Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread
Abstract
:1. Introduction
2. Lessons from Turnip Crinkle Virus, Satellite Tobacco Mosaic Virus, Groundnut Rosette Virus and Tobacco Mosaic Virus: Monopartite RNA Genomes
2.1. Positive-Strand RNA Packaging into Icosahedral Units
2.1.1. Autonomous Turnip Crinkle Virus
2.1.2. Helper Virus Replication-Dependent Satellite Tobacco Mosaic Virus
2.1.3. Umbraviruses Replicate Autonomously but Are Packaged in Trans
2.2. Positive-Strand RNA Packaging into Helical Units: Lessons from Tobacco Mosaic Virus
3. Multipartite RNA Genomes
3.1. Positive-Strand RNA Packaging into Icosahedral Units
3.1.1. Dianthovirus: Bipartite Genome
3.1.2. Brome Mosaic Virus: Tripartite Genome
3.2. Positive-Strand RNA Packaging into Helical Particle Units
3.2.1. Benyviruses: Up to Five Genomic RNAs
3.2.2. Potato Mop Top Virus: Tripartite, but Two Is Enough
4. Multipartite Positive-Strand RNA Genomes Have to Preserve Their Genome Integrity
5. Concluding Remarks: Lessons from Superinfection Exclusion Reports
Acknowledgments
Conflicts of Interest
References
- Gerber, M.; Isel, C.; Moules, V.; Marquet, R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014, 22, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Guu, T.S.; Zheng, W.; Tao, Y.J. Bunyavirus: Structure and replication. Adv. Exp. Med. Biol. 2012, 726, 245–266. [Google Scholar]
- Huiskonen, J.T.; de Haas, F.; Bubeck, D.; Bamford, D.H.; Fuller, S.D.; Butcher, S.J. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 2006, 14, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 2006, 44, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.G.; Makarov, V.V. Helical capsids of plant viruses: Architecture with structural lability. J. Gen. Virol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Waigmann, E.; Ueki, S.; Trutnyeva, K.; Citovsky, V. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 2004, 23, 195–250. [Google Scholar] [CrossRef]
- Roberts, A.G.; Cruz, S.S.; Roberts, I.M.; Prior, D.; Turgeon, R.; Oparka, K.J. Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell 1997, 9, 1381–1396. [Google Scholar] [CrossRef] [PubMed]
- Dykeman, E.C.; Grayson, N.E.; Toropova, K.; Ranson, N.A.; Stockley, P.G.; Twarock, R. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J. Mol. Biol. 2011, 408, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Building a viral capsid in the presence of genomic RNA. Phys. Rev. E Stat. Nonlin Soft Matter Phys. 2013, 87, 022717. [Google Scholar] [CrossRef] [PubMed]
- Rolfsson, O.; Middleton, S.; Manfield, I.W.; White, S.J.; Fan, B.; Vaughan, R.; Ranson, N.A.; Dykeman, E.; Twarock, R.; Ford, J.; et al. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016, 428, 431–448. [Google Scholar] [CrossRef] [PubMed]
- Sorger, P.K.; Stockley, P.G.; Harrison, S.C. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J. Mol. Biol. 1986, 191, 639–658. [Google Scholar] [CrossRef]
