De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Infection
2.2. Total RNA Isolation
2.3. Library Construction, Sequencing and Data Analysis for RNA-Seq
2.4. De Novo Assembly, Functional Annotation and Gene Ontology Classification
2.5. Identification of Differentially Expressed Genes
2.6. Data Access
2.7. Validation of RNA-Seq Data and Virus Quantification
3. Results
3.1. Gene Ontology Classification and KEGG Pathway Analysis
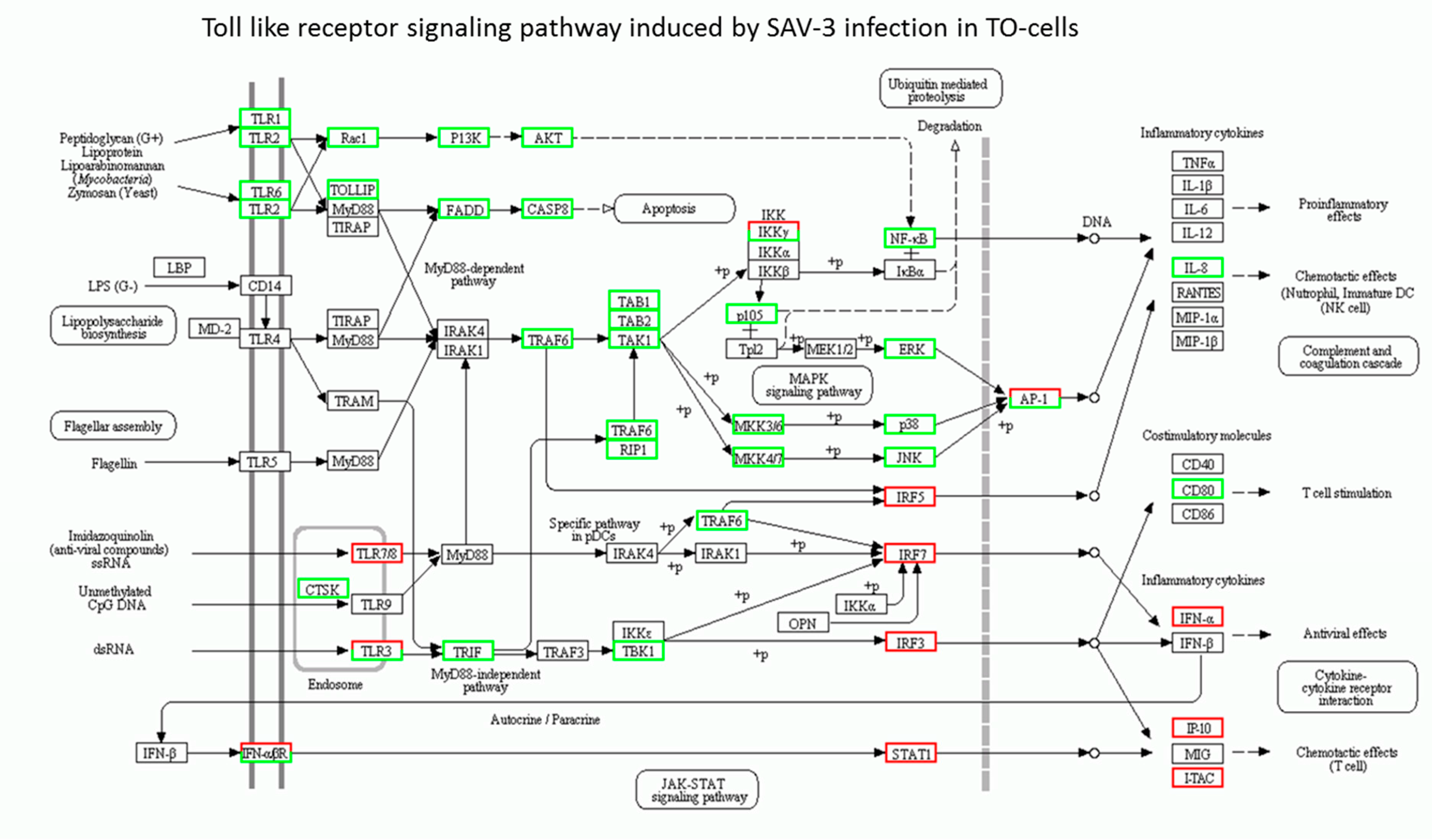
3.2. Toll-Like Receptor Signaling Pathways
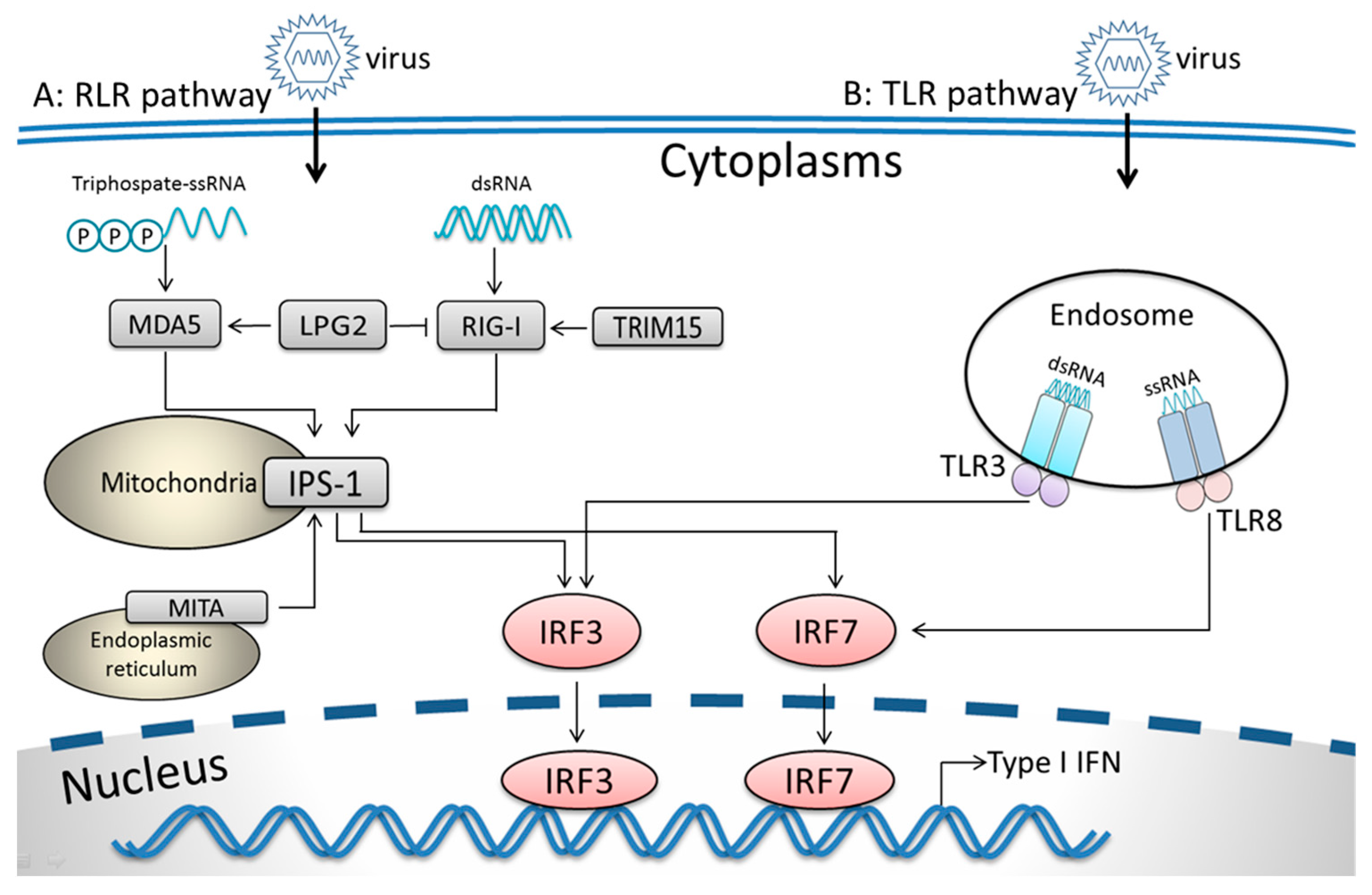
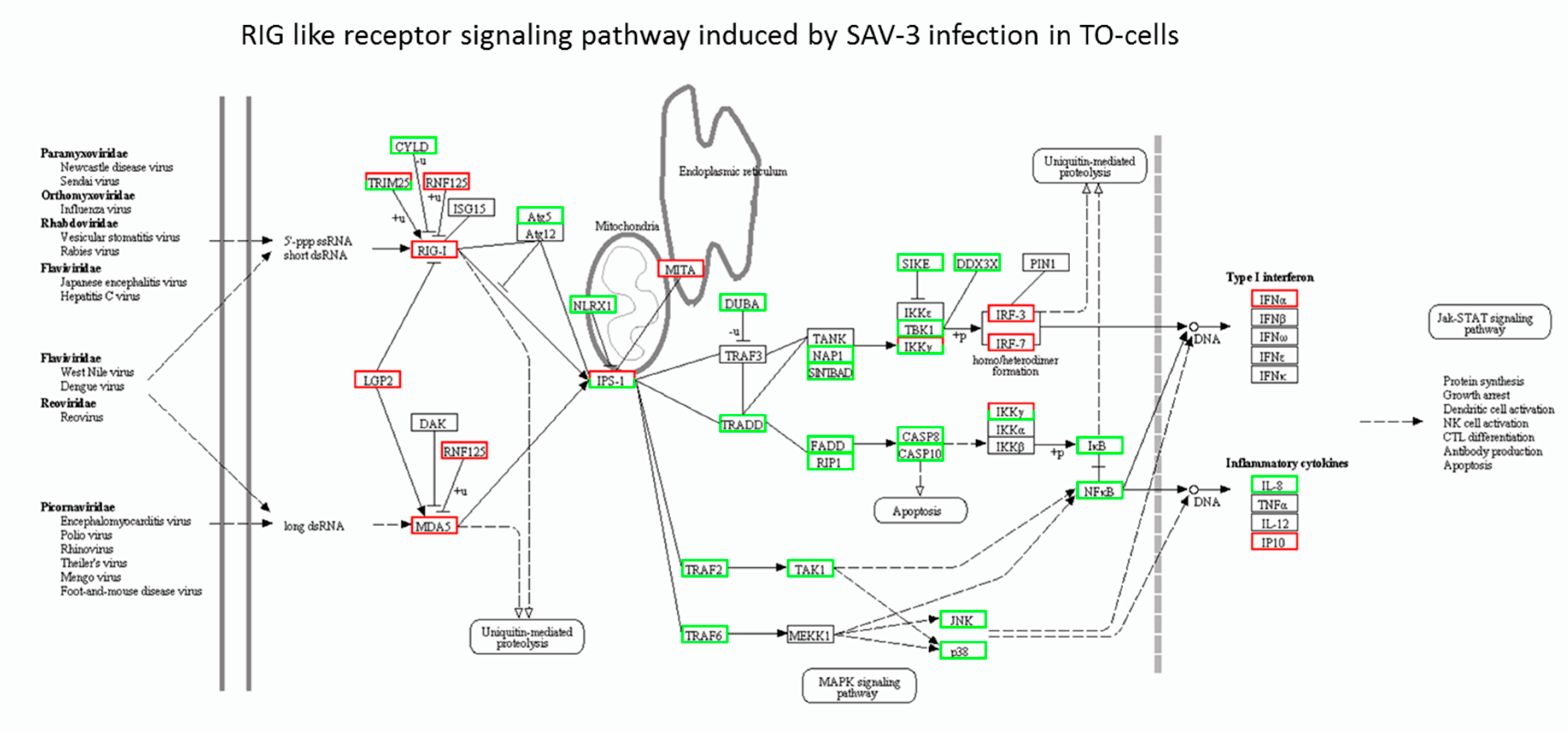
3.3. RIG-I-Like Receptor Signaling Pathway
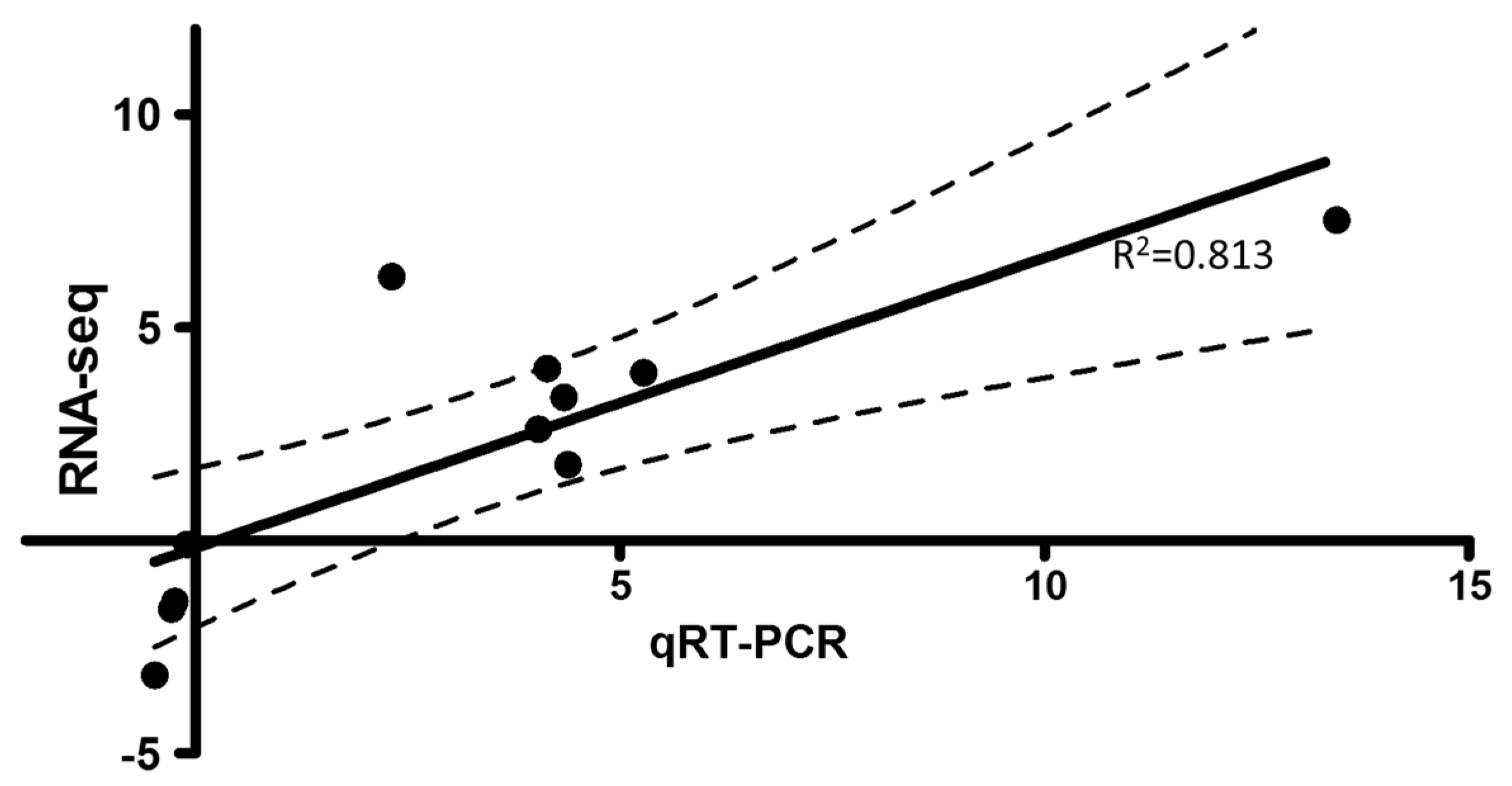
3.4. Quantitative Real-Time Polymerase Chain Reaction Test and Virus Quantification
4. Discussion
4.1. Transcriptome Signaling Pathway Analysis
4.2. Toll-Like Receptor Signaling Genes
4.3. RIG-I-Like Receptor Signaling Genes
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arancibia, S.A.; Beltran, C.J.; Aguirre, I.M.; Silva, P.; Peralta, A.L.; Malinarich, F.; Hermoso, M.A. Toll-like receptors are key participants in innate immune responses. Biol. Res. 2007, 40, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Munang’andu, H.M.; Evensen, Ø. A review of intra- and extracellular antigen delivery systems for virus vaccines of finfish. J. Immunol. Res. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; PrestonHurlburt, P.; Janeway, C.A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [PubMed]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Brunette, R.L.; Young, J.M.; Whitley, D.G.; Brodsky, I.E.; Malik, H.S.; Stetson, D.B. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 2012, 209, 1969–1983. [Google Scholar] [CrossRef] [PubMed]
