In Vivo Bactericidal Efficacy of GWH1 Antimicrobial Peptide Displayed on Protein Nanoparticles, a Potential Alternative to Antibiotics
Abstract
1. Introduction
2. Materials and Methods
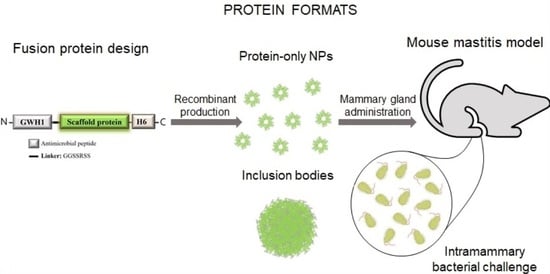
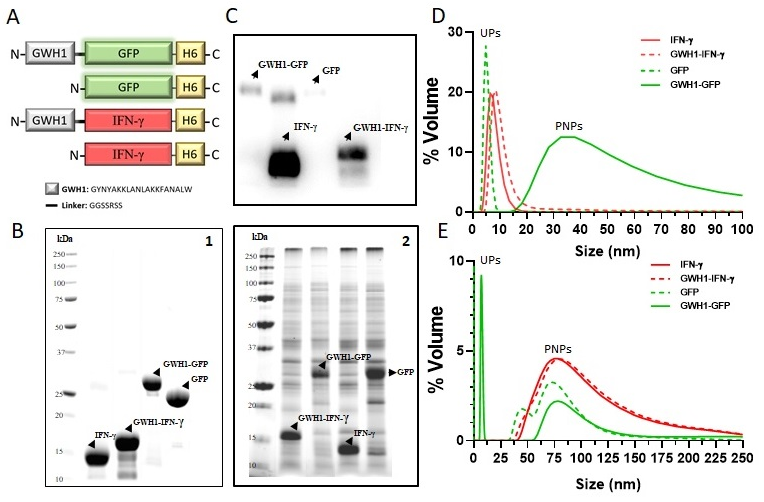
2.1. Protein Design and Production
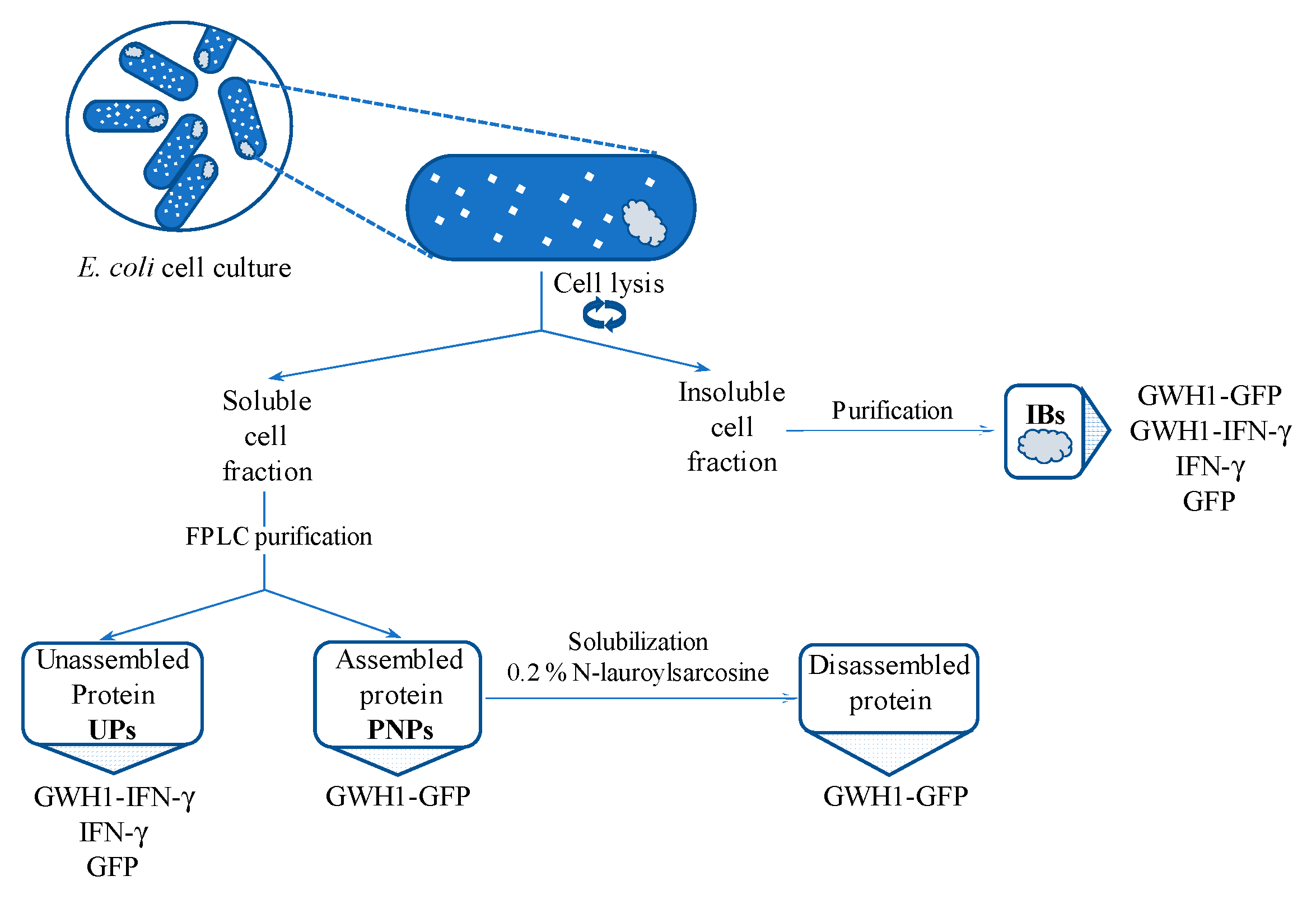
2.2. Protein Purification
2.3. Nanoparticle Size Characterization
2.4. Bactericidal Activity Assay
2.5. Disassembling of Soluble Nanoparticles
2.6. Protein Solubilization from IBs
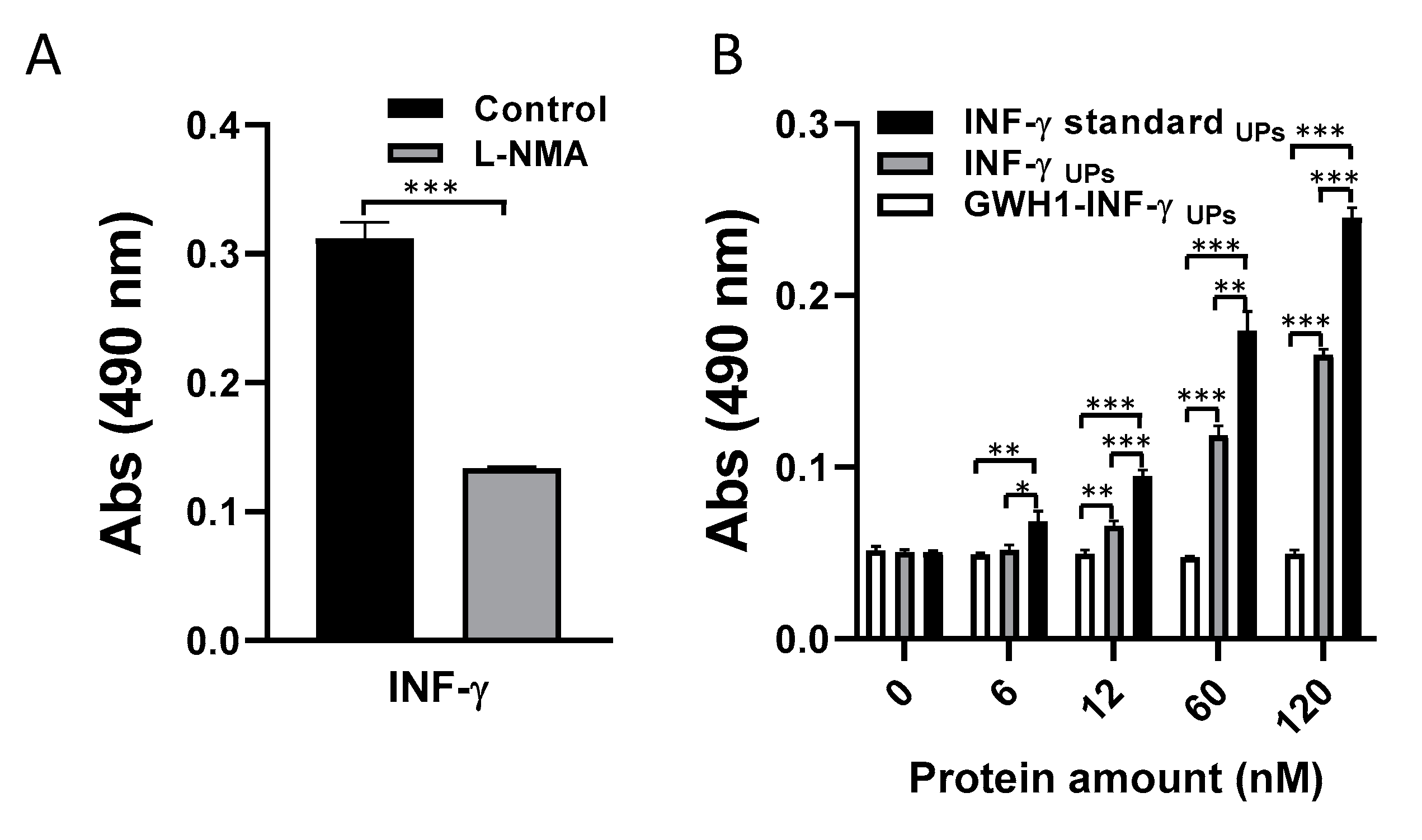
2.7. Determination of Murine IFN-γ Biological Activity
2.8. Mouse Model of Infectious Mastitis
2.9. Statistical Analysis
3. Results
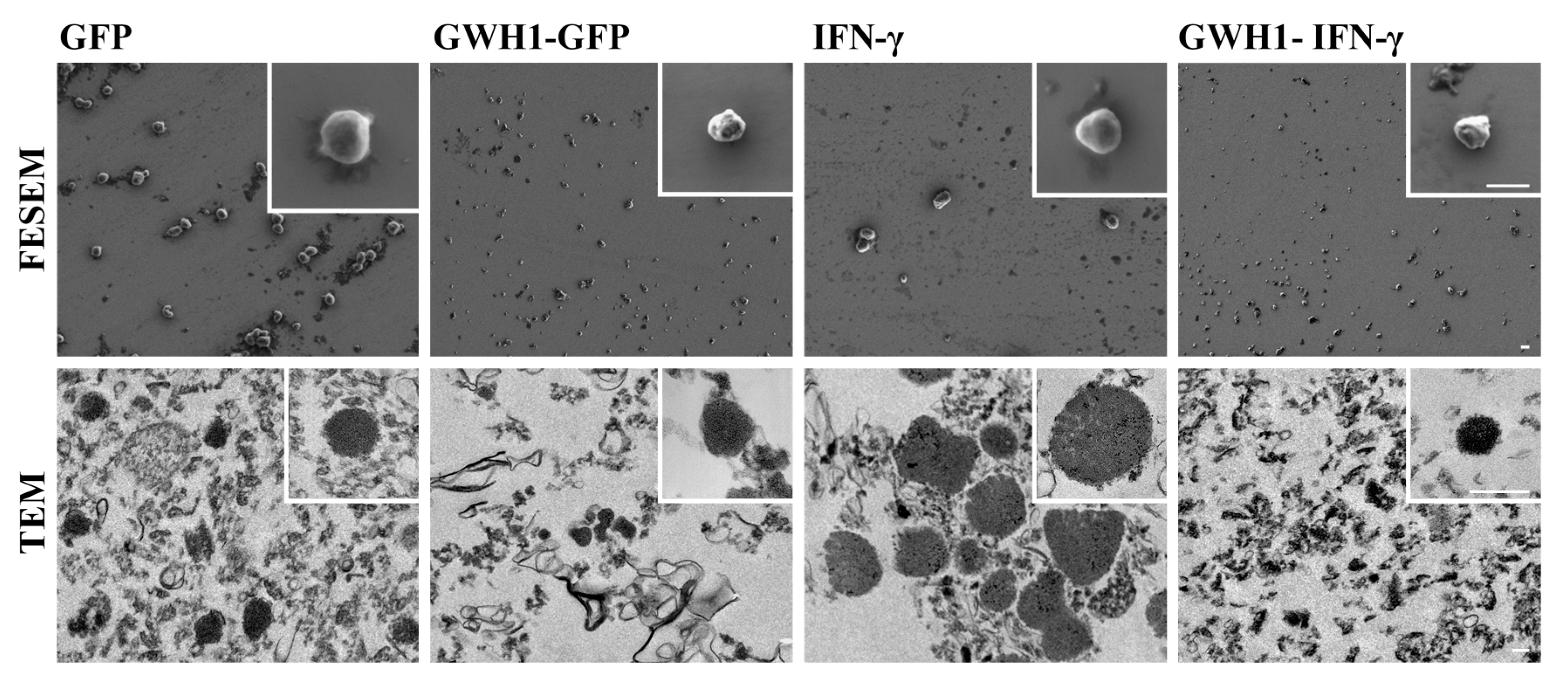
3.1. Characterization of Self-Assembling Nanoparticles
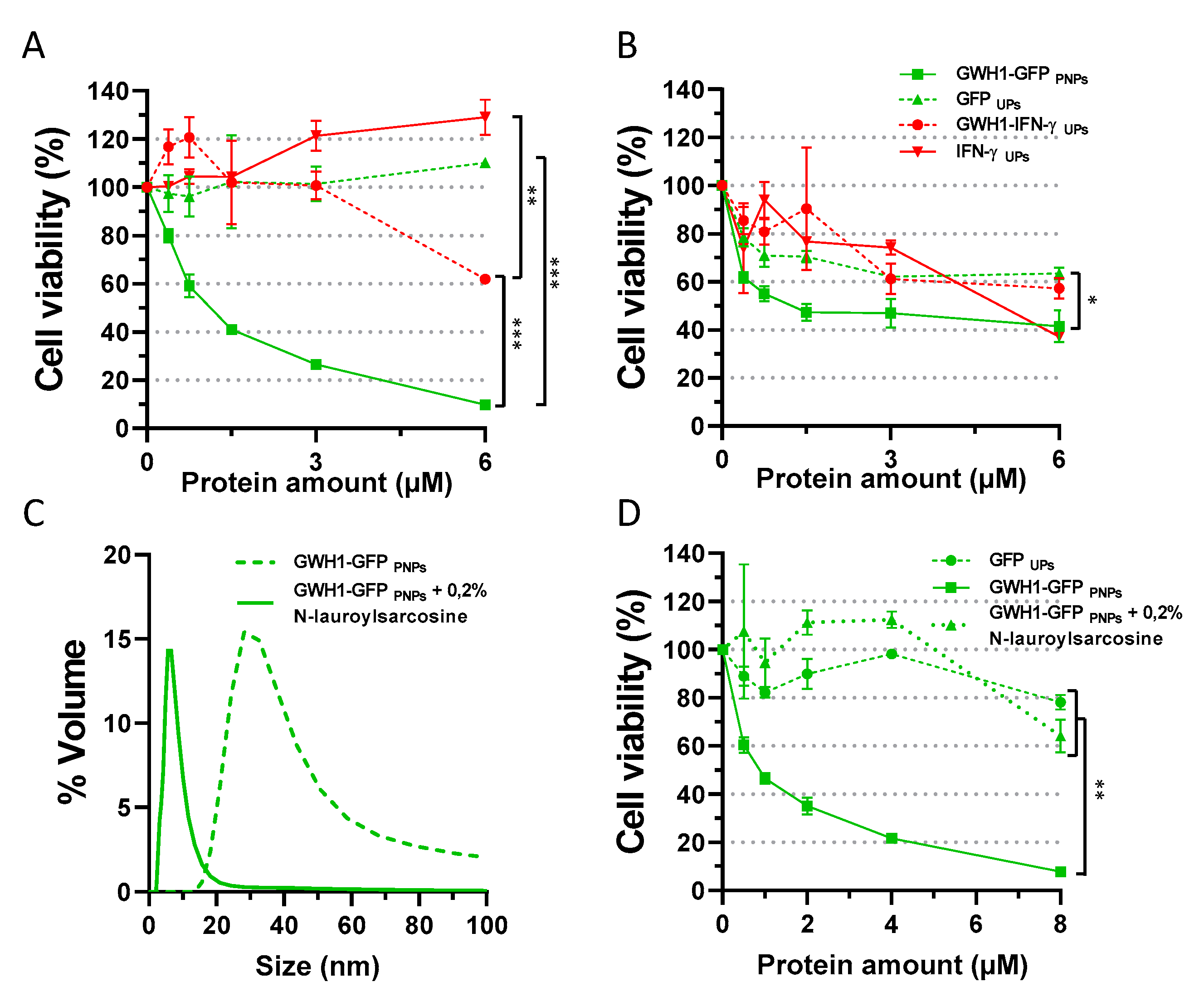
3.2. In Vitro Evaluation of Antibacterial Activity
3.3. Importance of Nanoparticle Format on Antibacterial Activity
3.4. Bioactivity Evaluation of IFN-γ-Based Proteins
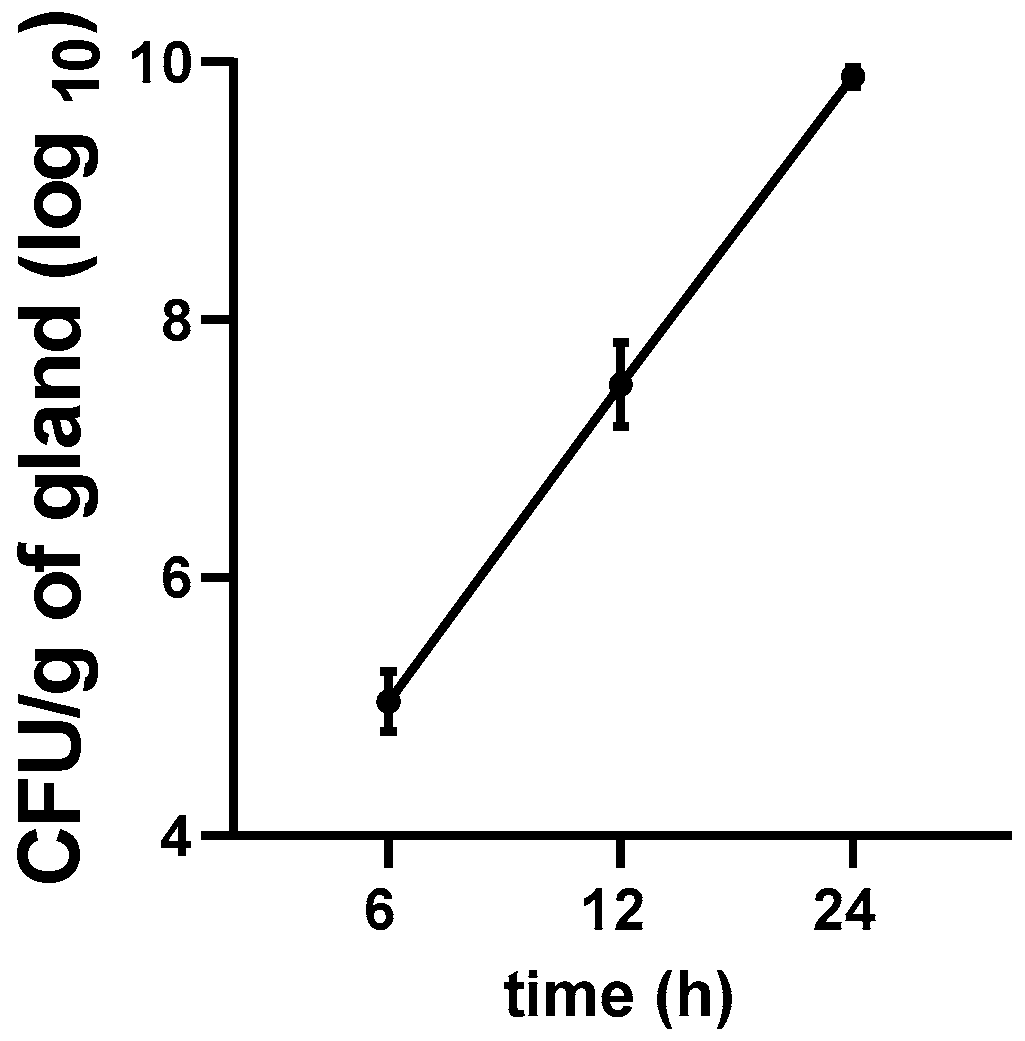
3.5. Growth Kinetics of E. coli in the Mastitis Mice Model
3.6. Characterization of IBs
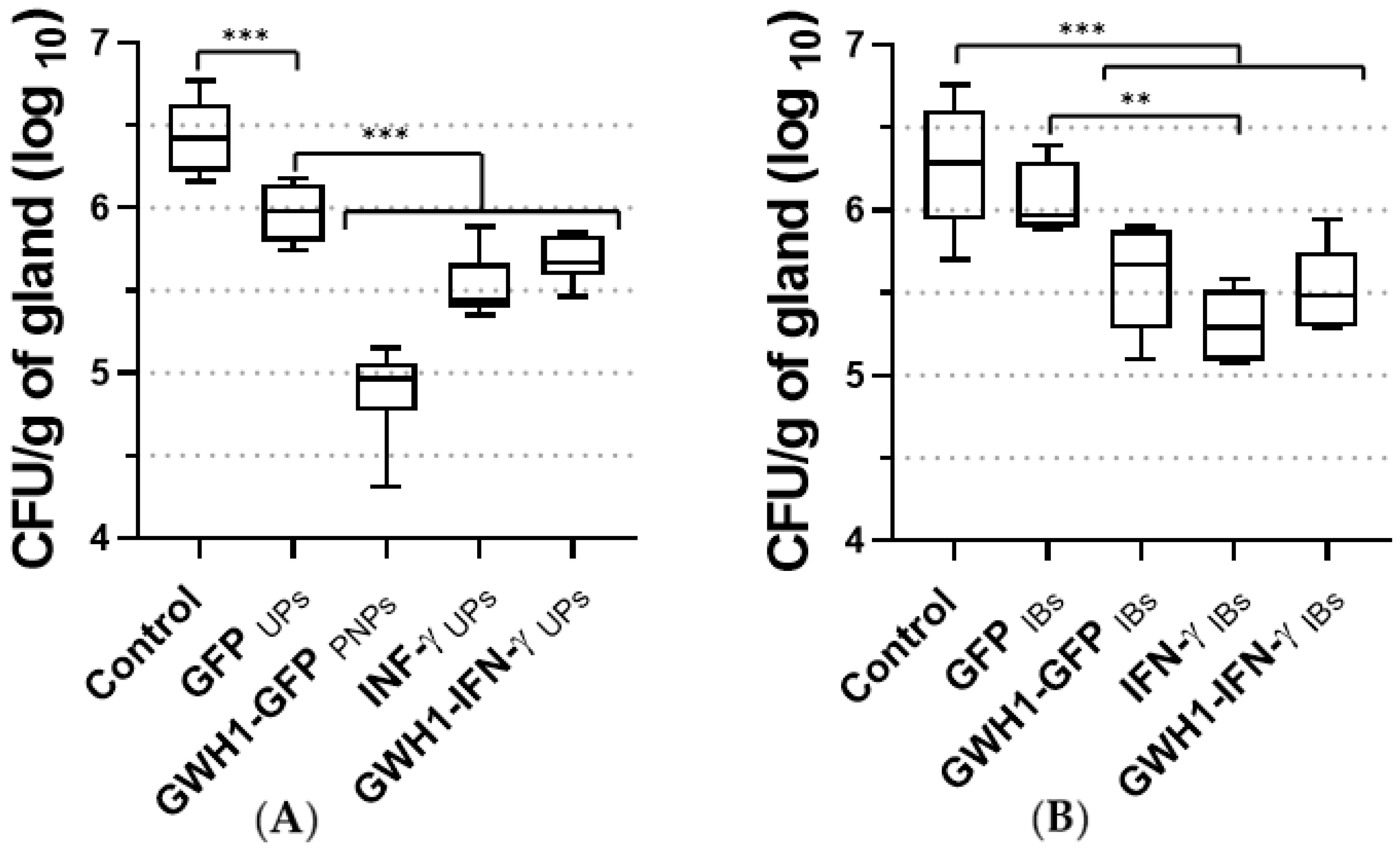
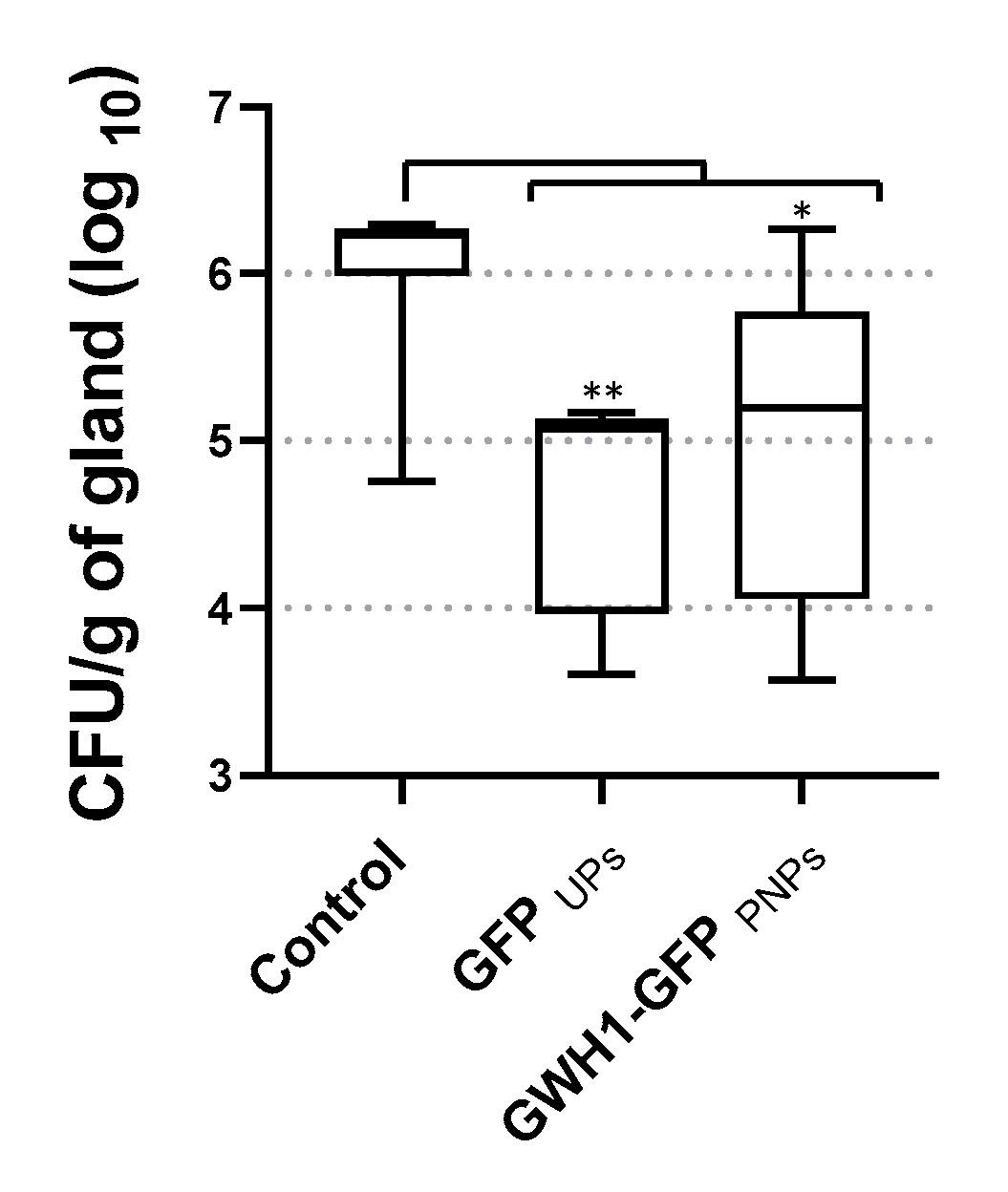
3.7. Therapeutic Effect of PNPs and IBs in the Mouse Mastitis Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.D.; Won, H.S.; Kim, J.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides for therapeutic applications: A review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.L.; Corl, C.M.; Sordillo, L.M. Immunopathology of mastitis: Insights into disease recognition and resolution. J. Mammary Gland Biol. Neoplasia 2011, 16, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.M.; Babiuk, L.A. Controlling acute Escherichia coli mastitis during the periparturient period with recombinant bovine interferon gamma. Vet. Microbiol. 1991, 28, 189–198. [Google Scholar] [CrossRef]
- Pighetti, G.; Sordillo, L. Specific immune responses of dairy cattle after primary inoculation with recombinant bovine interferon-gamma as an adjuvant when vaccinating against mastitis. Am. J. Vet. Res. 1996, 57, 819–824. [Google Scholar]