- Qu, F.; Morris, T.J. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 1997, 71, 1428–1435. [Google Scholar] [PubMed]
- Cohen, Y.; Gisel, A.; Zambryski, P.C. Cell-to-cell and systemic movement of recombinant green fluorescent protein-tagged turnip crinkle viruses. Virology 2000, 273, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Ren, T.; Morris, T.J. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 2003, 77, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Zeng, Y.; Heitsch, C.E. The icosahedral RNA virus as a grotto: Organizing the genome into stalagmites and stalactites. J. Biol. Phys. 2013, 39, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Archer, E.J.; Simpson, M.A.; Watts, N.J.; O'Kane, R.; Wang, B.; Erie, D.A.; McPherson, A.; Weeks, K.M. Long-range architecture in a viral RNA genome. Biochemistry 2013, 52, 3182–3190. [Google Scholar] [CrossRef] [PubMed]
- Valverde, R.A.; Heick, J.A.; Dodds, J.A. Interactions between satellite tobacco mosaic virus, helper tobamovirus, and their hosts. Phytopathology 1991, 81, 99–104. [Google Scholar] [CrossRef]
- Robinson, D.J.; Ryabov, E.V.; Raj, S.K.; Roberts, I.M.; Taliansky, M.E. Satellite RNA is essential for encapsidation of groundnut rosette umbravirus RNA by groundnut rosette assistor luteovirus coat protein. Virology 1999, 254, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.E.; Robinson, D.J. Molecular biology of umbraviruses: Phantom warriors. J. Gen. Virol. 2003, 84, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.R.; Butler, P.J. Essential features of the assembly origin of tobacco mosaic virus RNA as studied by directed mutagenesis. Nucleic Acids Res. 1986, 14, 9229–9242. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiao, J.; Niu, Z.; Wang, Q. Natural supramolecular building blocks: From virus coat proteins to viral nanoparticles. Chem. Soc. Rev. 2012, 41, 6178–6194. [Google Scholar] [CrossRef] [PubMed]
- Guenoune-Gelbart, D.; Elbaum, M.; Sagi, G.; Levy, A.; Epel, B.L. Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol. Plant Microbe Interact. 2008, 21, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Dorokhov, Y.L.; Alexandrova, N.M.; Miroshnichenko, N.A.; Atabekov, J.G. Isolation and analysis of virus-specific ribonucleoprotein of tobacco mosaic virus-infected tobacco. Virology 1983, 127, 237–252. [Google Scholar] [CrossRef]
- Saito, T.; Yamanaka, K.; Okada, Y. Long-distance movement and viral assembly of tobacco mosaic virus mutants. Virology 1990, 176, 329–336. [Google Scholar] [CrossRef]
- Ding, X.; Shintaku, M.H.; Carter, S.A.; Nelson, R.S. Invasion of minor veins of tobacco leaves inoculated with tobacco mosaic virus mutants defective in phloem-dependent movement. Proc. Natl. Acad. Sci. USA 1996, 93, 11155–11160. [Google Scholar] [CrossRef] [PubMed]
- Dawson, W.O.; Bubrick, P.; Grantham, G.L. Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement and symptomatology. Phytopathology 1988, 78, 783–789. [Google Scholar] [CrossRef]