- Germain, R.N. An innately interesting decade of research in immunology. Nat. Med. 2004, 10, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009, 458, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Shevach, E.M.; Trinchieri, G.; Mellor, A.L.; Munn, D.H.; Gordon, S.; Libby, P.; Hansson, G.K.; Shortman, K.; Dong, C.; et al. Highlights of 10 years of immunology in Nature Reviews Immunology. Nat. Rev. Immunol. 2011, 11, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Chtarbanova, S.; Imler, J.L. Microbial sensing by Toll receptors: A historical perspective. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Creagh, E.M.; O’Neill, L.A.J. TLRs, NLRs and RLRs: A trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006, 27, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y. Toll-like receptors in bony fish: From genomics to function. Dev. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Rebl, A.; Goldammer, T.; Seyfert, H.M. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 2010, 134, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Laing, K.J.; Purcell, M.K.; Winton, J.R.; Hansen, J.D. A genomic view of the NOD-like receptor family in teleost fish: Identification of a novel NLR subfamily in zebrafish. Bmc Evol. Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Collet, B.; Nie, P.; Lester, K.; Campbell, S.; Secombes, C.J.; Zou, J. Expression and functional characterization of the RIG-I-Like receptors MDA5 and LGP2 in rainbow trout (Oncorhynchus mykiss). J. Virol. 2011, 85, 8403–8412. [Google Scholar] [CrossRef] [PubMed]