- Lei, R.; Hou, J.; Chen, Q.; Yuan, W.; Cheng, B.; Sun, Y.; Jin, Y.; Ge, L.; Ben-Sasson, S.A.; Chen, J.; et al. Self-assembling myristoylated human α-defensin 5 as a next-generation nanobiotics potentiates therapeutic efficacy in bacterial infection. ACS Nano 2018, 12, 5284–5296. [Google Scholar] [CrossRef]
- Serna, N.; Sánchez-García, L.; Sánchez-Chardi, A.; Unzueta, U.; Roldán, M.; Mangues, R.; Vázquez, E.; Villaverde, A. Protein-only, antimicrobial peptide-containing recombinant nanoparticles with inherent built-in antibacterial activity. Acta Biomater. 2017, 60, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Serna, N.; Céspedes, M.V.; Saccardo, P.; Xu, Z.; Unzueta, U.; Álamo, P.; Pesarrodona, M.; Sánchez-Chardi, A.; Roldán, M.; Mangues, R.; et al. Rational engineering of single-chain polypeptides into protein-only, BBB-targeted nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Serna, N.; Céspedes, M.V.; Sánchez-García, L.; Unzueta, U.; Sala, R.; Sánchez-Chardi, A.; Cortés, F.; Ferrer-Miralles, N.; Mangues, R.; Vázquez, E.; et al. Peptide-based nanostructured materials with intrinsic proapoptotic activities in CXCR4+ solid tumors. Adv. Funct. Mater. 2017, 27, 1–9. [Google Scholar] [CrossRef]
- Unzueta, U.; Ferrer-Miralles, N.; Cedano, J.; Zikung, X.; Pesarrodona, M.; Saccardo, P.; García-Fruitós, E.; Domingo-Espín, J.; Kumar, P.; Gupta, K.C.; et al. Non-amyloidogenic peptide tags for the regulatable self-assembling of protein-only nanoparticles. Biomaterials 2012, 33, 8714–8722. [Google Scholar] [CrossRef]
- Chou, H.; Kuo, T.; Chiang, J.; Pei, M.; Yang, W.; Yu, H.; Lin, S.; Chen, W. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. Int. J. Antimicrob. Agents 2008, 32, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Carratalá, J.V.; Cano-garrido, O.; Sánchez, J.; Membrado, C.; Pérez, E.; Conchillo-solé, O.; Daura, X.; Sánchez-chardi, A. Aggregation-prone peptides modulate activity of bovine interferon gamma released from naturally occurring protein nanoparticles. N. Biotechnol. 