- Nelson, R.S.; Li, G.; Hodgson, R.A.; Beachy, R.N.; Shintaku, M.H. Impeded phloem-dependent accumulation of the masked strain of tobacco mosaic virus. Mol. Plant Microbe Interact. 1993, 6, 45–54. [Google Scholar] [CrossRef]
- Ding, X.S.; Shintaku, M.H.; Arnold, S.A.; Nelson, R.S. Accumulation of mild and severe strains of tobacco mosaic virus in minor veins of tobacco. Mol. Plant Microbe Interact. 1995, 8, 32–40. [Google Scholar] [CrossRef]
- Heinlein, M. Plant virus replication and movement. Virology 2015, 479–480, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Doering-Saad, C.; Newbury, H.J.; Bale, J.S.; Pritchard, J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 2002, 53, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Kragler, F. Rna in the phloem: A crisis or a return on investment? Plant Sci. 2010, 178, 99–104. [Google Scholar] [CrossRef]
- Sasaki, T.; Chino, M.; Hayashi, H.; Fujiwara, T. Detection of several mRNA species in rice phloem sap. Plant Cell Physiol. 1998, 39, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Robinson, D.J.; Taliansky, M.E. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. USA 1999, 96, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Taliansky, M.; Roberts, I.M.; Kalinina, N.; Ryabov, E.V.; Raj, S.K.; Robinson, D.J.; Oparka, K.J. An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 2003, 77, 3031–3040. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Sheng, J.; Hind, G.; Handa, A.K.; Citovsky, V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000, 19, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Citovsky, V. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 2003, 35, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Fenczik, C.A.; Padgett, H.S.; Holt, C.A.; Casper, S.J.; Beachy, R.N. Mutational analysis of the movement protein of odontoglossum ringspot virus to identify a host-range determinant. Mol. Plant Microbe Interact. 1995, 8, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lommel, S.A. Identification and analysis of the site of -1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology 1994, 200, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Osman, T.A.M.; Buck, K.W. Double-stranded RNAs isolated from plant tissue infected with red clover necrotic mosaic virus correspond to genomic and subgenomic single-stranded rRNA. J. Gen. Virol. 1990, 71, 945–948. [Google Scholar] [CrossRef]
- Xiong, Z.; Kim, K.H.; Giesman-Cookmeyer, D.; Lommel, S.A. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 1993, 192, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Lommel, S.A. The complete nucleotide sequence and genome organization of red clover necrotic mosaic virus RNA-1. Virology 1989, 171, 543–554. [Google Scholar] [CrossRef]