- Iliev, D.B.; Sobhkhez, M.; Fremmerlid, K.; Jorgensen, J.B. MyD88 interacts with interferon regulatory factor (IRF) 3 and IRF7 in Atlantic Salmon (Salmo salar) transgenic SsMyD88 modulates the IRF-Induced type I interferon response and accumulates in aggresomes. J. Biol. Chem. 2011, 286, 42715–42724. [Google Scholar] [CrossRef] [PubMed]
- Skjaeveland, I.; Iliev, D.B.; Strandskog, G.; Jorgensen, J.B. Identification and characterization of TLR8 and MyD88 homologs in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 2009, 33, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Evensen, O.; Munang’andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and Salmonid alphavirus subtype-3 infection. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Ingerslev, H.C.; Stavang, V.; Egenberg, M.; Wergeland, H.I. A highly phagocytic cell line TO from Atlantic salmon is CD83 positive and M-CSFR negative, indicating a dendritic-like cell type. Fish Shellfish Immunol. 2008, 25, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Wergeland, H.I.; Jakobsen, R.A. A salmonid cell line (TO) for production of infectious salmon anaemia virus (ISAV). Dis. Aquat. Org. 2001, 44, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Munro, A.L.S.; Ellis, A.E.; Mcvicar, A.H.; Mclay, H.A.; Needham, E.A. An exocrine pancreas disease of farmed atlantic salmon in Scotland. Helgol. Meeresunters. 1984, 37, 571–586. [Google Scholar]
- Murphy, T.M.; Rodger, H.D.; Drinan, E.M.; Gannon, F.; Kruse, P.; Korting, W. The sequential pathology of pancreas disease in atlantic salmon farms in Ireland. J. Fish Dis. 1992, 15, 401–408. [Google Scholar] [CrossRef]
- Weston, J.H.; Welsh, M.D.; McLoughlin, M.F.; Todd, D. Salmon pancreas disease virus, an alphavirus infecting farmed Atlantic salmon, Salmo salar L. Virology 1999, 256, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Rheme, C.; Ehrengruber, M.U.; Grandgirard, D. Alphaviral cytotoxicity and its implication in vector development. Exp. Physiol. 2005, 90, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [PubMed]