2020, 57, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Pesarrodona, M.; Jauset, T.; Díaz-Riascos, Z.V.; Sánchez-Chardi, A.; Beaulieu, M.-E.; Seras-Franzoso, J.; Sánchez-García, L.; Baltà-Foix, R.; Mancilla, S.; Fernández, Y.; et al. Targeting antitumoral proteins to breast cancer by local administration of functional inclusion bodies. Adv. Sci. 2019, 6, 1900849. [Google Scholar] [CrossRef] [PubMed]
- Céspedes, M.V.; Cano-Garrido, O.; Álamo, P.; Sala, R.; Gallardo, A.; Serna, N.; Falgàs, A.; Voltà-Durán, E.; Casanova, I.; Sánchez-Chardi, A.; et al. Engineering secretory amyloids for remote and highly selective destruction of metastatic foci. Adv. Mater. 2020, 32, e1907348. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Miralles, N.; Saccardo, P.; Corchero, J.L.; Xu, Z.; García-Fruitós, E. General introduction: Recombinant protein production and purification of insoluble proteins. Methods Mol. Biol. 2015, 1258, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Seras-Franzoso, J.; Sánchez-Chardi, A.; Garcia-Fruitós, E.; Vázquez, E.; Villaverde, A. Cellular uptake and intracellular fate of protein releasing bacterial amyloids in mammalian cells. Soft Matter 2016, 12, 3451–3460. [Google Scholar] [CrossRef] [PubMed]
- Cano-Garrido, O.; Sánchez-Chardi, A.; Parés, S.; Giró, I.; Tatkiewicz, W.I.; Ferrer-Miralles, N.; Ratera, I.; Natalello, A.; Cubarsi, R.; Veciana, J.; et al. Functional protein-based nanomaterial produced in microorganisms recognized as safe: A new platform for biotechnology. Acta Biomater. 2016, 43, 230–239. [Google Scholar] [CrossRef]
- Céspedes, M.V.; Fernández, Y.; Unzueta, U.; Mendoza, R.; Seras-Franzoso, J.; Sánchez-Chardi, A.; Álamo, P.; Toledo-Rubio, V.; Ferrer-Miralles, N.; Vázquez, E.; et al. Bacterial mimetics of endocrine secretory granules as immobilized in vivo depots for functional protein drugs. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Genís, S.; Sánchez-Chardi, A.; Bach, À.; Fàbregas, F.; Arís, A. A combination of lactic acid bacteria regulates Escherichia coli infection and inflammation of the bovine endometrium. J. Dairy Sci. 2017, 100, 479–492. [Google Scholar] [CrossRef]
