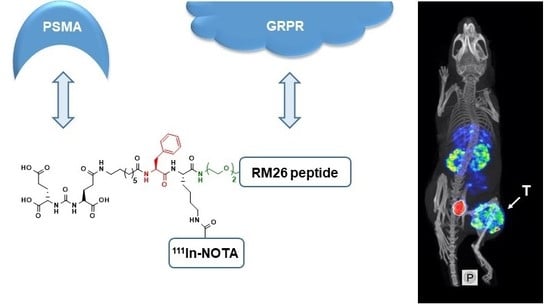
Heterodimeric Radiotracer Targeting PSMA and GRPR for Imaging of Prostate Cancer—Optimization of the Affinity towards PSMA by Linker Modification in Murine Model
Abstract
1. Introduction
2. Materials and Methods
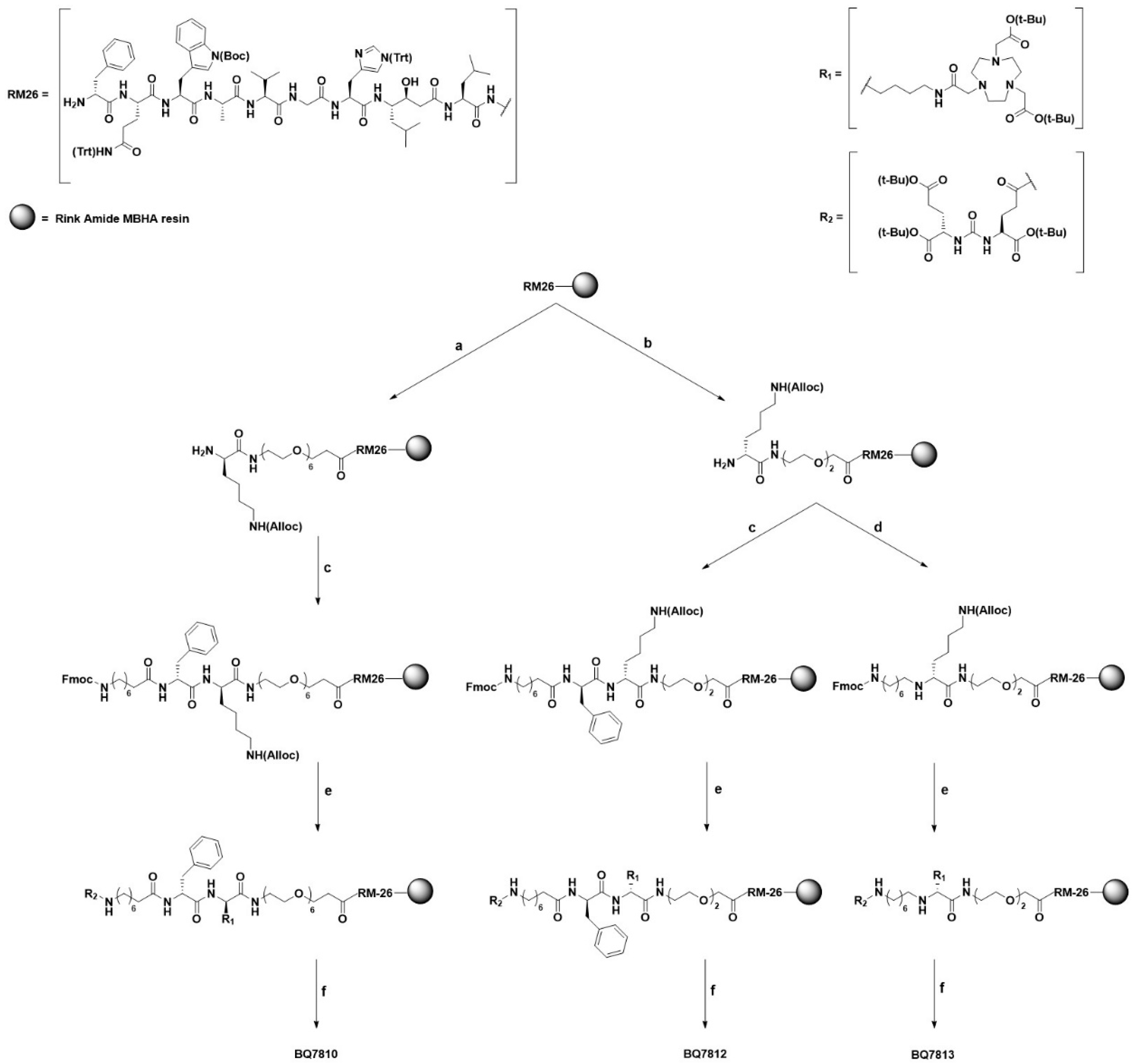
2.1. Synthesis
2.1.1. (S)-5-(tert-butoxy)-4-(3-((S)-1,5-di-tert-butoxy-1,5-dioxopentan-2-yl)ureido)-5-oxopentanoic acid (R2-OH)
2.1.2. On-Resin RM26
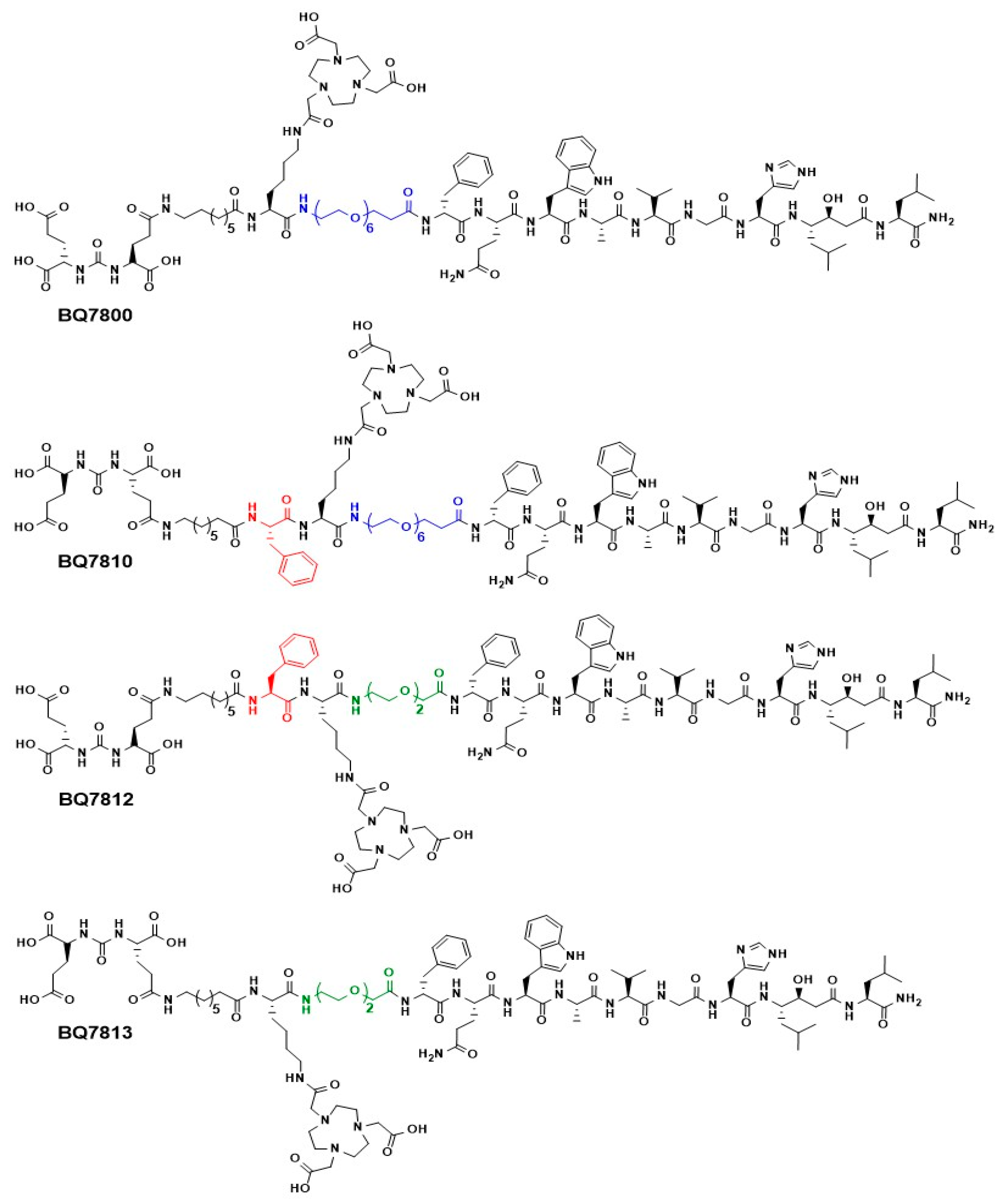
2.1.3. BQ7810, BQ7812 and BQ7813
2.2. Radiolabeling
2.3. Distribution Coefficient (LogD)
2.4. In Vitro Experiments
2.4.1. In Vitro Specificity Test
2.4.2. In Vitro Cellular Processing
2.4.3. In Vitro Competitive Binding Assay (IC50)
2.5. In Vivo Experiments
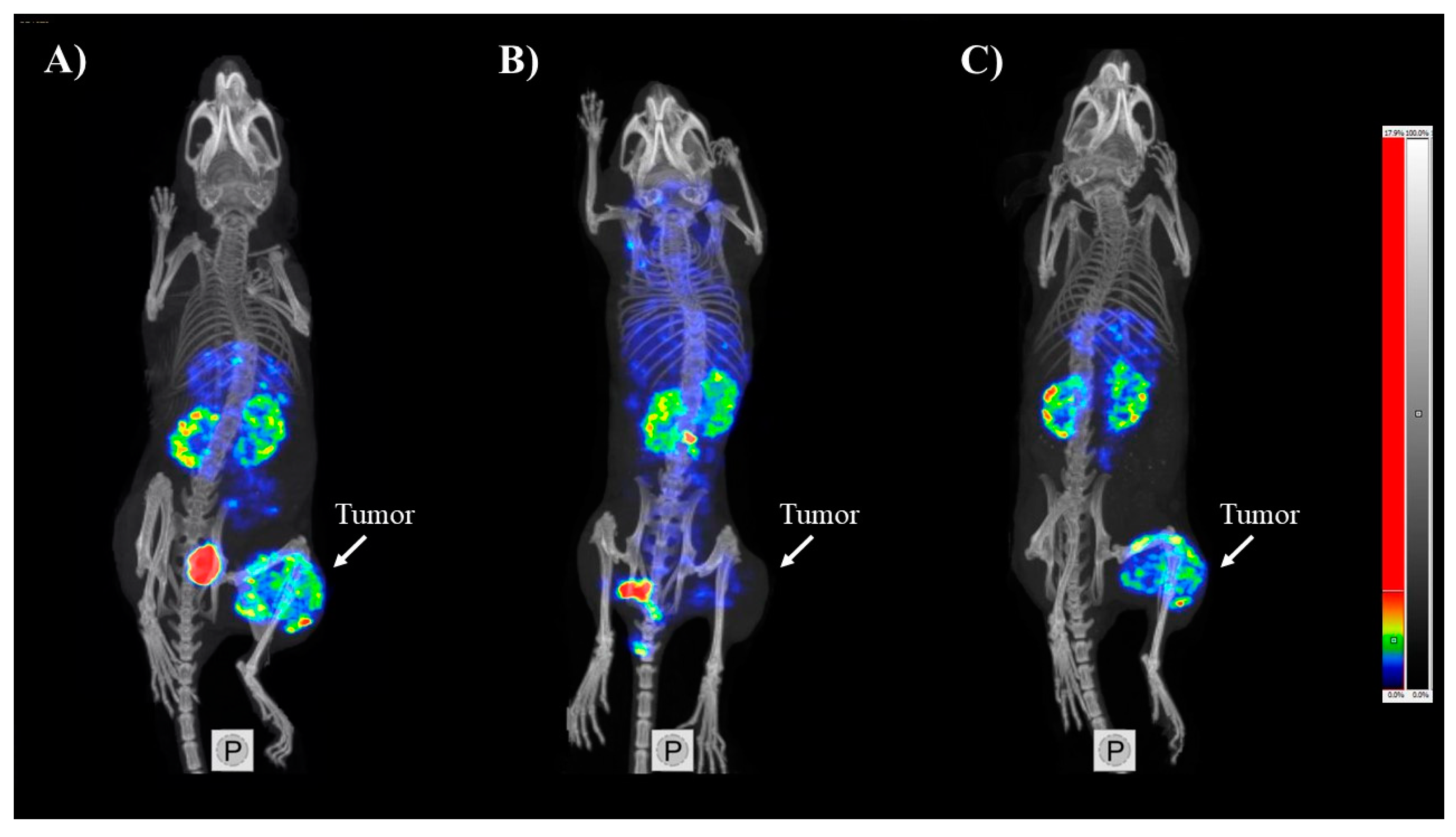
SPECT/CT Imaging
3. Results and Discussion
3.1. Synthesis and Radiolabeling
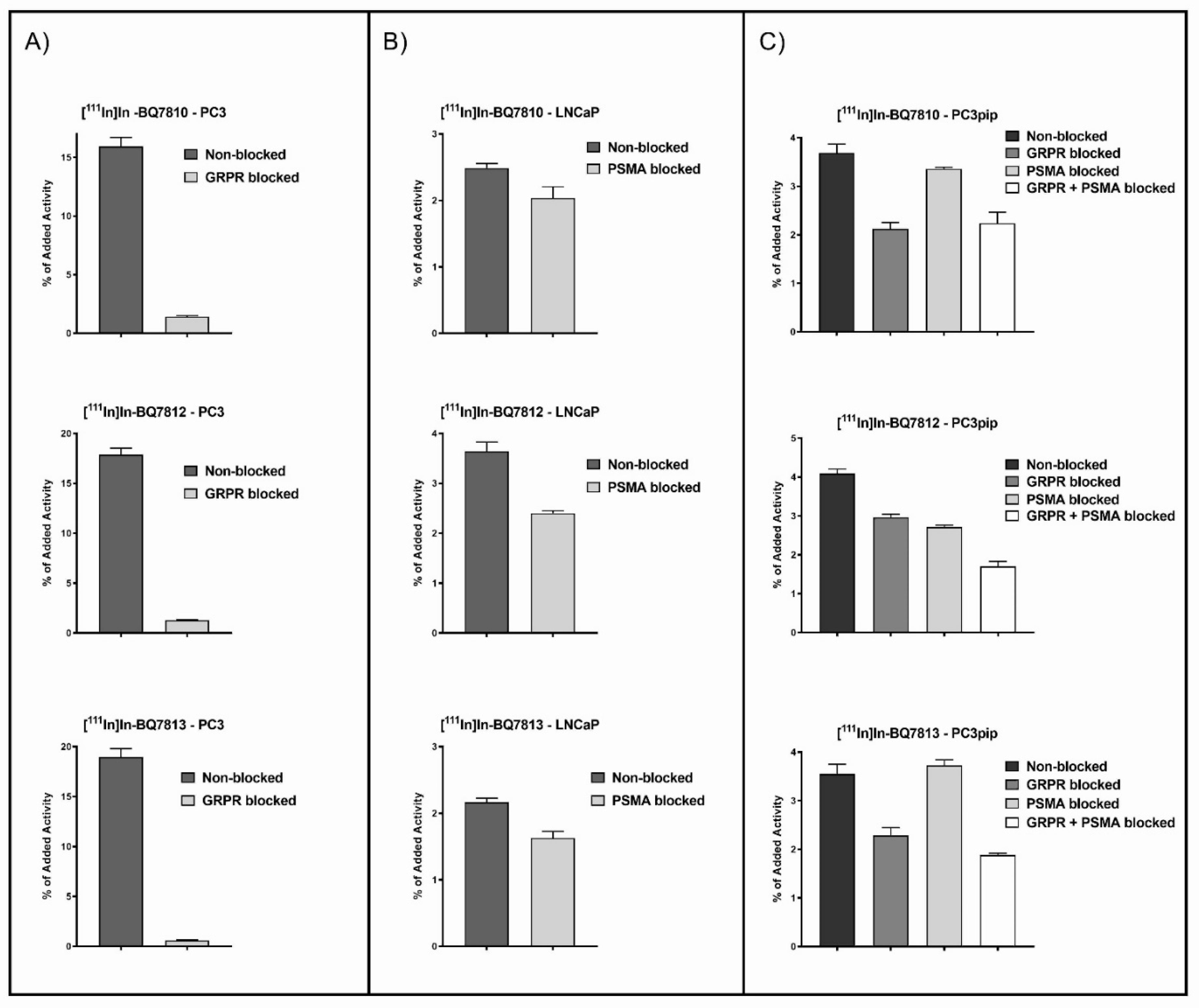
3.2. In Vitro Characterization
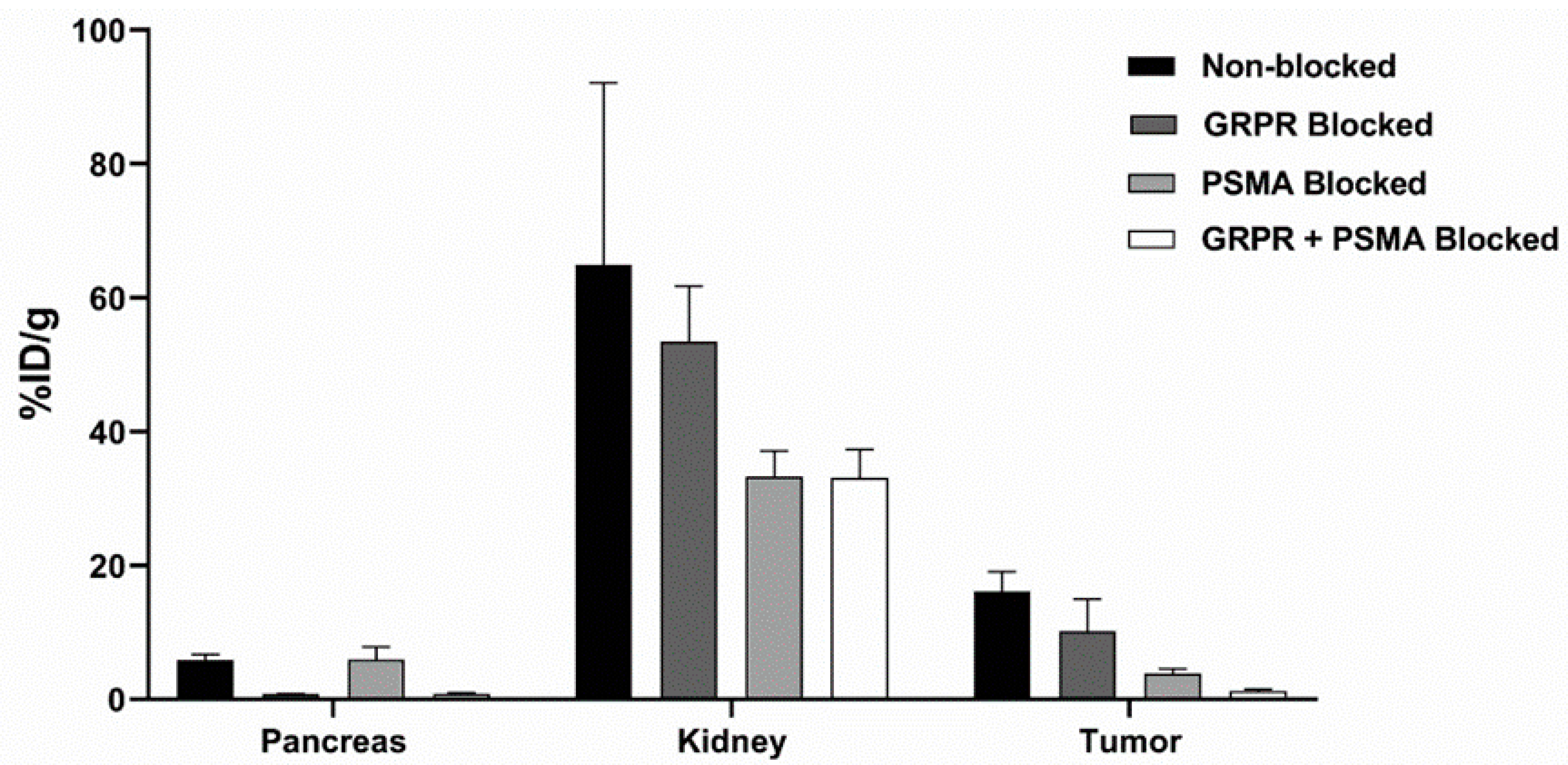
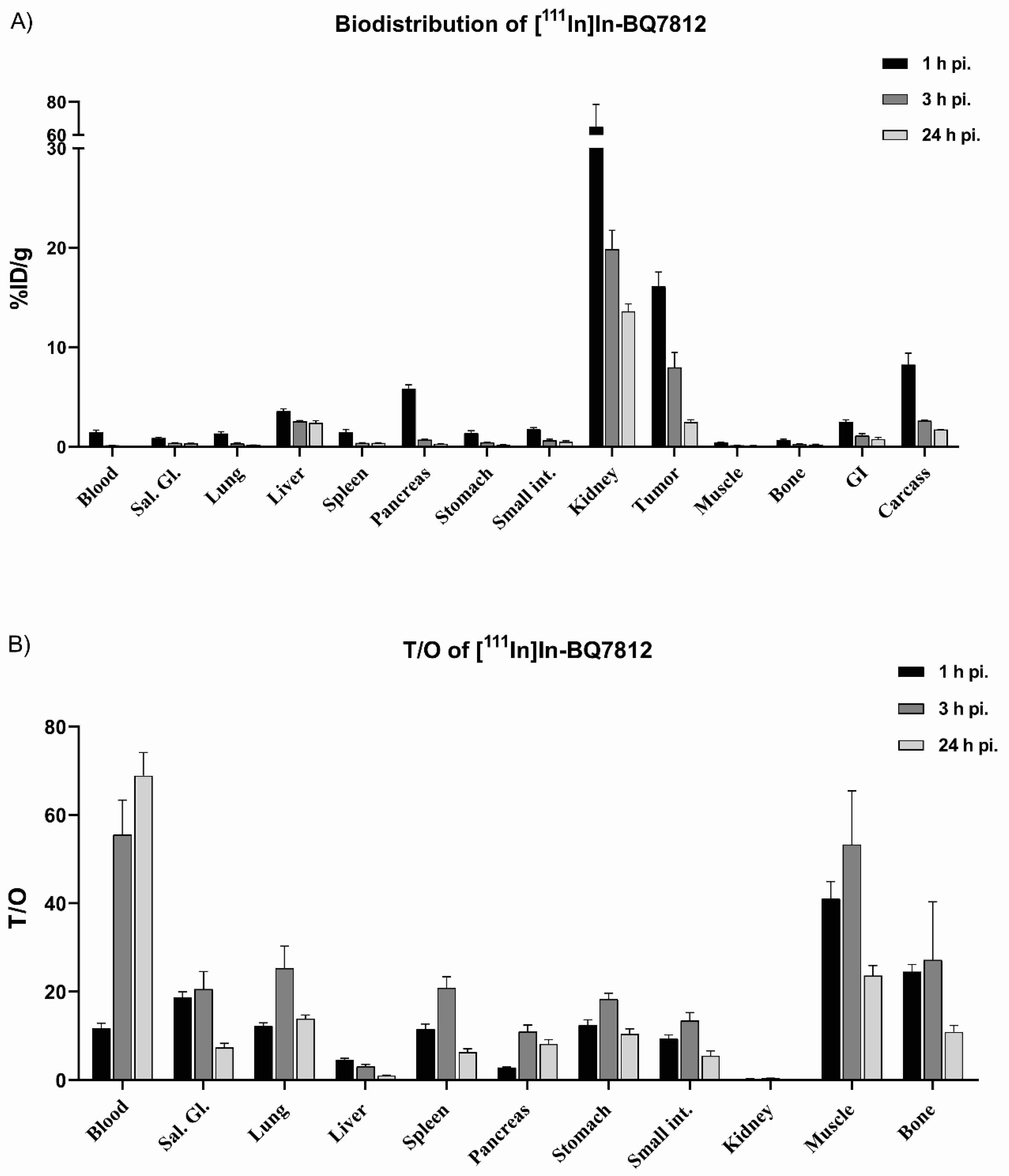
3.3. In Vivo Characterization
3.4. Comparison of [111In]In-BQ7800 and [111In]In-BQ7812 Heterodimers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Saad, F. Quality of life in men with prostate cancer. Lancet Oncol. 2019, 20, 325–326. [Google Scholar] [CrossRef]