- Bernard, M.A.; Han, X.B.; Inderbitzin, S.; Agbim, I.; Zhao, H.; Koziel, H.; Tachado, S.D. HIV-derived ssRNA binds to TLR8 to induce inflammation-driven macrophage foam cell formation. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Nikonov, A.; Molder, T.; Sikut, R.; Kiiver, K.; Mannik, A.; Toots, U.; Lulla, A.; Lulla, V.; Utt, A.; Merits, A.; et al. RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Hato, S.V.; Langereis, M.A.; Zoll, J.; Virgen-Slane, R.; Peisley, A.; Hur, S.; Semler, B.L.; van Rij, R.P.; van Kuppeveld, F.J.M. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012, 2, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, K.; Vakakis, E.; Kar, S.; Richer, E.; Evans, G.L.; Triantafilou, M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J. Cell Sci. 2012, 125 Pt 20, 4761–4769. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Howley, P.M. Fundamental Virology, 4th ed.; Lippincott Williams & Wilkins: New York, NY, USA, 2001. [Google Scholar]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Marjara, I.S.; Evensen, O. α interferon and not γ interferon inhibits salmonid alphavirus subtype 3 replication In Vitro. J. Virol. 2010, 84, 8903–8912. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.P.; Zhang, Y.; Song, J.; Zhao, L.J.; Wang, Z.Z. De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 2011, 98, 272–279. [Google Scholar]
- Conesa, M.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar]
- GEO Accession viewer – NCBI. Available online: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64095 (accessed on 18 April 2016).
