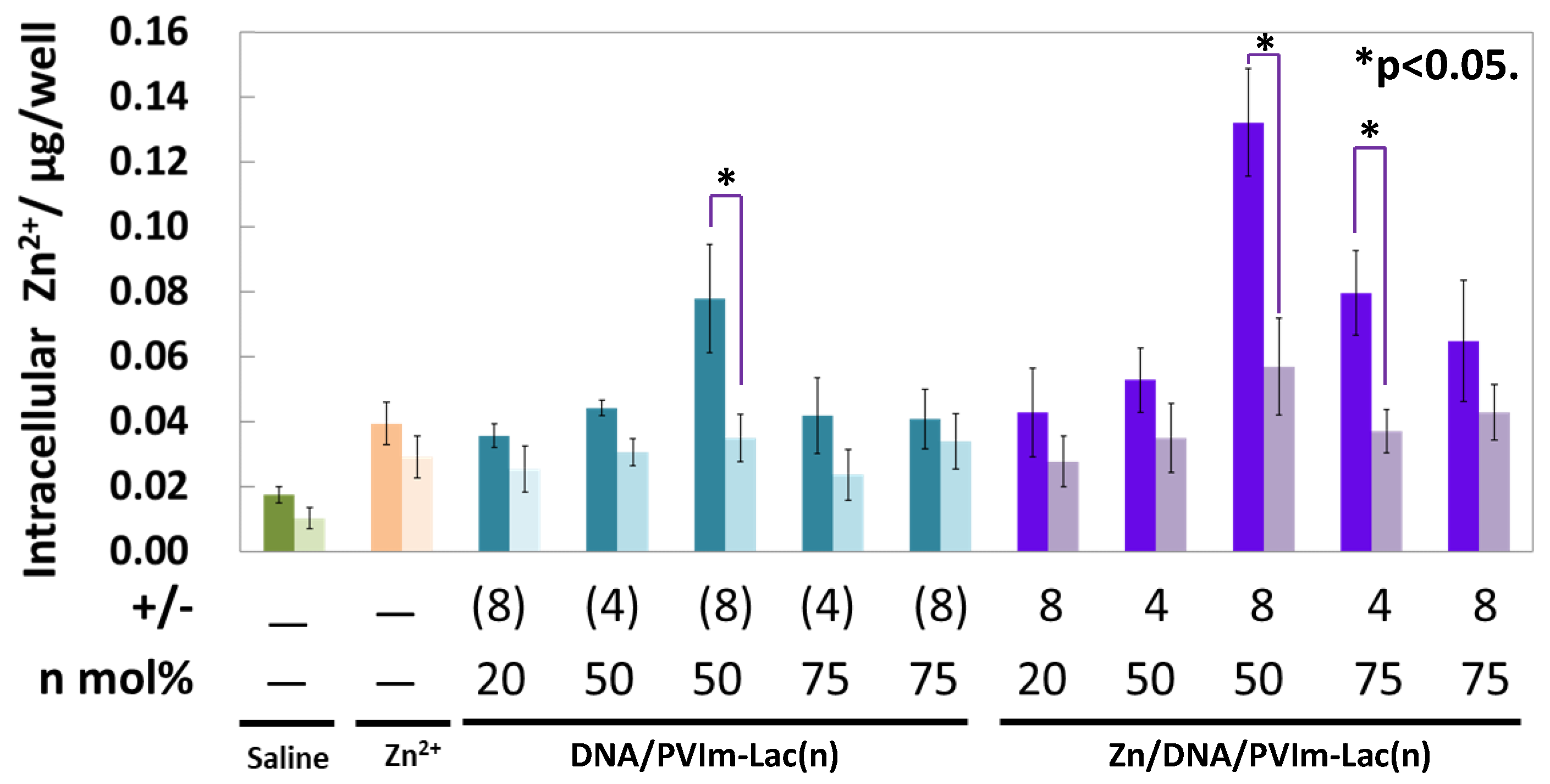
3.4. Intracellular Delivery of Zn2+ Ions by Zn/DNA/PVIm-Lac Complexes
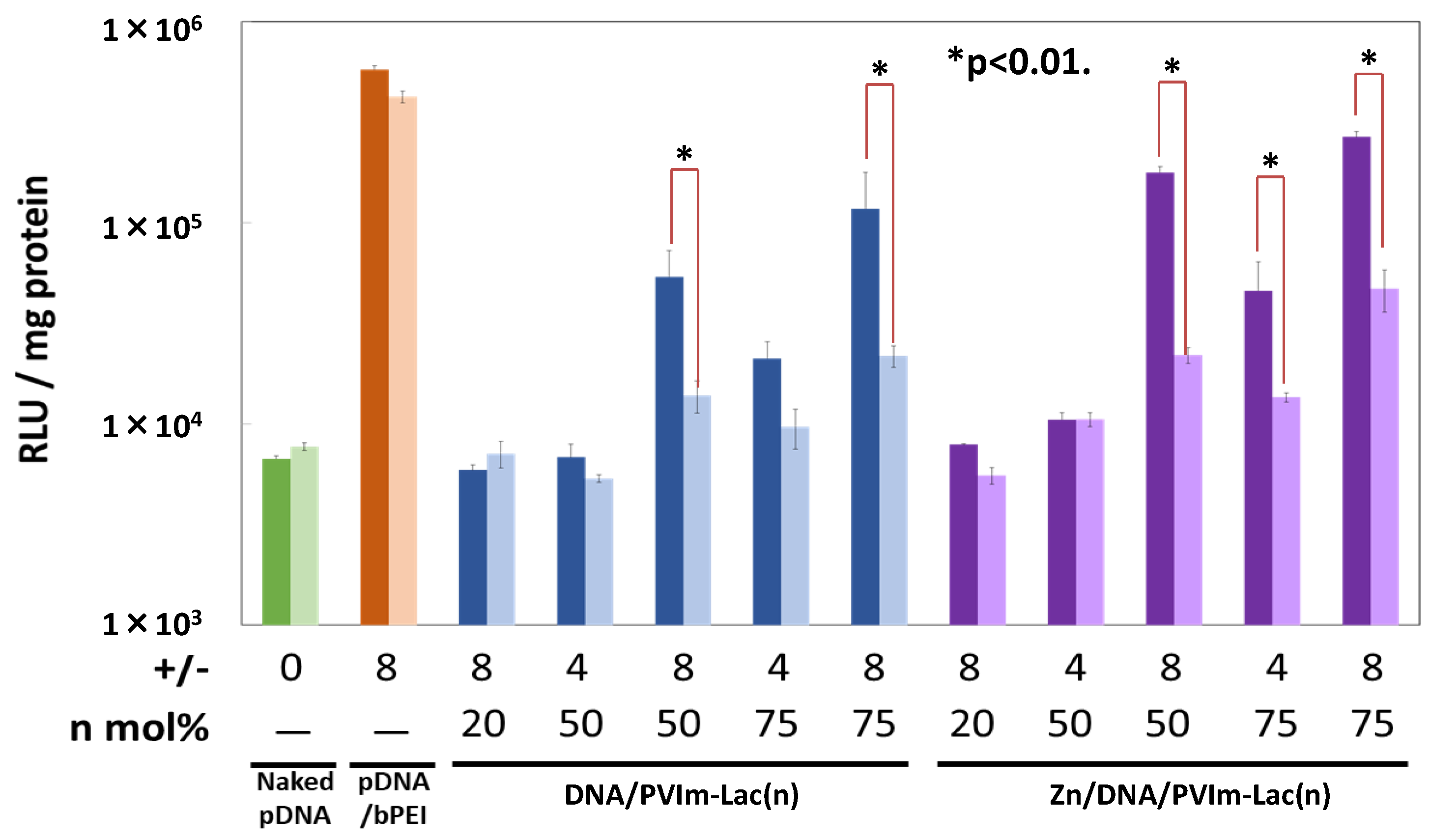
3.5. Gene Expression of pDNA Delivered by Zn/DNA/PVIm-Lac Complexes
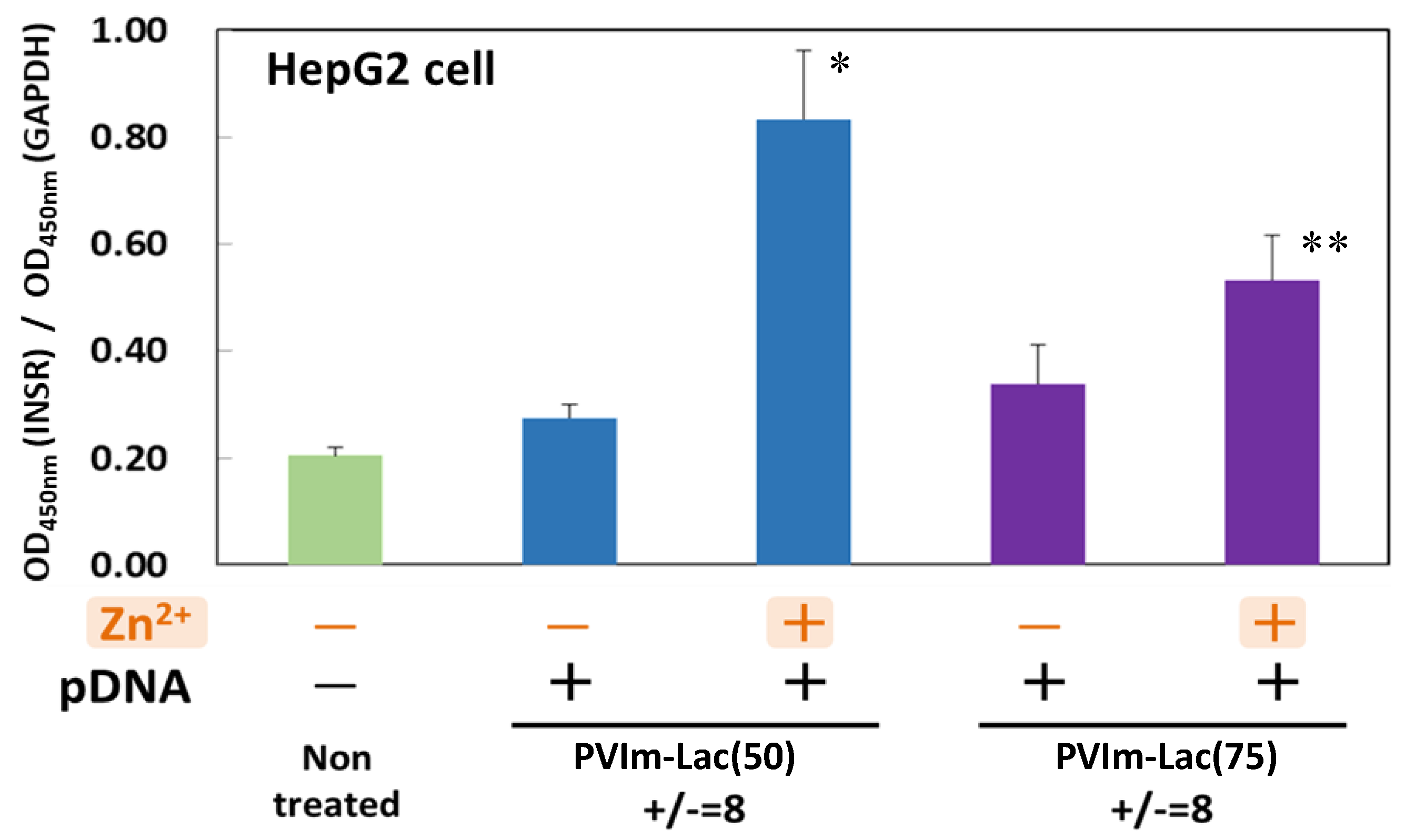
3.6. Suppression of Insulin Receptor Internalization by Zn/DNA/PVIm-Lac Complexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korichneva, I.; Hoyos, B.; Chua, R.; Levi, E.; Hammerling, U. Zinc Release from Protein Kinase C as the Common Event during Activation by Lipid Second Messenger or Reactive Oxygen. J. Biol. Chem. 2002, 277, 44327–44331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, S.; Sakata-Sogawa, K.; Hasegawa, A.; Suzuki, T.; Kabu, K.; Sato, E.; Kurosaki, T.; Yamashita, S.; Tokunaga, M.; Nishida, K.; et al. Zinc is a novel intracellular second messenger. J. Cell Biol. 2007, 177, 637–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Boillat, A.; Huang, D.; Liang, C.; Peers, C.; Gamper, N. Intracellular zinc activates KCNQ channels by reducing their dependence on phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA 2017, 114, E6410–E6419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, S.F.; Kilfoil, V.J.; Schmidt, M.H.; Amann, B.T.; Berg, J. Metal binding and folding properties of a minimalist Cys2His2 zinc finger peptide. Proc. Natl. Acad. Sci. USA 1992, 89, 4796–4800. [Google Scholar] [CrossRef] [Green Version]