- Attard, G.; Parker, C.; Eeles, R.A.; Schröder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; De Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Lavery, A.; Kirby, R.S.; Chowdhury, S. Prostate cancer. Medicine (Baltimore) 2016, 44, 47–51. [Google Scholar] [CrossRef]
- Mottet, N.; Bellmunt, J.; Bolla, M.; Briers, E.; Cumberbatch, M.G.; De Santis, M.; Fossati, N.; Gross, T.; Henry, A.M.; Joniau, S.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2017, 71, 618–629. [Google Scholar] [CrossRef]
- Rybalov, M.; Ananias, H.J.; Hoving, H.D.; Van der Poel, H.G.; Rosati, S.; de Jong, I.J. PSMA, EpCAM, VEGF and GRPR as imaging targets in locally recurrent prostate cancer after radiotherapy. Int. J. Mol. Sci. 2014, 15, 6046–6061. [Google Scholar] [CrossRef]
- Barve, A.; Jin, W.; Cheng, K. Prostate cancer relevant antigens and enzymes for targeted drug delivery. J. Control. Release 2014, 187, 118–132. [Google Scholar] [CrossRef]
- Kiess, A.P.; Banerjee, S.R.; Mease, R.C.; Rowe, S.P.; Rao, A.; Foss, C.A.; Chen, Y.; Yang, X.; Cho, S.Y.; Nimmagadda, S.; et al. Prostate-specific membrane antigen as a target for cancer imaging and therapy. Q. J. Nucl. Med. Mol. Imaging 2015, 59, 241–268. [Google Scholar] [CrossRef]