- Munang’andu, H.M.; Fredriksen, B.N.; Mutoloki, S.; Dalmo, R.A.; Evensen, O. The kinetics of CD4+ and CD8+ T-cell gene expression correlate with protection in Atlantic salmon (Salmo salar L) vaccinated against infectious pancreatic necrosis. Vaccine 2013, 31, 1956–1963. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Evensen, O. Gene expression studies of host response to Salmonid alphavirus subtype 3 experimental infections in Atlantic salmon. Vet. Res. 2012, 43, 78. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.T.; Zou, J.; Holland, J.W.; Martin, S.A.; Kanellos, T.; Secombes, C.J. Identification and characterization of TLR7, TLR8a2, TLR8b1 and TLR8b2 genes in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 2013, 41, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Lomax, J. Get ready to GO! A biologist’s guide to the Gene Ontology. Brief. Bioinform. 2005, 6, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, D.R.; Li, G.G.; Wang, H.H.; Li, X.W.; Zhang, W.; Wu, Y.L.; Chen, L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J. Gastroenterol. 2015, 21, 6215–6228. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Sun, G. Identification of key genes and pathways in renal cell carcinoma through expression profiling data. Kidney Blood Press Res. 2015, 40, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differen. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.; Bowie, A.G. TLR3 in antiviral immunity: Key player or bystander? Trends Immunol. 2005, 26, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.F.; Wiens, G.D.; Purcell, M.K.; Palti, Y. Characterization of Toll-like receptor 3 gene in rainbow trout (Oncorhynchus mykiss). Immunogenetics 2005, 57, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Abid, A.; Davies, S.J.; Waines, P.; Emery, M.; Castex, M.; Gioacchini, G.; Carnevali, O.; Bickerdike, R.; Romero, J.; Merrifield, D.L. Dietary synbiotic application modulates Atlantic salmon (Salmo salar) intestinal microbial communities and intestinal immunity. Fish Shellfish Immunol. 2013, 35, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Munang’andu, H.M.; Mutoloki, S.; Evensen, O. A review of the immunological mechanisms following mucosal vaccination of finfish. Front. Immunol. 2015, 6, 427. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y.; Rodriguez, M.F.; Gahr, S.A.; Purcell, M.K.; Rexroad, C.E., III; Wiens, G.D. Identification, characterization and genetic mapping of TLR1 loci in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2010, 28, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef] [PubMed]
- Smerdou, C.; Liljestrom, P. Non-viral amplification systems for gene transfer: Vectors based on alphaviruses. Curr. Opin. Mol. Ther. 1999, 1, 244–251. [Google Scholar] [PubMed]
- Knudsen, M.L.; Johansson, D.X.; Kostic, L.; Nordstron, E.K.L.; Tegerstedt, K.; Pasetto, A.; Applequist, S.E.; Ljungberg, K.; Sirard, J.C.; Liljestrom, P. The adjuvant activity of alphavirus replicons is enhanced by incorporating the microbial molecule flagellin into the replicon. PLoS ONE 2013, 8, e65964. [Google Scholar] [CrossRef] [PubMed]
- Rayner, J.O.; Dryga, S.A.; Kamrud, K.I. Alphavirus vectors and vaccination. Rev. Med. Virol. 2002, 12, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Vander Veen, R.L.; Harris, D.L.; Kamrud, K.I. Alphavirus replicon vaccines. Anim. Health Res. Rev. 2012, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, J.L.; Weinerman, B.; Basole, C.; Salazar, J.C. TLR8: The forgotten relative revindicated. Cell. Mol. Immunol. 2012, 9, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Thomsen, A.R. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012, 86, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Renn, C.N.; Sanchez, D.J.; Ochoa, M.T.; Legaspi, A.J.; Oh, C.K.; Liu, P.T.; Krutzik, S.R.; Sieling, P.A.; Cheng, G.H.; Modlin, R.L. TLR activation of langerhans cell-like dendritic cells triggers an antiviral immune response. J. Immunol. 