- Zavriev, S.K.; Hickey, C.M.; Lommel, S.A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 1996, 216, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Lommel, S.A.; Weston-Fina, M.; Xiong, Z.; Lomonossoff, G.P. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res. 1988, 16, 8587–8602. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.G.; Sit, T.L.; Heinsohn, C.; George, C.G.; Kim, K.H.; Lommel, S.A. The red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology 2008, 381, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Wang, Y.; Giesman-Cookmeyer, D.; Lommel, S.A.; Lucas, W.J. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology 1998, 245, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Basnayake, V.R.; Sit, T.L.; Lommel, S.A. The red clover necrotic mosaic virus origin of assembly is delimited to the RNA-2 trans-activator. Virology 2009, 384, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Osman, T.A.M.; Buck, K.W. Replication of red clover necrotic mosaic virus RNA in cowpea protoplasts: RNA 1 replicates independently of RNA 2. J. Gen. Virol. 1987, 68, 289–296. [Google Scholar] [CrossRef]
- Paje-Manalo, L.L.; Lommel, S.A. Independent replication of red clover necrotic mosaic virus RNA-1 in electroporated host and nonhost nicotiana species protoplasts. Phytopathology 1989, 79, 457–461. [Google Scholar] [CrossRef]
- Sit, T.L.; Vaewhongs, A.A.; Lommel, S.A. RNA-mediated trans-activation of transcription from a viral RNA. Science 1998, 281, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.; Guenther, R.H.; Basnayake, V.R.; Lommel, S.A.; Franzen, S. Controlled encapsidation of gold nanoparticles by a viral protein shell. J. Am. Chem. Soc. 2006, 128, 4502–4503. [Google Scholar] [CrossRef] [PubMed]
- Basnayake, V.R.; Sit, T.L.; Lommel, S.A. The genomic RNA packaging scheme of red clover necrotic mosaic virus. Virology 2006, 345, 532–539. [Google Scholar] [CrossRef]
- Vaewhongs, A.A.; Lommel, S.A. Virion formation is required for the long-distance movement of red clover necrotic mosaic virus in movement protein transgenic plants. Virology 1995, 212, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Lane, L.C. Bromoviruses. In Handbook of Plant Virus Infections and Comparative Diagnosis; Kurstak, E., Ed.; Elsevier/North Holland Biomedical Press: Amsterdam, The Netherlands, 1981; pp. 333–375. [Google Scholar]
- Choi, Y.G.; Rao, A.L. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003, 77, 9750–9757. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, P.; Rao, A.L. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3′ tRNA-like structure. J. Virol. 2007, 81, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Damayanti, T.A.; Tsukaguchi, S.; Mise, K.; Okuno, T. Cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 2003, 77, 9979–9986. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, P.; Rao, A.L. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 2006, 80, 10096–10108. [Google Scholar] [CrossRef] [PubMed]