- Fierke, C.A.; Thompson, R.B. Fluorescence-based biosensing of zinc using carbonic anhydrase. BioMetals 2001, 14, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.; McCall, K.; Kramer, M.; Blankman, E.; Winge, D.R.; Eide, D.J. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 2003, 22, 5137–5146. [Google Scholar] [CrossRef] [Green Version]
- Hojyo, S.; Fukuda, T. Zinc transporters and signaling in physiology and pathogenesis. Arch. Biochem. Biophys. 2016, 611, 43–50. [Google Scholar] [CrossRef]
- Baltaci, A.K.; Yuce, K. Zinc Transporter Proteins. Neurochem. Res. 2018, 43, 517–530. [Google Scholar] [CrossRef]
- Chimienti, F.; Devergnas, S.; Favier, A.; Seve, M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes 2004, 53, 2330–2337. [Google Scholar] [CrossRef] [Green Version]
- Sladek, R.; Rocheleau, G.; Rung, J.; Dina, C.; Shen, L.; Serre, D.; Boutin, P.; Vincent, D.R.; Belisle, A.; Hadjadj, S.; et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007, 445, 881–885. [Google Scholar] [CrossRef]
- Chabosseau, P.; Rutter, G.A. Zinc and diabetes. Arch. Biochem. Biophys. 2016, 611, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamaki, M.; Fujitani, Y.; Hara, A.; Uchida, T.; Tamura, Y.; Takeno, K.; Kawaguchi, M.; Watanabe, T.; Ogihara, T.; Fukunaka, A.; et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J. Clin. Investig. 2013, 123, 4513–4524. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Park, S.; Lin, X.; Copps, K.; Yi, X.; White, M.F. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J. Clin. Investig. 2006, 116, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Nagakura, T.; Kawakami, H.; Asayama, S. Synthesis and characterization of lactosylated poly(1-vinylimidazole) for cell-specific plasmid DNA carrier. Intern. Med. Care 2020, 4, 1–5. [Google Scholar] [CrossRef]