2006, 177, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Blasius, A.L.; Beutler, B. Intracellular Toll-like receptors. Immunity 2010, 32, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. TLR signaling pathways. Semin. Immunol. 2004, 16, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Uematsu, S.; Matsui, K.; Tsujimura, T.; Takeda, K.; Fujita, T.; Takeuchi, O.; et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2005, 23, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U. Mechanisms of RIG-I-Like receptor activation and manipulation by viral pathogens. J. Virol. 2014, 88, 5213–5216. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.W.; Amarasinghe, G.K. Structural insights into RNA recognition and activation of RIG-I-like receptors. Curr. Opin. Struct. Biol. 2012, 22, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Ramos, H.J.; Gale, M. RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr. Opin. Virol. 2011, 1, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Pichmair, A.; Gorna, M.W.; Superti-Furga, G.; Nagar, B. Structural basis for viral 5′-ppp-RNA recognition by human IFIT proteins. Nature 2013, 494, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Lassnig, C.; Eberle, C.A.; Gorna, M.W.; Baumann, C.L.; Burkard, T.R.; Burckstummer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.J.; Takeuchi, O.; Akira, S.; Chen, Z.J.; Inoue, S.S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Horner, S.M.; Liu, H.M.; Park, H.S.; Briley, J.; Gale, M. Mitochondrial-associated ER membranes form MAVS-anchored innate immune synapses that are targeted by hepatitis C virus. Cytokine 2011, 56, 65. [Google Scholar] [CrossRef]
- Hou, F.J.; Sun, L.J.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Peisley, A.; Jo, M.H.; Lin, C.; Wu, B.; Orme-Johnson, M.; Walz, T.; Hohng, S.; Hur, S. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. USA 2012, 109, E3340–E3349. [Google Scholar] [CrossRef] [PubMed]
- Skjesol, A.; Skjaeveland, I.; Elnaes, M.; Timmerhaus, G.; Fredriksen, B.N.; Jorgensen, S.M.; Krasnov, A.; Jorgensen, J.B. IPNV with high and low virulence: Host immune responses and viral mutations during infection. Virol. J. 2011, 8, 396. [Google Scholar] [CrossRef] [PubMed]
- Munang’andu, H.M.; Fredriksen, B.N.; Mutoloki, S.; Dalmo, R.A.; Evensen, O. Antigen dose and humoral immune response correspond with protection for inactivated infectious pancreatic necrosis virus vaccines in Atlantic salmon (Salmo salar L). Vet. Res. 2013, 44, 7. [Google Scholar] [CrossRef] [PubMed]
- Childs, K.S.; Randall, R.E.; Goodbourn, S. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.R.; Bruns, A.M.; Horvath, C.M. MDA5 and LGP2: Accomplices and antagonists of antiviral signal transduction. J. Virol. 2014, 88, 8194–8200. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Fornek, J.; Crochet, N.; Bajwa, G.; Perwitasari, O.; Martinez-Sobrido, L.; Akira, S.; Gill, M.A.; Garcia-Sastre, A.; Katze, M.G.; et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008, 82, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Hidmark, A.S.; McInerney, G.M.; Nordstrom, E.K.L.; Douagi, I.; Werner, K.M.; Liljestrom, P.; Hedestam, G.B.K. Early α/β interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88. J. Virol. 2005, 79, 10376–10385. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Sequence | GeneBank Accession No. |
|---|---|---|
| SAV-3 E2-F | CAGTGAAATTCGATAAGAAGTGCAA | EF675594 |
| SAV-3 E2-R | TGGGAGTCGCTGGTAAAGGT | |
| β-Actin-F | CCAGTCCTGCTCACTGAGGC | AF012125 |
| β-Actin-R | GGTCTCAAACATGATCTGGGTCA | |
| IP-10-F | TGCCAGAACATGGAGATCAT | EF619047 |
| IP-10-R | TTTACTGCACACTCCTTTGGTT | |
| TLR3-F | TTTGATGAGTCTCCGCCAACTCCA | KP231342 |
| TLR3-R | AATCTGCGAGGGACACAAAGGTCT | |
| TLR8-F | ACAAGAAAGAATGCCTCAATGTCA | NM_001161693 |
| TLR8-R | CACCCAGTCTGACACCAACA | |
| IRF3-F | TGGACCAATCAGGAGCGAAC | FJ517643 |
| IRF3-R | AGCCCACGCCTTGAAAATAA | |