- Mandahar, C.L. Infection by and uncoating of virus particles. In Multiplication of RNA Plant Viruses; Mandahar, C.L., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2006; p. 339. [Google Scholar]
- Brisco, M.; Hull, R.; Wilson, T.M. Swelling of isometric and of bacilliform plant virus nucleocapsids is required for virus-specific protein synthesis in vitro. Virology 1986, 148, 210–217. [Google Scholar] [CrossRef]
- Seo, J.K.; Kwon, S.J.; Rao, A.L. A physical interaction between viral replicase and capsid protein is required for genome-packaging specificity in an RNA virus. J. Virol. 2012, 86, 6210–6221. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Fanning, S.W.; Horn, J.R.; Bujarski, J.J. Mutations in the coat protein-binding cis-acting RNA motifs debilitate RNA recombination of brome mosaic virus. Virus Res. 2012, 170, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Kaido, M.; Okuno, T.; Mise, K. Coat protein-independent cell-to-cell movement of bromoviruses expressing brome mosaic virus movement protein with an adaptation-related amino acid change in the central region. Arch. Virol. 2005, 150, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Nakamura, W.; Sasaki, N.; Goto, K.; Kaido, M.; Okuno, T.; Mise, K. Natural isolates of brome mosaic virus with the ability to move from cell to cell independently of coat protein. J. Gen. Virol. 2005, 86, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, K.; Kao, C.C. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 2007, 19, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.M.; Zambryski, P.C. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001, 125, 1802–1812. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.; Ratti, C. Benyvirus. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 1133–1138. [Google Scholar]
- Jupin, I.; Richards, K.; Jonard, G.; Guilley, H.; Pleij, C.W. Mapping sequences required for productive replication of beet necrotic yellow vein virus RNA 3. Virology 1990, 178, 273–280. [Google Scholar] [CrossRef]
- Gilmer, D.; Allmang, C.; Ehresmann, C.; Guilley, H.; Richards, K.; Jonard, G.; Ehresmann, B. The secondary structure of the 5′-noncoding region of beet necrotic yellow vein virus RNA 3: Evidence for a role in viral RNA replication. Nucleic Acids Res. 1993, 21, 1389–1395. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.; Richards, K.; Jonard, G.; Guilley, H. Cis-active sequences near the 5′-termini of beet necrotic yellow vein virus RNAs 3 and 4. Virology 1992, 190, 55–67. [Google Scholar] [CrossRef]
- Ziegler, V.; Richards, K.; Guilley, H.; Jonard, G.; Putz, C. Cell-free translation of beet necrotic yellow vein virus: Readthrough of the coat protein cistron. J. Gen. Virol. 1985, 66, 2079–2087. [Google Scholar] [CrossRef]