- Mease, R.C.; Foss, C.A.; Pomper, M.G. PET Imaging in Prostate Cancer: Focus on Prostate-Specific Membrane Antigen. Curr. Top. Med. Chem. 2014, 13, 951–962. [Google Scholar] [CrossRef]
- Donin, N.M.; Reiter, R.E. Why Targeting PSMA Is a Game Changer in the Management of Prostate Cancer. J. Nucl. Med. 2017, 59, 177–182. [Google Scholar] [CrossRef]
- Mannweiler, S.; Amersdorfer, P.; Trajanoski, S.; Terrett, J.A.; King, D.; Mehes, G. Heterogeneity of prostate-specific membrane antigen (PSMA) expression in prostate carcinoma with distant metastasis. Pathol. Oncol. Res. 2009, 15, 167–172. [Google Scholar] [CrossRef]
- Baratto, L.; Jadvar, H.; Iagaru, A. Prostate Cancer Theranostics Targeting Gastrin-Releasing Peptide Receptors. Mol. Imaging Biol. 2017, 20, 501–509. [Google Scholar] [CrossRef]
- Cheng, S.; Lang, L.; Wang, Z.; Jacobson, O.; Yung, B.C.; Zhu, G.; Gu, D.; Ma, Y.; Zhu, X.; Niu, G.; et al. PET Imaging of Prostate Cancer with Ga-68 Labeled GRPR Agonist BBN7-14 and Antagonist RM26. Bioconjugate Chem. 2017, 29, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Millar, J.B.; Rozengurt, E. Chronic desensitization to bombesin by progressive down-regulation of bombesin receptors in Swiss 3T3 cells. Distinction from acute desensitization. J. Biol. Chem. 1990, 265, 12052–12058. [Google Scholar] [PubMed]