| IRF7-F | GAGGAGTGGGCAGAGAACTA | NM_001171850 |
| IRF7-R | TTCTGGGAGACTGGCTGGG | |
| STAT1-F | CGGGCCCTGTCACTGTTC | GQ325309 |
| STAT1-R | GGCATACAGGGCTGTCTCT | |
| RIG-I-F | GACGGTCAGCAGGGTGTACT | NM_001163699 |
| RIG-I-R | CCCGTGTCCTAACGAACAGT | |
| MDA5-F | AGAGCCCGTCCAAAGTGAAGT | NM_001195179 |
| MDA5-R | GTTCAGCATAGTCAAAGGCAGGTA | |
| LGP2-F | GTGGCAGGCAATGGGGAATG | FN396358 |
| LGP2-R | CCTCCAGTGTAATAGCGTATCAATCC | |
| TOLLIP-F | ACCATTAGCACCCAACGAG | BT045489 |
| TOLLIP-R | TGGGAGTAATACGCAGGAAG | |
| RAC1-F | GACAGGAAGACTACGACAGAC | NM_001160673 |
| RAC1-R | TCAAAGGAGGCAGGACTCAC | |
| TRAF6-F | ACAGACTGTCCAAAGGCTC | – |
| TRAF6-R | TCATTGCGCTGCATCATC | |
| P38-F | TCCACGCCAAGAGAACCTAC | NM_001123715 |
| P38-R | ACATCATTGAACTCCTCCAGAC |
| Parameters | Toll Like Receptor | RIG-I-Like Receptor | NOD Like Receptor |
|---|---|---|---|
| Pathway ID | Ko04620 | Ko04622 | ko04621 |
| Pathway significance | 0.058 | 0.024 | 0.9101 |
| Pathway enrichment | 4.865157 × 10−1 | 3.011733 × 10−1 | 1.00000 × 100 |
| Total KO genes | 20115 | 20115 | 20115 |
| All genes with pathway annotation | 9315 | 9315 | 9315 |
| All genes in each pathway | 216 | 144 | 212 |
| DEGs | 112 | 79 | 89 |
| Gene Name | Abbr. | NCBI | Unig | KO | Reg | Log2 ratio | p-Value |
|---|---|---|---|---|---|---|---|
| Toll like receptor 3 | TLR3 | |DAA64469.1| | Unig9113 | K05401 | Up | 2.6140 | 7.4290 × 10−71 |
| Toll like receptor 8 | TLR8 | |NP_001155165.1| | Unig2363 | K10170 | Up | 4,0462 | 3.4201 × 10−5 |
| Signal transducer and activator of transcription 1 | STAT1 | |NP_001134757.1| | CL2066.2 | K11220 | Up | 6.27213 | 3.0554 × 10−68 |
| Interferon regulatory factor 3 | IRF3 | |ACL68544.1| | Unig4271 | K05411 | Up | 3.3644 | 5.3137 × 10−135 |
| Interferon regulatory factor 7 | IRF7 | |NP_001165321.1| | Unig10251 | K09447 | Up | 3.1970 | 1,13523 × 10−22 |
| Interferon α | IFN-a2 | |NP_001117042.1| | Unig5589 | K05414 | Up | 7.6042 | – |
| Interferon α receptor 1 | IFNAR1 | |NP_001268239.1| | Unig34816 | K05130 | Up | 1.8640 | 8.9299 × 10−66 |
| IFNγ induced protein 10 | IP-10 | |ACI69209.1| | Unig8163 | K12671 | Up | 7.5233 | 2.2267 × 10−112 |
| IFN-inducible T-cell α chemoattractant | I-TAC | |NM_0011412293.1| | Unig1740 | K12762 | Up | 9,55672 | – |
| Gene Name | Abbr. | NCBI | Unigene | KO | Reg | Log2 ratio | p-Value |
|---|---|---|---|---|---|---|---|
| Receptor interacting serine/threonine protein kinase 1 | RIP1 | |NP_001036815.1| | Unig17924 | K02861 | Down | −1.3056 | 2.0987 × 10−8 |
| Caspase 8 | CASP8 | |XP_001335163| | CL4461.1 | K04398 | Down | −1.1800 | 2.4337 × 10−142 |
| Toll like receptor 1 | TLR1 | |ACV92064.1| | Unig41380 | K05398 | Down | −4.2514 | 3.3298 × 10−5 |
| Toll like receptor 2 | TLR2 | |CCK73195.1| | Unig9045 | K10159 | Down | −16589 | 1.7562 × 10−41 |
| Transcription factor AP-1 | AP-1 | |XP_004369047.1| | CL3191.1 | K04448 | Down | −2.2025 | 3.9706 × 10−9 |
| Extracellular signal-regulated kinase | ERK | |BAD23843.1| | Unig24550 | K04371 | Down | −1.8016 | 1.5449 × 10−5 |
| NF-kappa-B inhibitor α | NFκBα | |ACI67986.1| | CL8473.1 | K04735 | Down | −1.3923 | 1.6765 × 10−13 |
| TANK-binding kinase 1 | TBK1 | |JF241943.1| | Unig5544 | K05410 | Down | −1.2619 | 1.0212 × 10−124 |
| TNF receptor associated factor 6 | TRAF6 | – | Unig40008 | K03175 | Down | −3.1583 | 3.552 × 10−4 |
| Interleukin 8 | IL-8 | |NP_001134182.1| | Unig7278 | K10030 | Down | −1.7368 | 6.5401 × 10−93 |
| Kinase 1-binding protein 1 | TAB1 | |XP_002662286.2| | Unig1972 | K04403 | Down | −1.5614 | 7.0199 × 10−66 |
| Kinase 1-binding protein 2 | TAB2 | |XP_003971436.1| | CL4395 | K04404 | Down | −2.07925 | 9.2399 × 10−28 |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase | PI3K | |XP_003455769.1| | CL120 | K02649 | Down | −1,9295 | 5,7289 × 10−39 |
| RAC-α serine/threonine-protein kinase (AkT) | AkT | |ACH70834.1| | CL5806 | K04456 | Down | −1.43478 | 2.9777 × 10−19 |