- Asayama, S.; Nishinohara, S.; Kawakami, H. Zinc-chelated imidazole groups for DNA polyion complex formation. Metallomics 2011, 3, 680–682. [Google Scholar] [CrossRef]
- Asayama, S.; Nishinohara, S.; Kawakami, H. Zinc-Chelated Poly(1-vinylimidazole) and a Carbohydrate Ligand Polycation Form DNA Ternary Complexes for Gene Delivery. Bioconjugate Chem. 2011, 22, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Asayama, S.; Matsuda, K.; Negishi, Y.; Kawakami, H. Intracellular co-delivery of zinc ions and plasmid DNA for enhancing gene transfection activity. Metallomics 2014, 6, 82–87. [Google Scholar] [CrossRef]
- Asayama, S.; Sakata, M.; Kawakami, H. Structure-activity relationship between Zn2+-chelated alkylated poly(1-vinylimidazole) and gene transfection. J. Inorg. Biochem. 2017, 173, 120–125. [Google Scholar] [CrossRef]
- Endo, A.; Asayama, S. Preparation of Zn2+-Chelated Carboxymethyl Poly(1-Vinylimidazole) for Intracellular Zn2+ Delivery. J. Miner. Mater. Charact. Eng. 2019, 7, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Udenfriend, S.; Stein, S.; Böhlen, P.; Dairman, W.; Leimgruber, W.; Weigele, M. Fluorescamine: A Reagent for Assay of Amino Acids, Peptides, Proteins, and Primary Amines in the Picomole Range. Science 1972, 178, 871–872. [Google Scholar] [CrossRef]
- Unsworth, J.M.; Rose, F.R.A.J.; Wright, E.; Scotchford, C.A.; Shakesheff, K.M. Seeding cells into needled felt scaffolds for tissue engineering applications. J. Biomed. Mater. Res. 2003, 