Viroporins, Examples of the Two-Stage Membrane Protein Folding Model
Abstract
:1. Introduction
2. Membrane Architecture

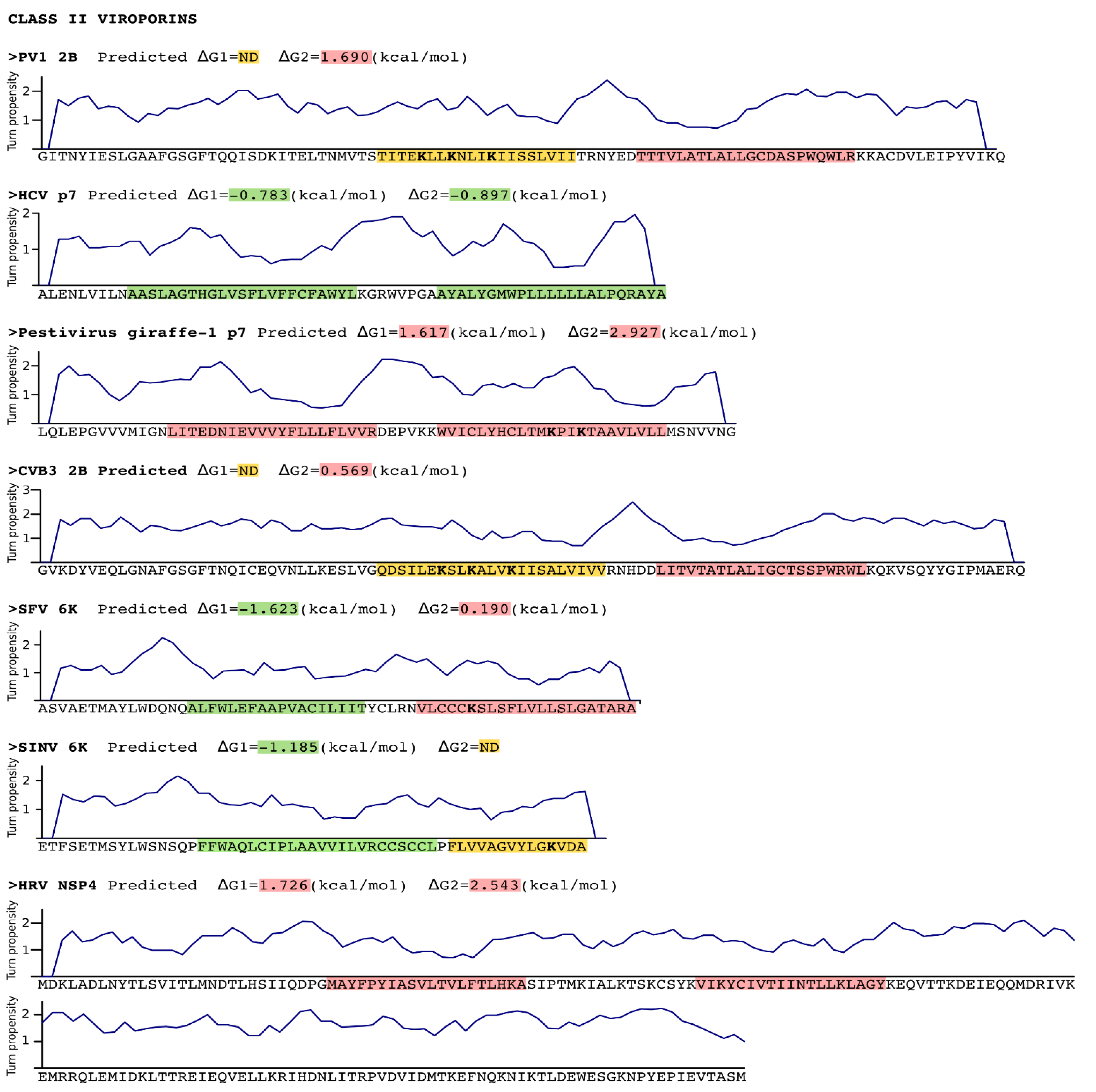
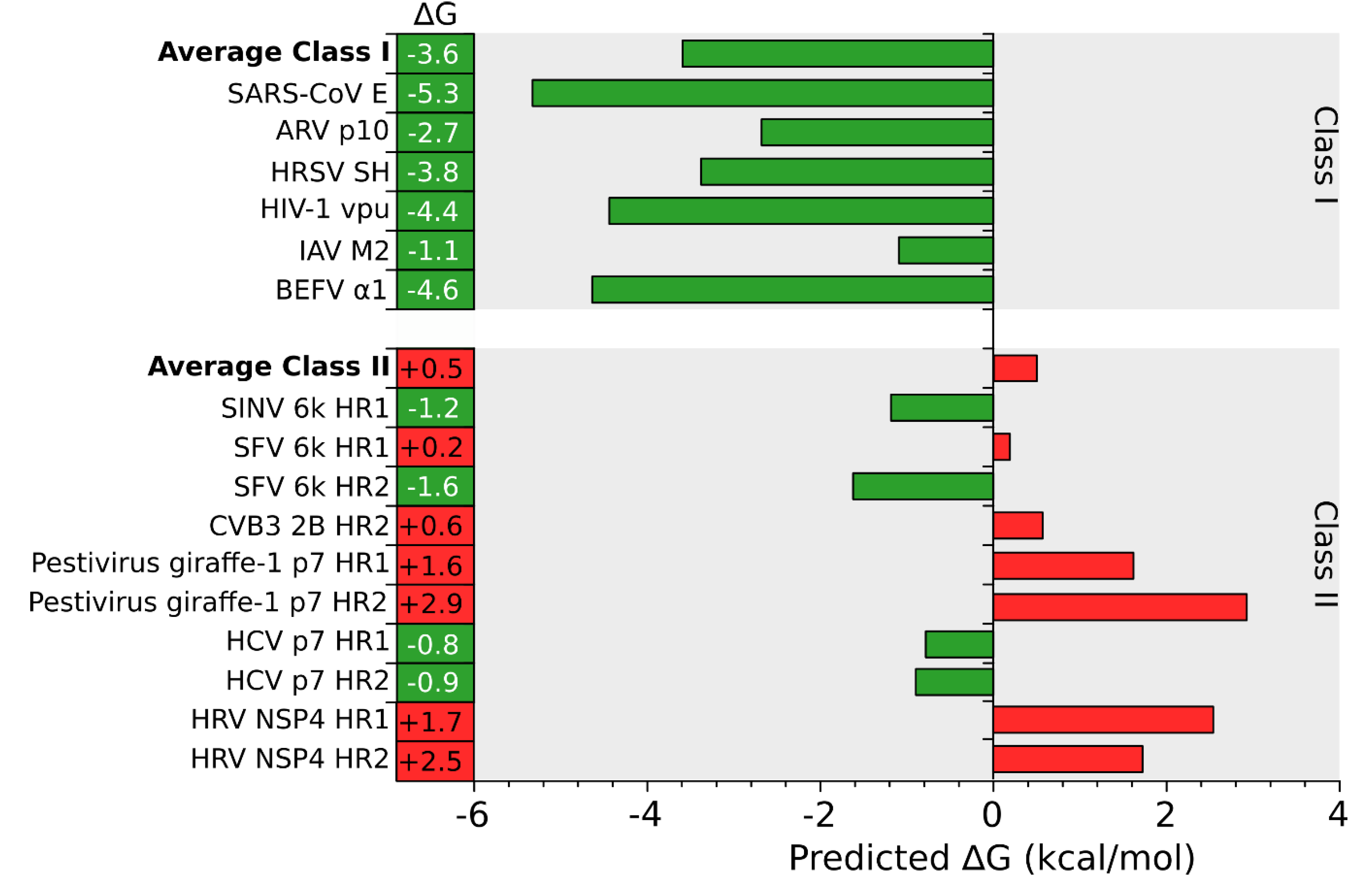
3. Stage I: Protein Insertion into the Membrane
3.1. Targeting the ER Membrane
3.2. Insertion into the ER Membrane
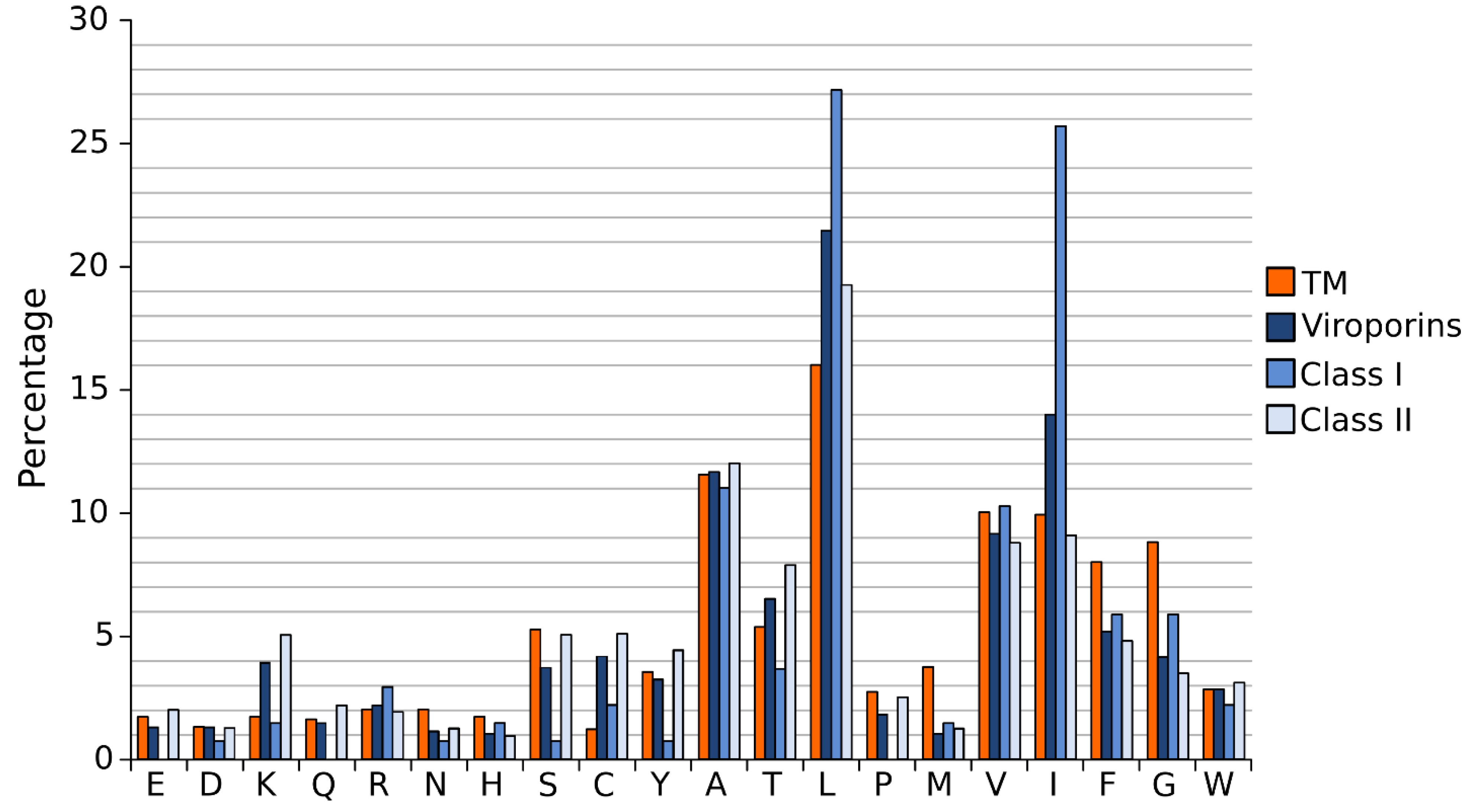
4. Stage II: Transmembrane Helix-Helix Interactions
5. Influenza Matrix Protein 2: Example of Quaternary Structure
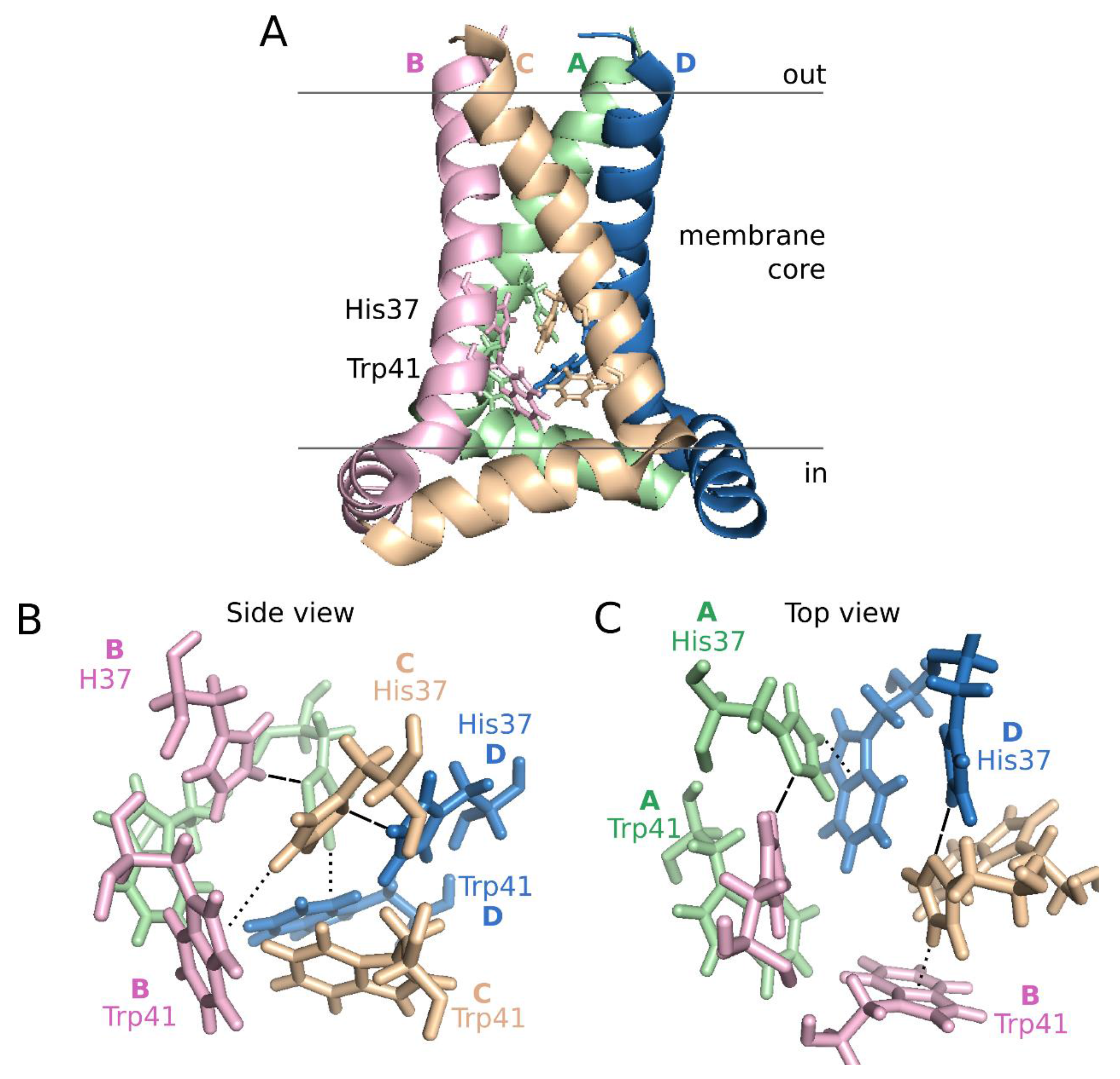
6. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carrasco, L. Membrane leakiness after viral infection and a new