| Mitogen-activated protein kinase kinase 6 | MKK6 | |AAV52830| | Unig80 | K04433 | Down | −1.6590 | 7.5296 × 10−169 |
| Mitogen-activated protein kinase kinase 4 | MKK4 | |ACI33552.1| | CL292.2 | KO4430 | Down | −1.80397 | 1.9570 × 10−19 |
| p38b1 mitogen activated protein kinase | p38 | |EF123660.1| | Unig10574 | K04441 | Down | −1.6142 | 7.29346 × 10−9 |
| Gene Name | Abbr. | NCBI | Unigene | KO | Reg | Log2ratio | p-Value |
|---|---|---|---|---|---|---|---|
| Retinoic acid-inducible gene-I | RIG-I | |NP_001157171.1| | Unig7848 | K12646 | Up | 3.9377 | 3.171 × 10−102 |
| Melanoma differentiation associated gene 5 | MDA5 | |NP_001182108.1| | Unig6816 | K12647 | Up | 1.7788 | 1.8065 × 10−172 |
| Laboratory of genetics and physiology 2 | LPG2 | |NP_001133649.1| | CL8555 | K12649 | Up | 5.5128 | 0 |
| Interferon promoter stimulating protein 1 | IPS-1 | |NP_001161824.1| | Uni12389 | K12648 | Up | 1.5588 | 3.0551 × 10−18 |
| Tripartite motif-containing protein 25 | TRIM25 | |ACN11060.1| | CL8518.2 | K10652 | UP | 4.2080 | 1.8646 × 10−5 |
| IFNγ induced protein 10 | IP-10 | |ACI69209.1| | Unig8163 | K12671 | Up | 7.5233 | 2.2267 × 10−112 |
| Optineurin | Optn | |NP_001133761.1| | CL4866 | K07210 | Up | 1.9853 | 0 |
| Interferon regulatory factor 3 | IRF3 | |ACL68544.1| | Unig4271 | K05411 | Up | 3.3644 | 5.3137 × 10−135 |
| Interferon regulatory factor 7 | IRF7 | |NP_001165321.1| | Unig8533 | K09447 | Up | 11.055 | 1.1354 × 10−22 |
| Interferon a2 | IFN-a2 | |NP_001117042.1| | Unig5589 | K05414 | Up | 7.6042 | 0 |
| Gene Name | Abbr. | NCBI | Unigene | KO | Reg | Log2ratio | p-Value |
|---|---|---|---|---|---|---|---|
| NLR family member X1 | NLRX1 | |AFY26970.1| | Unig11078 | K12653 | Down | −1.6500 | 1.5422 × 10−58 |
| Autophagy protein 5 | Atg5 | |ACN11274.1| | Unig5014 | K08339 | Down | −1.3748 | 7.961 × 10−10 |
| Interleukin 8 | IL-8 | |ABA86669.1| | Unig7278 | K10030 | Down | −1.7368 | 6.5401 × 10−93 |
| Ubiquitin carboxyl-terminal hydrolase CYLD | CyLD | |XP_004068277.1| | Unig18460 | K08601 | Down | −1.9826 | 4.4252 × 10−11 |
| Suppressor of IKK-epsilon | SIKE | |ACI33887.1| | Unig21218 | K12656 | Down | −1.5344 | 2.5131 × 10−7 |
| TNF receptor type 1-associated death domain | TRADD | |Q1M161.1| | CL8304 | K03171 | Down | −1.6665 | 1.8945 × 10−20 |
| TGF-β-activated kinase 1 | TAK1 | |AAT07829.1| | CL7020.1 | K04427 | Down | −1.1643 | 3.2707 × 10−14 |
| Nuclear factor κ-B | NFκβ | |HM771267| | CL8473.1 | K04735 | Down | −1.3923 | 1.6765 × 10−13 |
| TANK-binding kinase 1 | TBK1 | |JF241943.1| | Unig5544 | K05410 | Down | −1.2619 | 1.0212 × 10−124 |
| TNF receptor-associated factor 2 | TRAF2 | |NP_001167255.1| | Unig16762 | K03173 | Down | −1.1445 | 1.0105 × 10−4 |
| NF-κ-B inhibitor | IκB | |CAC85086.1| | Unig31637 | K02581 | Down | −2.8369 | 1.9042 × 10−4 |
| Caspase 8 | CASP8 | |XP_001335163| | CL4461.1 | K04398 | Down | −1.1800 | 2.4337 × 10−142 |
| Caspase 10 | CASP10 | |CAE51933.1| | CL7349 | K04400 | Down | −1.2252 | 2.8250 × 10−23 |
| Receptor-interacting threonine-protein kinase 1 | RIPK1 | |NP_001036815.1| | Unig17924 | K02861 | Down | −1.3056 | 2.0987 × 10−8 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Evensen, Ø.; Munang’andu, H.M. De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells. Viruses 2016, 8, 114. https://doi.org/10.3390/v8040114
Xu C, Evensen Ø, Munang’andu HM. De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells. Viruses. 2016; 8(4):114. https://doi.org/10.3390/v8040114
Chicago/Turabian StyleXu, Cheng, Øystein Evensen, and Hetron Mweemba Munang’andu. 2016. "De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells" Viruses 8, no. 4: 114. https://doi.org/10.3390/v8040114
APA StyleXu, C., Evensen, Ø., & Munang’andu, H. M. (2016). De Novo Transcriptome Analysis Shows That SAV-3 Infection Upregulates Pattern Recognition Receptors of the Endosomal Toll-Like and RIG-I-Like Receptor Signaling Pathways in Macrophage/Dendritic Like TO-Cells. Viruses, 8(4), 114. https://doi.org/10.3390/v8040114