- De Pinho Favaro, M.T.; Sánchez-García, L.; Sánchez-Chardi, A.; Roldán, M.; Unzueta, U.; Serna, N.; Cano-Garrido, O.; Azzoni, A.R.; Ferrer-Miralles, N.; Villaverde, A.; et al. Protein nanoparticles are nontoxic, tuneable cell stressors. Nanomedicine 2018, 13, 255–268. [Google Scholar] [CrossRef]
- Cano-Garrido, O.; Garcia-Fruitós, E.; Villaverde, A.; Sánchez-Chardi, A. Improving Biomaterials Imaging for Nanotechnology: Rapid Methods for Protein Localization at Ultrastructural Level. Biotechnol. J. 2018, 13, 1–8. [Google Scholar] [CrossRef]
- Unzueta, U.; Cespedes, M.V.; Sala, R.; Alamo, P.; Sánchez-Chardi, A.; Pesarrodona, M.; Sánchez-García, L.; Cano-Garrido, O.; Villaverde, A.; Vázquez, E.; et al. Release of targeted protein nanoparticles from functional bacterial amyloids: A death star-like approach. J. Control. Release 2018, 279, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, J.M.; López-Laguna, H.; Álamo, P.; Serna, N.; Sánchez-Chardi, A.; Nolan, V.; Cano-Garrido, O.; Casanova, I.; Unzueta, U.; Vazquez, E.; et al. Artificial inclusion bodies for clinical development. Adv. Sci. 2019, 1902420, 1–7. [Google Scholar] [CrossRef]
- Malu, S.; Srinivasan, S.; Maiti, P.K.; Rajagopal, D.; John, B.; Nandi, D. IFN-γ bioassay: Development of a sensitive method by measuring nitric oxide production by peritoneal exudate cells from C57BL/6 mice. J. Immunol. Methods 2003, 272, 55–65. [Google Scholar] [CrossRef]
- Brouillette, E.; Malouin, F. The pathogenesis and control of Staphylococcus aureus-induced mastitis: Study models in the mouse. Microbes Infect. 2005, 7, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Asli, A.; Brouillette, E.; Ster, C.; Ghinet, M.G.; Brzezinski, R.; Lacasse, P.; Jacques, M.; Malouin, F. Antibiofilm and antibacterial effects of specific chitosan molecules on Staphylococcus aureus isolates associated with bovine mastitis. PLoS ONE 2017, 12, e0176988. [Google Scholar] [CrossRef] [PubMed]