- Haeberle, A.M.; Stussi-Garaud, C.; Schmitt, C.; Garaud, J.C.; Richards, K.E.; Guilley, H.; Jonard, G. Detection by immunogold labelling of p75 readthrough protein near an extremity of beet necrotic yellow vein virus particles. Arch. Virol. 1994, 134, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.; Balmori, E.; Jonard, G.; Richards, K.E.; Guilley, H. In vitro mutagenesis of biologically active transcripts of beet necrotic yellow vein virus RNA 2: Evidence that a domain of the 75-kda readthrough protein is important for efficient virus assembly. Proc. Natl. Acad. Sci. USA 1992, 89, 5715–5719. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Schmitt, C.; Saito, M.; Guilley, H.; Richards, K.; Jonard, G. High resolution analysis of the readthrough domain of beet necrotic yellow vein virus readthrough protein: A KTER motif is important for efficient transmission of the virus by Polymyxa betae. J. Gen. Virol. 1996, 77, 1359–1367. [Google Scholar] [CrossRef] [PubMed]
- Pakdel, A.; Mounier, C.; Klein, E.; Hleibieh, K.; Monsion, B.; Mutterer, J.; Erhardt, M.; Bouzoubaa, S.; Ratti, C.; Gilmer, D. On the interaction and localization of the beet necrotic yellow vein virus replicase. Virus Res. 2015, 196, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Erhardt, M.; Dunoyer, P.; Guilley, H.; Richards, K.; Jonard, G.; Bouzoubaa, S. Beet necrotic yellow vein virus particles localize to mitochondria during infection. Virology 2001, 286, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Valentin, C.; Dunoyer, P.; Vetter, G.; Schalk, C.; Dietrich, A.; Bouzoubaa, S. Molecular basis for mitochondrial localization of viral particles during beet necrotic yellow vein virus infection. J. Virol. 2005, 79, 9991–10002. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Shirako, Y.; Abe, H.; Saito, M.; Kigushi, T.; Harada, T. Production and pathogenicity of isolates of beet necrotic yellow vein virus with different numbers of RNA components. J. Gen. Virol. 1989, 70, 3399–3409. [Google Scholar] [CrossRef]
- Quillet, L.; Guilley, H.; Jonard, G.; Richards, K. In vitro synthesis of biologically active beet necrotic yellow vein virus RNA. Virology 1989, 172, 293–301. [Google Scholar] [PubMed]
- Rahim, M.D.; Andika, I.B.; Han, C.; Kondo, H.; Tamada, T. RNA4-encoded p31 of beet necrotic yellow vein virus is involved in efficient vector transmission, symptom severity and silencing suppression in roots. J. Gen. Virol. 2007, 88, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Abe, H. Evidence that beet necrotic yellow vein virus RNA-4 is essential for transmission by the fungus Polymyxa betae. J. Gen. Virol. 1989, 70, 3391–3398. [Google Scholar] [CrossRef]