approach to the development of antiviral agents. Nature 1978, 272, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, L. Modification of membrane permeability by animal viruses. Adv. Virus Res. 1995, 45, 61–112. [Google Scholar] [PubMed]
- Griffin, S.D.C.; Beales, L.P.; Clarke, D.S.; Worsfold, O.; Evans, S.D.; Jaeger, J.; Harris, M.P.G.; Rowlands, D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003, 535, 34–38. [Google Scholar] [CrossRef]
- wart, G.D.; Mills, K.; Cox, G.B.; Gage, P.W. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 2002, 31, 26–35. [Google Scholar]
- Król, E.; Rychłowska, M.; Szewczyk, B. Antivirals—Current trends in fighting influenza. Acta Biochim. Pol. 2014, 61, 495–504. [Google Scholar] [PubMed]
- Moorthy, N.S.H.N.; Poongavanam, V.; Pratheepa, V. Viral M2 ion channel protein: A promising target for anti-influenza drug discovery. Mini Rev. Med. Chem. 2014, 14, 819–830. [Google Scholar] [PubMed]
- Watanabe, S.; Watanabe, T.; Kawaoka, Y. Influenza A virus lacking M2 protein as a live attenuated vaccine. J. Virol. 2009, 83, 5947–5950. [Google Scholar] [CrossRef] [PubMed]
- OuYang, B.; Chou, J.J. The minimalist architectures of viroporins and their therapeutic implications. Biochim. Biophys. Acta 2014, 1838, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.L.; Engelman, D.M. Membrane protein folding and oligomerization: The two-stage model. Biochemistry 1990, 29, 4031–4037. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.E.; Carrasco, L. Viroporins. FEBS Lett. 2003, 552, 28–34. [Google Scholar] [CrossRef]
- Yuan, X.; Li, J.; Shan, Y.; Yang, Z.; Zhao, Z.; Chen, B.; Yao, Z.; Dong, B.; Wang, S.; Chen, J.; Cong, Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005, 109, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Prediction of ΔG for TM helix insertion. Available online: http://dgpred.cbr.su.se/ (accessed on 30 April 2015).