- Mordmuang, A.; Brouillette, E.; Voravuthikunchai, S.P.; Malouin, F. Evaluation of a Rhodomyrtus tomentosa ethanolic extract for its therapeutic potential on Staphylococcus aureus infections using in vitro and in vivo models of mastitis. Vet. Res. 2019, 50, 49. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 1–10. [Google Scholar] [CrossRef]
- Boireau, C.; Cazeau, G.; Jarrige, N.; Calavas, D.; Madec, J.-Y.; Leblond, A.; Haenni, M.; Gay, É. Antimicrobial resistance in bacteria isolated from mastitis in dairy cattle in France, 2006–2016. J. Dairy Sci. 2018, 101, 9451–9462. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured antimicrobial peptides: The last push towards clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef]
- Bray, B.L. Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov. 2003, 2, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Meiyalaghan, S.; Latimer, J.M.; Kralicek, A.V.; Shaw, M.L.; Lewis, J.G.; Conner, A.J.; Barrell, P.J. Expression and purification of the antimicrobial peptide GSL1 in bacteria for raising antibodies. BMC Res. Notes 2014, 7, 777. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Wibowo, D.; Sainsbury, F.; Zhao, C.-X. Design and production of a novel antimicrobial fusion protein in Escherichia coli. Appl. Microbiol. Biotechnol. 2018, 102, 8763–8772. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, H.; Gao, D.; Gao, C.; Qi, Q. Secretory production of antimicrobial peptides in Escherichia coli using the catalytic domain of a cellulase as fusion partner. J. Biotechnol. 2015, 214, 77–82. [Google Scholar] [CrossRef]
- Soundrarajan, N.; Cho, H.-S.; Ahn, B.; Choi, M.; Thong, L.M.; Choi, H.; Cha, S.-Y.; Kim, J.-H.; Park, C.-K.; Seo, K.; et al. Green fluorescent protein as a scaffold for high efficiency production of functional bacteriotoxic proteins in Escherichia coli. Sci. Rep. 2016, 6, 20661. [Google Scholar] [CrossRef]
- Zhou, Q.-F.; Luo, X.-G.; Ye, L.; Xi, T. High-level production of a novel antimicrobial peptide perinerin in Escherichia coli by fusion expression. Curr. Microbiol. 2007, 54, 366–370. [Google Scholar] [CrossRef]
- Li, J.F.; Zhang, J.; Zhang, Z.; Kang, C.T.; Zhang, S.Q. SUMO mediating fusion expression of antimicrobial peptide CM4 from two joined genes in Escherichia coli. Curr. Microbiol. 2011, 62, 296–300. [Google Scholar] [CrossRef]