- Tamada, T.; Uchino, H.; Kusume, T.; Saito, M. RNA 3 deletion mutants of beet necrotic yellow vein virus do not cause rhizomania disease in sugar beets. Phytopathology 1999, 89, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Kondo, H.; Tamada, T. Evidence that RNA silencing-mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol. Plant Microbe Interact. 2005, 18, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Chiba, S.; Hleibieh, K.; Delbianco, A.; Klein, E.; Ratti, C.; Ziegler-Graff, V.; Bouzoubaa, S.; Gilmer, D. The benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing-suppression activity. Mol. Plant Microbe Interact. 2013, 26, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Delbianco, A.; Lanzoni, C.; Klein, E.; Rubies Autonell, C.; Gilmer, D.; Ratti, C. Agroinoculation of beet necrotic yellow vein virus cDNA clones results in plant systemic infection and efficient Polymyxa betae transmission. Mol. Plant Pathol. 2013, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.; Bouzoubaa, S.; Hehn, A.; Guilley, H.; Richards, K.; Jonard, G. Efficient cell-to-cell movement of beet necrotic yellow vein virus requires 3′ proximal genes located on RNA 2. Virology 1992, 189, 40–47. [Google Scholar] [CrossRef]
- Adams, M.J.; Heinze, C.; Jackson, A.O.; Kreuze, J.F.; Macfarlane, S.; Torrance, L. Family Virgaviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 1139–1162. [Google Scholar]
- Kassanis, B.; Woods, R.D.; White, R.F. Some properties of potato mop-top virus and its serological relationship to tobacco mosaic virus. J. Gen. Virol. 1972, 14, 123–132. [Google Scholar] [CrossRef]
- Cowan, G.H.; Torrance, L.; Reavy, B. Detection of potato mop-top virus capsid readthrough protein in virus particles. J. Gen. Virol. 1997, 78 Pt 7, 1779–1783. [Google Scholar] [CrossRef] [PubMed]
- Reavy, B.; Arif, M.; Cowan, G.H.; Torrance, L. Association of sequences in the coat protein/readthrough domain of potato mop-top virus with transmission by Spongospora subterranea. J. Gen. Virol. 1998, 79 Pt 10, 2343–2347. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Lukhovitskaya, N.I.; Schepetilnikov, M.V.; Cowan, G.H.; Ziegler, A.; Savenkov, E.I. Unusual long-distance movement strategies of potato mop-top virus RNAs in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2009, 22, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Savenkov, E.I.; Germundsson, A.; Zamyatnin, A.A., Jr.; Sandgren, M.; Valkonen, J.P.T. Potato mop-top virus: The coat protein-encoding RNA and the gene for cysteine-rich protein are dispensable for systemic virus movement in Nicotiana benthamiana. J. Gen. Virol. 2003, 84, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.M.; Cowan, G.H.; Lukhovitskaya, N.I.; Tilsner, J.; Roberts, A.G.; Savenkov, E.I.; Torrance, L. The N-terminal domain of PMTV TGB1 movement protein is required for nucleolar localization, microtubule association, and long-distance movement. Mol. Plant Microbe Interact. 2010, 23, 1486–1497. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Macfarlane, S.; Kalinina, N.O.; Rakitina, D.V.; Ryabov, E.V.; Gillespie, T.; Haupt, S.; Brown, J.W.; Taliansky, M. Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA 2007, 104, 11115–11120. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Richter, A.S.; Backofen, R. Intarna: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.R.; Georg, J.; Mann, M.; Sorescu, D.A.; Richter, A.S.; Lott, S.; Kleinkauf, R.; Hess, W.R.; Backofen, R. Coprarna and intarna: Predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014, 42, W119–W123. [Google Scholar] [CrossRef] [PubMed]