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, J.; Andersson, H.; Nilsson, I.; White, S.H.; von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Hessa, T.; Meindl-Beinker, N.M.; Bernsel, A.; Kim, H.; Sato, Y.; Lerch-Bader, M.; Nilsson, I.; White, S.H.; von Heijne, G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 2007, 450, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, L.; Bañó-Polo, M.; Redondo, N.; Sánchez-Martínez, S.; Nieva, J.L.; Carrasco, L.; Mingarro, I. Membrane integration of poliovirus 2B viroporin. J. Virol. 2011, 85, 11315–11324. [Google Scholar] [CrossRef] [PubMed]
- Bañó-Polo, M.; Martínez-Gil, L.; Wallner, B.; Nieva, J.L.; Elofsson, A.; Mingarro, I. Charge pair interactions in transmembrane helices and turn propensity of the connecting sequence promote helical hairpin insertion. J. Mol. Biol. 2013, 425, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Baeza-Delgado, C.; Marti-Renom, M.A.; Mingarro, I. Structure-based statistical analysis of transmembrane helices. Eur. Biophys. J. 2013, 42, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Monné, M.; Nilsson, I.; Elofsson, A.; von Heijne, G. Turns in transmembrane helices: Determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J. Mol. Biol. 1999, 293, 807–814. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, D.M.; Bernardi, P.; Chieco-Bianchi, L.; Ciminale, V. Mitochondria as functional targets of proteins coded by human tumor viruses. Adv. Cancer Res. 2005, 94, 87–142. [Google Scholar] [PubMed]
- Madan, V.; Castelló, A.; Carrasco, L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 2008, 10, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.M.; Longen, S.; Weckbecker, D.; Depuydt, M. Biogenesis of mitochondrial proteins. Adv. Exp. Med. Biol. 2012, 748, 41–64. [Google Scholar] [PubMed]
- Saller, M.J.; Wu, Z.C.; de Keyzer, J.; Driessen, A.J.M. The YidC/Oxa1/Alb3 protein family: Common principles and distinct features. Biol. Chem. 2012, 393, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Dalbey, R.E. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 2011, 1808, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim. Biophys. Acta 1988, 947, 307–333. [Google Scholar] [CrossRef]
- Lakkaraju, A.K.K.; Mary, C.; Scherrer, A.; Johnson, A.E.; Strub, K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 2008, 133, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.J.; Chen, J.-C.; Miao, Y.; Shao, Y.; Lin, J.; Bock, P.E.; Johnson, A.E. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003, 278, 18628–18637. [Google Scholar] [CrossRef] [PubMed]
- Abell, B.M.; Pool, M.R.; Schlenker, O.; Sinning, I.; High, S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004, 23, 2755–2764. [Google Scholar] [CrossRef] [PubMed]
- Rabu, C.; Wipf, P.; Brodsky, J.L.; High, S. A Precursor-specific Role for Hsp40/Hsc70 during Tail-anchored Protein Integration at the Endoplasmic Reticulum. J. Biol. Chem. 2008, 283, 27504–27513. [Google Scholar] [CrossRef] [PubMed]
- Brambillasca, S.; Yabal, M.; Makarow, M.; Borgese, N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006, 175, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Hull, J.D.; Gilmore, R.; Lamb, R.A. Integration of a small integral membrane protein, M2, of influenza virus into the endoplasmic reticulum: Analysis of the internal signal-anchor domain of a protein with an ectoplasmic NH2 terminus. J. Cell Biol. 1988, 106, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Whitley, P.; Mingarro, I. Stitching proteins into membranes, not sew simple. Biol. Chem. 2014, 395, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Santolini, E.; Pacini, L.; Fipaldini, C.; Migliaccio, G.; Monica, N. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J. Virol. 1995, 69, 7461–7471. [Google Scholar] [PubMed]
- Alvisi, G.; Madan, V.; Bartenschlager, R. Hepatitis C virus and host cell lipids: An intimate connection. RNA Biol. 2011, 8, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Hijikata, M.; Kato, N.; Ootsuyama, Y.; Nakagawa, M.; Shimotohno, K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 1991, 88, 5547–5551. [Google Scholar] [CrossRef] [PubMed]
- Vieyres, G.; Dubuisson, J.; Pietschmann, T. Incorporation of hepatitis C virus E1 and E2 glycoproteins: The keystones on a peculiar virion. Viruses 2014, 6, 1149–1187. [Google Scholar] [CrossRef] [PubMed]
- SignalP 4.1 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 30 April 2015).