- Wei, X.; Wu, R.; Zhang, L.; Ahmad, B.; Si, D.; Zhang, R. Expression, Purification, and Characterization of a Novel Hybrid Peptide with Potent Antibacterial Activity. Molecules 2018, 23, 1491. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, J.H.; Hwang, S.W.; Lee, W.J.; Yoon, H.K.; Lee, H.S.; Hong, S.S. High-level expression of antimicrobial peptide mediated by a fusion partner reinforcing formation of inclusion bodies. Biochem. Biophys. Res. Commun. 2000, 277, 575–580. [Google Scholar] [CrossRef]
- Sánchez-García, L.; Serna, N.; Álamo, P.; Sala, R.; Céspedes, M.V.; Roldan, M.; Sánchez-Chardi, A.; Unzueta, U.; Casanova, I.; Mangues, R.; et al. Self-assembling toxin-based nanoparticles as self-delivered antitumoral drugs. J. Control. Release 2018, 274, 81–92. [Google Scholar] [CrossRef]
- Gray, P.W.; Goeddel, D.V. Cloning and expression of murine immune interferon cDNA. Proc. Natl. Acad. Sci. USA 1983, 80, 5842–5846. [Google Scholar] [CrossRef]
- Ahmed, C.M.I.; Burkhart, M.A.; Subramaniam, P.S.; Mujtaba, M.G.; Johnson, H.M. Peptide Mimetics of Gamma Interferon Possess Antiviral Properties against Vaccinia Virus and Other Viruses in the Presence of Poxvirus B8R Protein. J. Virol. 2005, 79, 5632–5639. [Google Scholar] [CrossRef] [PubMed]
- Magazine, H.I.; Carter, J.M.; Russell, J.K.; Torres, B.A.; Dunn, B.M.; Johnson, H.M. Use of synthetic peptides to identify an N-terminal epitope on mouse γ interferon that may be involved in function. Proc. Natl. Acad. Sci. USA 1988, 85, 1237–1241. [Google Scholar] [CrossRef]
- Torcato, I.M.; Huang, Y.-H.; Franquelim, H.G.; Gaspar, D.; Craik, D.J.; Castanho, M.A.R.B.; Troeira Henriques, S. Design and characterization of novel antimicrobial peptides, R-BP100 and RW-BP100, with activity against Gram-negative and Gram-positive bacteria. Biochim. Biophys. Acta 2013, 1828, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A.R.B. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammadi, M.; Haine, D.; Kelton, D.F.; Barkema, H.W.; Hogeveen, H.; Keefe, G.P.; Dufour, S. Herd-Level Mastitis-Associated Costs on Canadian Dairy Farms. Front. Vet. Sci. 2018, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Apparao, M.D.; Ruegg, P.L.; Lago, A.; Godden, S.; Bey, R.; Leslie, K. Relationship between in vitro susceptibility test results and treatment outcomes for gram-positive mastitis pathogens following treatment with cephapirin sodium. J. Dairy Sci. 2009, 92, 2589–2597. [Google Scholar] [CrossRef]
- Demon, D.; Ludwig, C.; Breyne, K.; Guédé, D.; Dörner, J.-C.; Froyman, R.; Meyer, E. The intramammary efficacy of first generation cephalosporins against Staphylococcus aureus mastitis in mice. Vet. Microbiol. 2012, 160, 141–150. [Google Scholar] [CrossRef]
- Li, L.; Wang, L.; Gao, Y.; Wang, J.; Zhao, X. Effective antimicrobial activity of plectasin-derived antimicrobial peptides against Staphylococcus aureus infection in mammary glands. Front. Microbiol. 2017, 8, 2386. [Google Scholar] [CrossRef]
- Wei, W.; Dejie, L.; Xiaojing, S.; Tiancheng, W.; Yongguo, C.; Zhengtao, Y.; Naisheng, Z. Magnolol inhibits the inflammatory response in mouse mammary epithelial cells and a mouse mastitis model. Inflammation 2015, 38, 16–26. [Google Scholar] [CrossRef]
- Schmelcher, M.; Powell, A.M.; Camp, M.J.; Pohl, C.S.; Donovan, D.M. Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl. Microbiol. Biotechnol. 2015, 99, 8475–8486. [Google Scholar] [CrossRef]
- Notebaert, S.; Demon, D.; Vanden Berghe, T.; Vandenabeele, P.; Meyer, E. Inflammatory mediators in Escherichia coli-induced mastitis in mice. Comp. Immunol. Microbiol. Infect. Dis. 2008, 31, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fang, J.T.; Sun, J.; Zheng, M.; Zhang, Q.; He, J.S.; Liao, X.P.; Liu, Y.H. Efficacy of cefquinome against Escherichia coli environmental mastitis assessed by pharmacokinetic and pharmacodynamic integration in lactating mouse model. Front. Microbiol. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fruitós, E. Inclusion bodies: A new concept. Microb. Cell Fact. 2010, 9, 80. [Google Scholar] [CrossRef]