- Kahkonen, E.; Jambor, I.; Kemppainen, J.; Lehtio, K.; Gronroos, T.J.; Kuisma, A.; Luoto, P.; Sipila, H.J.; Tolvanen, T.; Alanen, K.; et al. In vivo imaging of prostate cancer using [68Ga]-labeled bombesin analog BAY86-7548. Clin. Cancer Res. 2013, 19, 5434–5443. [Google Scholar] [CrossRef] [PubMed]
- Minamimoto, R.; Sonni, I.; Hancock, S.; Vasanawala, S.; Loening, A.; Gambhir, S.S.; Iagaru, A. Prospective evaluation of 68 Ga-RM2 PET/MRI in patients with biochemical recurrence of prostate cancer and negative findings on conventional imaging. J. Nucl. Med. 2018, 59, 803–808. [Google Scholar] [CrossRef]
- Nock, B.A.; Kaloudi, A.; Lymperis, E.; Giarika, A.; Kulkarni, H.R.; Klette, I.; Singh, A.; Krenning, E.P.; De Jong, M.; Maina, T.; et al. Theranostic perspectives in prostate cancer with the gastrin-releasing peptide receptor antagonist NeoBOMB1: Preclinical and first clinical results. J. Nucl. Med. 2017, 58, 75–80. [Google Scholar] [CrossRef]
- Iagaru, A. Will GRPR Compete with PSMA as a Target in Prostate Cancer? J. Nucl. Med. 2017, 58, 1883–1884. [Google Scholar] [CrossRef] [PubMed]
- Minamimoto, R.; Hancock, S.; Schneider, B.; Chin, F.T.; Jamali, M.; Loening, A.; Vasanawala, S.; Gambhir, S.S.; Iagaru, A. Pilot comparison of 68Ga-RM2 PET and 68Ga-PSMA-11 PET in patients with biochemically recurrent prostate cancer. J. Nucl. Med. 2016, 57, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Eder, M.; Schäfer, M.; Bauder-Wüst, U.; Haberkorn, U.; Eisenhut, M.; Kopka, K. Preclinical evaluation of a bispecific low-molecular heterodimer targeting both PSMA and GRPR for improved PET imaging and therapy of prostate cancer. Prostate 2014, 74, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Bandari, R.P.; Jiang, Z.; Reynolds, T.S.; Bernskoetter, N.E.; Szczodroski, A.F.; Bassuner, K.J.; Kirkpatrick, D.L.; Rold, T.L.; Sieckman, G.L.; Hoffman, T.J.; et al. Synthesis and biological evaluation of copper-64 radiolabeled [DUPA-6-Ahx-(NODAGA)-5-Ava-BBN(7-14)NH2], a novel bivalent targeting vector having affinity for two distinct biomarkers (GRPr/PSMA) of prostate cancer. Nucl. Med. Biol. 2014, 41, 355–363. [Google Scholar] [CrossRef]
- Liolios, C.; Schäfer, M.; Haberkorn, U.; Eder, M.; Kopka, K. Novel Bispecific PSMA/GRPr Targeting Radioligands with Optimized Pharmacokinetics for Improved PET Imaging of Prostate Cancer. Bioconjugate Chem. 2016, 27, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Castellanos, A.; Ocampo-García, B.; Morales-Ávila, E.; Luna-Gutiérrez, M.; Isaac-Olivé, K.; Ferro-Flores, G.; Santos-Cuevas, C. Synthesis and preclinical evaluation of the 177Lu-DOTA-PSMA(inhibitor)-Lys3-bombesin heterodimer designed as a radiotheranostic probe for prostate cancer. Nucl. Med. Commun. 