66A, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Pouton, C.W.; Lucas, P.; Thomas, B.J.; Uduehi, A.N.; Milroy, D.A.; Moss, S.H. Polycation-DNA complexes for gene delivery: A comparison of the biopharmaceutical properties of cationic polypeptides and cationic lipids. J. Control. Release 1998, 53, 289–299. [Google Scholar] [CrossRef]
- Shuai, X.; Merdan, T.; Unger, F.; Kissel, T. Supramolecular Gene Delivery Vectors Showing Enhanced Transgene Expression and Good Biocompatibility. Bioconjugate Chem. 2005, 16, 322–329. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Endo, A.; Asayama, S. Hepatocyte-Specific Co-Delivery of Zinc Ions and Plasmid DNA by Lactosylated Poly(1-vinylimidazole) for Suppression of Insulin Receptor Internalization. Pharmaceutics 2021, 13, 2084. https://doi.org/10.3390/pharmaceutics13122084
Endo A, Asayama S. Hepatocyte-Specific Co-Delivery of Zinc Ions and Plasmid DNA by Lactosylated Poly(1-vinylimidazole) for Suppression of Insulin Receptor Internalization. Pharmaceutics. 2021; 13(12):2084. https://doi.org/10.3390/pharmaceutics13122084
Chicago/Turabian StyleEndo, Akito, and Shoichiro Asayama. 2021. "Hepatocyte-Specific Co-Delivery of Zinc Ions and Plasmid DNA by Lactosylated Poly(1-vinylimidazole) for Suppression of Insulin Receptor Internalization" Pharmaceutics 13, no. 12: 2084. https://doi.org/10.3390/pharmaceutics13122084
APA StyleEndo, A., & Asayama, S. (2021). Hepatocyte-Specific Co-Delivery of Zinc Ions and Plasmid DNA by Lactosylated Poly(1-vinylimidazole) for Suppression of Insulin Receptor Internalization. Pharmaceutics, 13(12), 2084. https://doi.org/10.3390/pharmaceutics13122084