- Villaverde, A.; Corchero, J.L.; Seras-Franzoso, J.; Garcia-Fruitós, E. Functional protein aggregates: Just the tip of the iceberg. Nanomedicine 2015, 10, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Rinas, U.; Garcia-Fruitós, E.; Corchero, J.L.; Vázquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial inclusion bodies: Discovering their better half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef]
- Peternel, Š.; Grdadolnik, J.; Gaberc-Porekar, V.; Komel, R. Engineering inclusion bodies for non denaturing extraction of functional proteins. Microb. Cell Fact. 2008, 7, 1–9. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carratalá, J.V.; Brouillette, E.; Serna, N.; Sánchez-Chardi, A.; Sánchez, J.M.; Villaverde, A.; Arís, A.; Garcia-Fruitós, E.; Ferrer-Miralles, N.; Malouin, F. In Vivo Bactericidal Efficacy of GWH1 Antimicrobial Peptide Displayed on Protein Nanoparticles, a Potential Alternative to Antibiotics. Pharmaceutics 2020, 12, 1217. https://doi.org/10.3390/pharmaceutics12121217
Carratalá JV, Brouillette E, Serna N, Sánchez-Chardi A, Sánchez JM, Villaverde A, Arís A, Garcia-Fruitós E, Ferrer-Miralles N, Malouin F. In Vivo Bactericidal Efficacy of GWH1 Antimicrobial Peptide Displayed on Protein Nanoparticles, a Potential Alternative to Antibiotics. Pharmaceutics. 2020; 12(12):1217. https://doi.org/10.3390/pharmaceutics12121217
Chicago/Turabian StyleCarratalá, Jose V., Eric Brouillette, Naroa Serna, Alejandro Sánchez-Chardi, Julieta M. Sánchez, Antonio Villaverde, Anna Arís, Elena Garcia-Fruitós, Neus Ferrer-Miralles, and François Malouin. 2020. "In Vivo Bactericidal Efficacy of GWH1 Antimicrobial Peptide Displayed on Protein Nanoparticles, a Potential Alternative to Antibiotics" Pharmaceutics 12, no. 12: 1217. https://doi.org/10.3390/pharmaceutics12121217
APA StyleCarratalá, J. V., Brouillette, E., Serna, N., Sánchez-Chardi, A., Sánchez, J. M., Villaverde, A., Arís, A., Garcia-Fruitós, E., Ferrer-Miralles, N., & Malouin, F. (2020). In Vivo Bactericidal Efficacy of GWH1 Antimicrobial Peptide Displayed on Protein Nanoparticles, a Potential Alternative to Antibiotics. Pharmaceutics, 12(12), 1217. https://doi.org/10.3390/pharmaceutics12121217