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Castelló, A.; Alvarez, E.; Carrasco, L. The multifaceted poliovirus 2A protease: Regulation of gene expression by picornavirus proteases. BioMed Res. Int. 2011, 2011. [Google Scholar] [CrossRef]
- Madan, V.; Sánchez-Martínez, S.; Vedovato, N.; Rispoli, G.; Carrasco, L.; Nieva, J.L. Plasma membrane-porating domain in poliovirus 2B protein. A short peptide mimics viroporin activity. J. Mol. Biol. 2007, 374, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Egea, P.F.; Stroud, R.M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 17182–17187. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, L.; Saurí, A.; Marti-Renom, M.A.; Mingarro, I. Membrane protein integration into the endoplasmic reticulum. FEBS J. 2011, 278, 3846–3858. [Google Scholar] [CrossRef] [PubMed]
- Tsukazaki, T.; Mori, H.; Fukai, S.; Ishitani, R.; Mori, T.; Dohmae, N.; Perederina, A.; Sugita, Y.; Vassylyev, D.G.; Ito, K.; et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 2008, 455, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Martoglio, B.; Hofmann, M.W.; Brunner, J.; Dobberstein, B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell 1995, 81, 207–214. [Google Scholar] [CrossRef]
- Wimley, W.C.; White, S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996, 3, 842–848. [Google Scholar] [CrossRef] [PubMed]
- Jaud, S.; Fernández-Vidal, M.; Nilsson, I.; Meindl-Beinker, N.M.; Hübner, N.C.; Tobias, D.J.; von Heijne, G.; White, S.H. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA 2009, 106, 11588–11593. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; Kim, H.; Nilsson, I.; White, S.H.; von Heijne, G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc. Natl. Acad. Sci. USA 2008, 105, 15702–15707. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Kida, Y.; Hagiwara, M.; Morimoto, F.; Sakaguchi, M. Positive charges of translocating polypeptide chain retrieve an upstream marginal hydrophobic segment from the endoplasmic reticulum lumen to the translocon. Mol. Biol. Cell 2010, 21, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, S.; Broecker, J.; Keller, S. Protein folding in membranes. Cell. Mol. Life Sci. 2010, 67, 1779–1798. [Google Scholar] [CrossRef] [PubMed]
- Mingarro, I.; Nilsson, I.; Whitley, P.; von Heijne, G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 2000, 1. [Google Scholar] [CrossRef] [PubMed]
- White, S.H.; Wimley, W.C. Membrane protein folding and stability: Physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Hedin, L.E.; Ojemalm, K.; Bernsel, A.; Hennerdal, A.; Illergård, K.; Enquist, K.; Kauko, A.; Cristobal, S.; von Heijne, G.; Lerch-Bader, M.; et al. Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J. Mol. Biol. 2010, 396, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Meindl-Beinker, N.M.; Lundin, C.; Nilsson, I.; White, S.H.; von Heijne, G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006, 7, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Nir Ben-Tal, D.S. Free energy of amide hydrogen bond formation in vacuum, in water, and in liquid alkane solution. J. Phys. Chem. B 1997, 101, 450–457. [Google Scholar] [CrossRef]
- Heinrich, S.U.; Mothes, W.; Brunner, J.; Rapoport, T.A. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 2000, 102, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Saurí, A.; Saksena, S.; Salgado, J.; Johnson, A.E.; Mingarro, I. Double-spanning plant viral movement protein integration into the endoplasmic reticulum membrane is signal recognition particle-dependent, translocon-mediated, and concerted. J. Biol. Chem. 2005, 280, 25907–25912. [Google Scholar] [CrossRef] [PubMed]