2018, 40, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Figueroa, M.J.; Escudero-Castellanos, A.; Ramirez-Nava, G.J.; Ocampo-García, B.E.; Santos-Cuevas, C.L.; Ferro-Flores, G.; Pedraza-Lopez, M.; Avila-Rodriguez, M.A. Preparation and preclinical evaluation of 68Ga-iPSMA-BN as a potential heterodimeric radiotracer for PET-imaging of prostate cancer. J. Radioanal. Nucl. Chem. 2018, 318, 2097–2105. [Google Scholar] [CrossRef]
- Abouzayed, A.; Yim, C.-B.; Mitran, B.; Rinne, S.S.; Tolmachev, V.; Larhed, M.; Rosenström, U.; Orlova, A. Synthesis and Preclinical Evaluation of Radio-Iodinated GRPR/PSMA Bispecific Heterodimers for the Theranostics Application in Prostate Cancer. Pharmaceutics 2019, 11, 358. [Google Scholar] [CrossRef] [PubMed]
- Mitran, B.; Varasteh, Z.; Abouzayed, A.; Rinne, S.S.; Puuvuori, E.; De Rosa, M.; Larhed, M.; Tolmachev, V.; Orlova, A.; Rosenström, U. Bispecific GRPR-Antagonistic Anti-PSMA/GRPR Heterodimer for PET and SPECT Diagnostic Imaging of Prostate Cancer. Cancers 2019, 11, 1371. [Google Scholar] [CrossRef] [PubMed]
- Kopka, K.; Benešová, M.; Bařinka, C.; Haberkorn, U.; Babich, J. Glu-ureido-based inhibitors of prostate-specific membrane antigen: Lessons learned during the development of a novel class of low-molecular-weight theranostic radiotracers. J. Nucl. Med. 2017, 58, 17S–26S. [Google Scholar] [CrossRef]