- Sandgren, M.; Savenkov, E.I.; Valkonen, J.P. The readthrough region of potato mop-top virus (PMTV) coat protein encoding RNA, the second largest RNA of PMTV genome, undergoes structural changes in naturally infected and experimentally inoculated plants. Arch. Virol. 2001, 146, 467–477. [Google Scholar] [CrossRef] [PubMed]
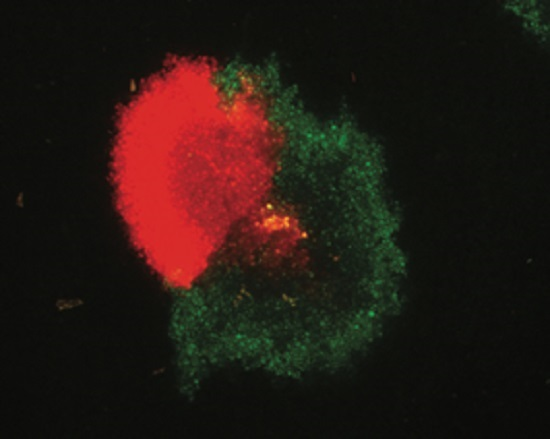
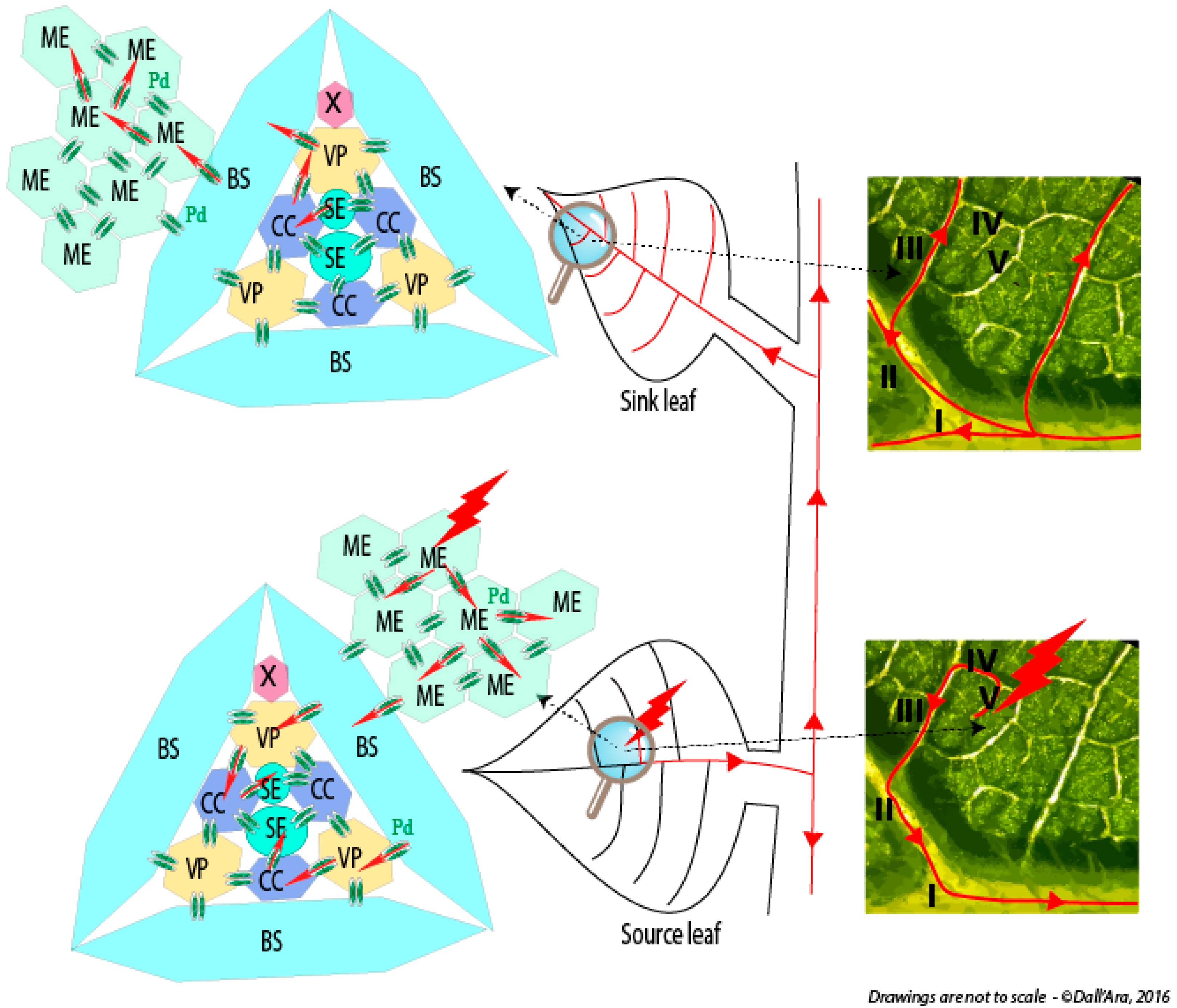
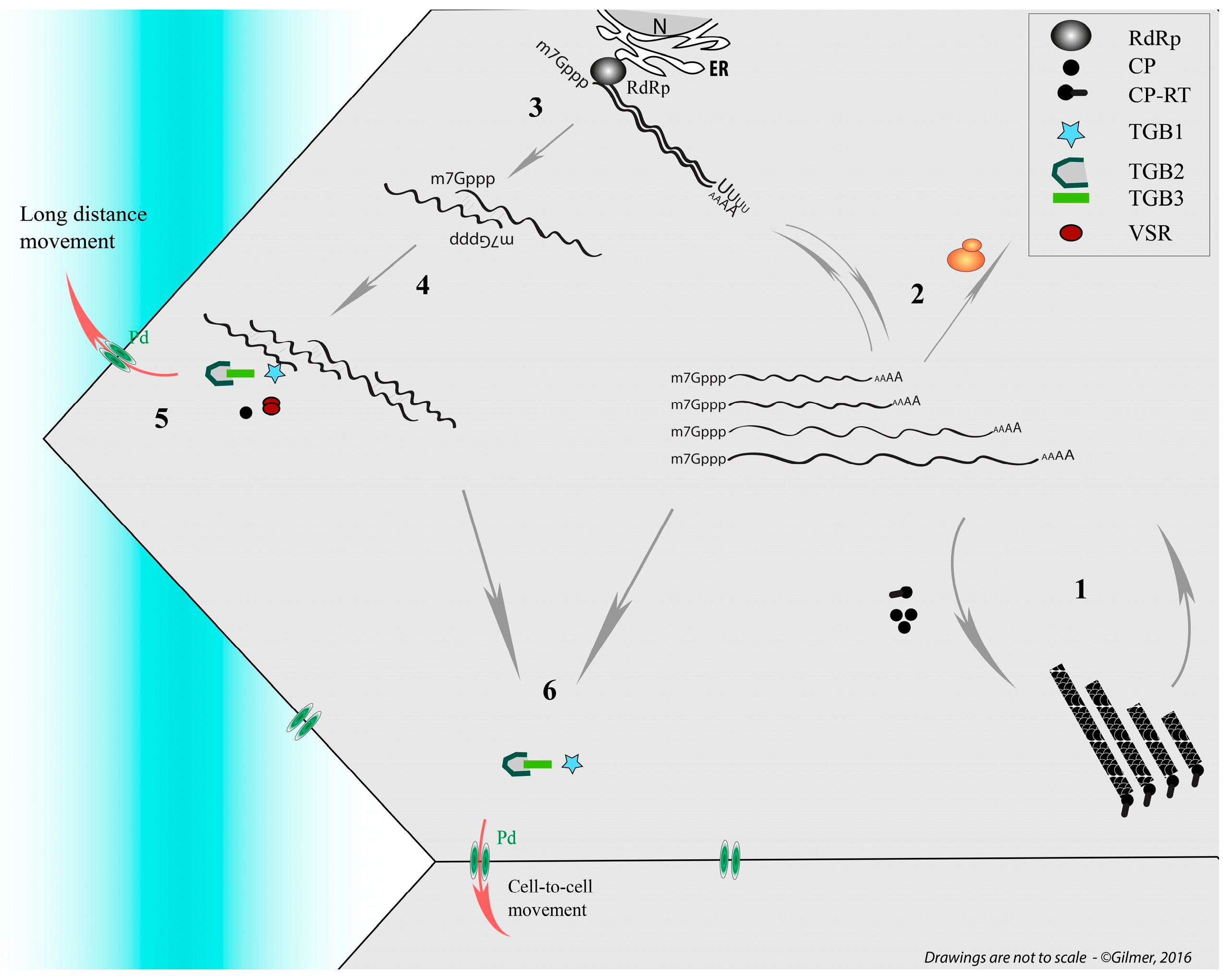
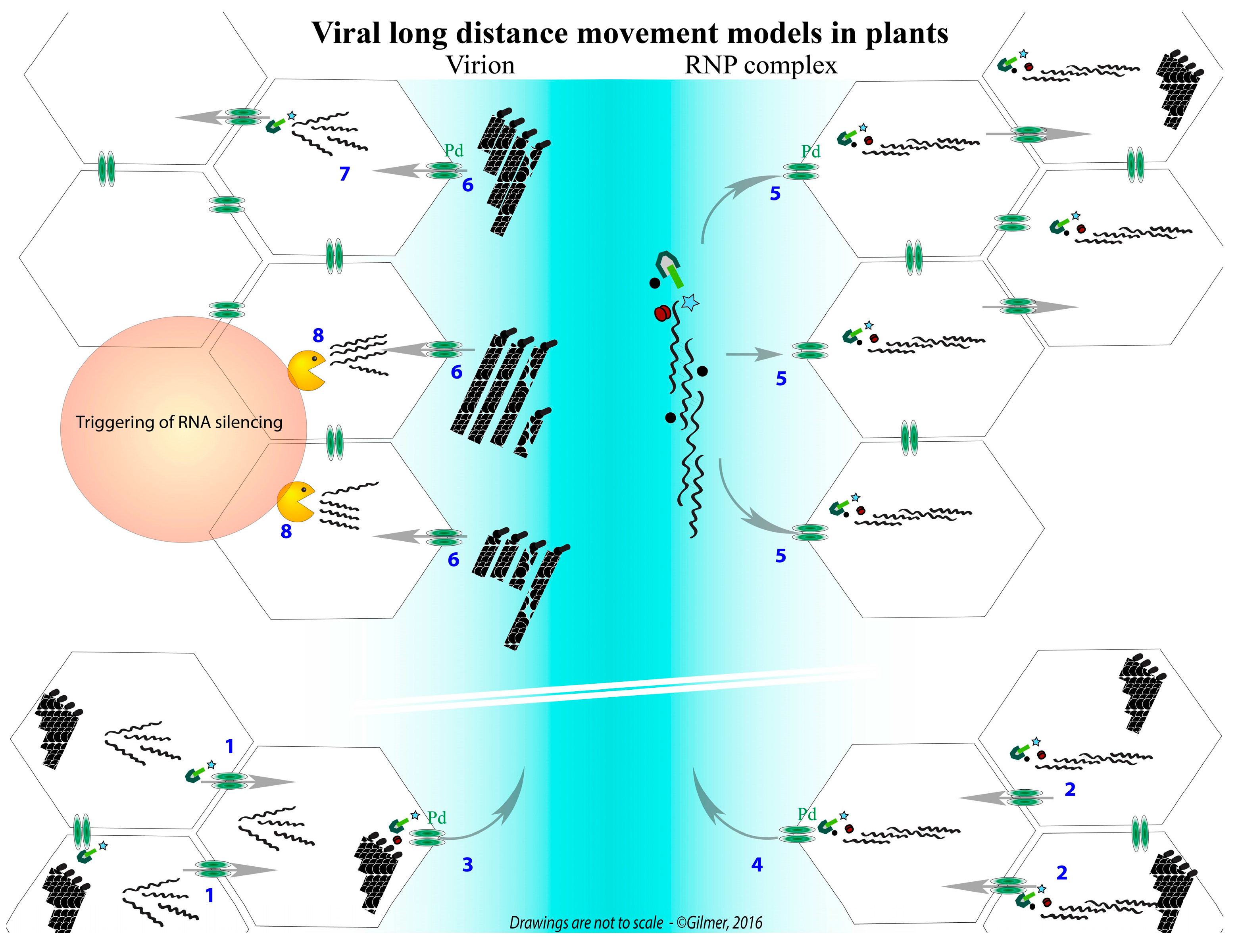
- Dall’Ara, M.; (University of Bologna, Bologna, Italy); Ratti, C.; (University of Bologna, Bologna, Italy); Bouzoubaa, S.E.; (University of Strasbourg, Strasbourg, France); Gilmer, D.; (University of Strasbourg, Strasbourg, France). Personal communication, 2016.
- Iranzo, J.; Manrubia, S.C. Evolutionary dynamics of genome segmentation in multipartite viruses. Proc. Biol. Sci. 2012, 279, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.J.; Henning, U. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 1994, 2, 137–139. [Google Scholar] [CrossRef]
- Roy, N.; Pacini, G.; Berlioz-Torrent, C.; Janvier, K. Mechanisms underlying HIV-1 Vpu-mediated viral egress. Front. Microbiol. 2014, 5, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tromas, N.; Zwart, M.P.; Lafforgue, G.; Elena, S.F. Within-host spatiotemporal dynamics of plant virus infection at the cellular level. PLoS Genet. 2014, 10, e1004186. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Rush, C. Evidence of cross-protection between beet soilborne mosaic virus and beet necrotic yellow vein virus in sugar beet. Plant Dis. 1999, 83, 521–526. [Google Scholar] [CrossRef]
- Ratti, C.; Hleibieh, K.; Bianchi, L.; Schirmer, A.; Autonell, C.R.; Gilmer, D. Beet soil-borne mosaic virus RNA-3 is replicated and encapsidated in the presence of BNYVV RNA-1 and -2 and allows long distance movement in Beta macrocarpa. Virology 2009, 385, 392–399. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dall’Ara, M.; Ratti, C.; Bouzoubaa, S.E.; Gilmer, D. Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread. Viruses 2016, 8, 228. https://doi.org/10.3390/v8080228
Dall’Ara M, Ratti C, Bouzoubaa SE, Gilmer D. Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread. Viruses. 2016; 8(8):228. https://doi.org/10.3390/v8080228
Chicago/Turabian StyleDall’Ara, Mattia, Claudio Ratti, Salah E. Bouzoubaa, and David Gilmer. 2016. "Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread" Viruses 8, no. 8: 228. https://doi.org/10.3390/v8080228
APA StyleDall’Ara, M., Ratti, C., Bouzoubaa, S. E., & Gilmer, D. (2016). Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread. Viruses, 8(8), 228. https://doi.org/10.3390/v8080228