- Saurí, A.; McCormick, P.J.; Johnson, A.E.; Mingarro, I. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J. Mol. Biol. 2007, 366, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Sadlish, H.; Pitonzo, D.; Johnson, A.E.; Skach, W.R. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005, 12, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, S.U.; Rapoport, T.A. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 2003, 22, 3654–3663. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Sakaguchi, M.; Hamasaki, N.; Mihara, K. Membrane integration of the second transmembrane segment of band 3 requires a closely apposed preceding signal-anchor sequence. J. Biol. Chem. 2000, 275, 29743–29748. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Crawshaw, S.G.; Cross, B.C.S.; Haagsma, A.C.; High, S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem. J. 2008, 411, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gil, L.; Pérez-Gil, J.; Mingarro, I. The surfactant peptide KL4 sequence is inserted with a transmembrane orientation into the endoplasmic reticulum membrane. Biophys. J. 2008, 95, L36–L38. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Kini, R.M.; Yuen, R.; Khoo, H.E. Haemolytic activity of stonustoxin from stonefish (Synanceja horrida) venom: Pore formation and the role of cationic amino acid residues. Biochem. J. 1997, 325, 685–691. [Google Scholar] [PubMed]
- Park, C.B.; Lee, J.H.; Park, I.Y.; Kim, M.S.; Kim, S.C. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 1997, 411, 173–178. [Google Scholar] [CrossRef]
- Vogel, H.; Jähnig, F. The structure of melittin in membranes. Biophys. J. 1986, 50, 573–582. [Google Scholar] [CrossRef]
- Gilbert, R.J.C.; Serra, M.D.; Froelich, C.J.; Wallace, M.I.; Anderluh, G. Membrane pore formation at protein-lipid interfaces. Trends Biochem. Sci. 2014, 39, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Atoom, A.M.; Taylor, N.G.A.; Russell, R.S. The elusive function of the hepatitis C virus p7 protein. Virology 2014, 462–463, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, E.; Pietschmann, T. Hepatitis C virus p7-a viroporin crucial for virus assembly and an emerging target for antiviral therapy. Viruses 2010, 2, 2078–2095. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U. Membrane protein folding: How important are hydrogen bonds? Curr. Opin. Struct. Biol. 2011, 21, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Joh, N.H.; Oberai, A.; Yang, D.; Whitelegge, J.P.; Bowie, J.U. Similar energetic contributions of packing in the core of membrane and water-soluble proteins. J. Am. Chem. Soc. 2009, 131, 10846–10847. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, J.M.; Bond, P.J.; Sansom, M.S.P. Transmembrane helix-helix interactions: Comparative simulations of the glycophorin a dimer. Biochemistry 2006, 45, 14298–14310. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.K.; Subramaniam, S.; Senes, A. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα-H hydrogen bonds. Proc. Natl. Acad. Sci. USA 2014, 111, E888–E895. [Google Scholar] [CrossRef] [PubMed]
- Bañó-Polo, M.; Baeza-Delgado, C.; Orzáez, M.; Marti-Renom, M.A.; Abad, C.; Mingarro, I. Polar/Ionizable residues in transmembrane segments: Effects on helix-helix packing. PLoS ONE 2012, 7, e44263. [Google Scholar] [CrossRef] [PubMed]
- Hong, H. Toward understanding driving forces in membrane protein folding. Arch. Biochem. Biophys. 2014, 564C, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Cross, T.A. Modeling the membrane environment has implications for membrane protein structure and function: Influenza a M2 protein. Protein Sci. Publ. Protein Soc. 2013, 22, 381–394. [Google Scholar] [CrossRef] [PubMed]
- DeGrado, W.F.; Gratkowski, H.; Lear, J.D. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. Publ. Protein Soc. 2003, 12, 647–665. [Google Scholar] [CrossRef] [PubMed]
- Largo, E.; Gladue, D.P.; Huarte, N.; Borca, M.V.; Nieva, J.L. Pore-forming activity of pestivirus p7 in a minimal model system supports genus-specific viroporin function. Antivir. Res. 2014, 101, 30–36. [Google Scholar] [CrossRef] [PubMed]
- OuYang, B.; Xie, S.; Berardi, M.J.; Zhao, X.; Dev, J.; Yu, W.; Sun, B.; Chou, J.J. Unusual architecture of the p7 channel from hepatitis C virus. Nature 2013, 498, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Fischer, W.B. Assembly of viral membrane proteins. J. Chem. Theory Comput. 2009, 5, 2503–2513. [Google Scholar] [CrossRef]