- Varasteh, Z.; Rosenström, U.; Velikyan, I.; Mitran, B.; Altai, M.; Honarvar, H.; Rosestedt, M.; Lindeberg, G.; Sörensen, J.; Larhed, M.; et al. The effect of mini-PEG-based spacer length on binding and pharmacokinetic properties of a 68Ga-labeled NOTA-conjugated antagonistic analog of bombesin. Molecules 2014, 19, 10455–10472. [Google Scholar] [CrossRef]
- Varasteh, Z.; Velikyan, I.; Lindeberg, G.; Sörensen, J.; Larhed, M.; Sandström, M.; Selvaraju, R.K.; Malmberg, J.; Tolmachev, V.; Orlova, A. Synthesis and characterization of a high-affinity NOTA-conjugated bombesin antagonist for GRPR-targeted tumor imaging. Bioconjugate Chem. 2013, 24, 1144–1153. [Google Scholar] [CrossRef]

| Parameter | BQ7800 | BQ7810 | BQ7812 | BQ7813 |
|---|---|---|---|---|
| IC50 (GRPR) | 4 ± 1 nM | 3 ± 1 nM | 6 ± 3 nM | 8 ± 2 nM |
| IC50 (PSMA) | 921 ± 363 nM | >1000 nM | 102 ± 80 nM | >1000 nM |
| LogD | −2.62 | −1.47 | −1.44 | −2.27 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lundmark, F.; Abouzayed, A.; Mitran, B.; Rinne, S.S.; Varasteh, Z.; Larhed, M.; Tolmachev, V.; Rosenström, U.; Orlova, A. Heterodimeric Radiotracer Targeting PSMA and GRPR for Imaging of Prostate Cancer—Optimization of the Affinity towards PSMA by Linker Modification in Murine Model. Pharmaceutics 2020, 12, 614. https://doi.org/10.3390/pharmaceutics12070614
Lundmark F, Abouzayed A, Mitran B, Rinne SS, Varasteh Z, Larhed M, Tolmachev V, Rosenström U, Orlova A. Heterodimeric Radiotracer Targeting PSMA and GRPR for Imaging of Prostate Cancer—Optimization of the Affinity towards PSMA by Linker Modification in Murine Model. Pharmaceutics. 2020; 12(7):614. https://doi.org/10.3390/pharmaceutics12070614
Chicago/Turabian StyleLundmark, Fanny, Ayman Abouzayed, Bogdan Mitran, Sara S. Rinne, Zohreh Varasteh, Mats Larhed, Vladimir Tolmachev, Ulrika Rosenström, and Anna Orlova. 2020. "Heterodimeric Radiotracer Targeting PSMA and GRPR for Imaging of Prostate Cancer—Optimization of the Affinity towards PSMA by Linker Modification in Murine Model" Pharmaceutics 12, no. 7: 614. https://doi.org/10.3390/pharmaceutics12070614
APA StyleLundmark, F., Abouzayed, A., Mitran, B., Rinne, S. S., Varasteh, Z., Larhed, M., Tolmachev, V., Rosenström, U., & Orlova, A. (2020). Heterodimeric Radiotracer Targeting PSMA and GRPR for Imaging of Prostate Cancer—Optimization of the Affinity towards PSMA by Linker Modification in Murine Model. Pharmaceutics, 12(7), 614. https://doi.org/10.3390/pharmaceutics12070614