- Li, L.-H.; Hsu, H.-J.; Fischer, W.B. Assembling viral channel forming proteins: Vpu from HIV-1. Biopolymers 2013, 99, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, N.M.; Palese, P. The Biology of influenza viruses. Vaccine 2008, 26, D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, R.J.; Bahadur, G.; Zambon, M.C.; Hall-Smith, M.; Douglas, A.R.; Hay, A.J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990, 9, 3469–3476. [Google Scholar] [PubMed]
- Schmidt, N.W.; Mishra, A.; Wang, J.; DeGrado, W.F.; Wong, G.C.L. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J. Am. Chem. Soc. 2013, 135, 13710–13719. [Google Scholar] [CrossRef] [PubMed]
- Park, E.K.; Castrucci, M.R.; Portner, A.; Kawaoka, Y. The M2 ectodomain is important for its incorporation into influenza a virions. J. Virol. 1998, 72, 2449–2455. [Google Scholar] [PubMed]
- Wang, J.; Qiu, J.X.; Soto, C.; DeGrado, W.F. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza a virus. Curr. Opin. Struct. Biol. 2011, 21, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Leser, G.P.; Jackson, D.; Lamb, R.A. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 2008, 82, 10059–10070. [Google Scholar] [CrossRef] [PubMed]
- Kochendoerfer, G.G.; Salom, D.; Lear, J.D.; Wilk-Orescan, R.; Kent, S.B.H.; DeGrado, W.F. Total chemical synthesis of the integral membrane protein influenza A Virus M2: Role of its C-terminal domain in tetramer assembly. Biochemistry 1999, 38, 11905–11913. [Google Scholar] [CrossRef] [PubMed]
- Duff, K.C.; Ashley, R.H. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology 1992, 190, 485–489. [Google Scholar] [CrossRef]
- Ma, C.; Polishchuk, A.L.; Ohigashi, Y.; Stouffer, A.L.; Schön, A.; Magavern, E.; Jing, X.; Lear, J.D.; Freire, E.; Lamb, R.A.; et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. USA 2009, 106, 12283–12288. [Google Scholar] [CrossRef] [PubMed]
- Acharya, R.; Carnevale, V.; Fiorin, G.; Levine, B.G.; Polishchuk, A.L.; Balannik, V.; Samish, I.; Lamb, R.A.; Pinto, L.H.; DeGrado, W.F.; et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza a virus. Proc. Natl. Acad. Sci. USA 2010, 107, 15075–15080. [Google Scholar] [CrossRef] [PubMed]
- Cady, S.D.; Schmidt-Rohr, K.; Wang, J.; Soto, C.S.; Degrado, W.F.; Hong, M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Asbury, T.; Achuthan, S.; Li, C.; Bertram, R.; Quine, J.R.; Fu, R.; Cross, T.A. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza a virus. Biophys. J. 2007, 92, 4335–4343. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza a virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Gao, P.F.; Pinto, L.H.; Lamb, R.A.; Cross, T.A. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. Publ. Protein Soc. 2003, 12, 2597–2605. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Yi, M.; Dong, H.; Qin, H.; Peterson, E.; Busath, D.D.; Zhou, H.-X.; Cross, T.A. Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer. Science 2010, 330, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Kawano, K.; Yano, Y.; Matsuzaki, K. A dimer is the minimal proton-conducting unit of the influenza a virus M2 channel. J. Mol. Biol. 2014, 426, 2679–2691. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Gil, L.; Mingarro, I. Viroporins, Examples of the Two-Stage Membrane Protein Folding Model. Viruses 2015, 7, 3462-3482. https://doi.org/10.3390/v7072781
Martinez-Gil L, Mingarro I. Viroporins, Examples of the Two-Stage Membrane Protein Folding Model. Viruses. 2015; 7(7):3462-3482. https://doi.org/10.3390/v7072781
Chicago/Turabian StyleMartinez-Gil, Luis, and Ismael Mingarro. 2015. "Viroporins, Examples of the Two-Stage Membrane Protein Folding Model" Viruses 7, no. 7: 3462-3482. https://doi.org/10.3390/v7072781
APA StyleMartinez-Gil, L., & Mingarro, I. (2015). Viroporins, Examples of the Two-Stage Membrane Protein Folding Model. Viruses, 7(7), 3462-3482. https://doi.org/10.3390/v7072781




