Aptamers in Diagnostics and Treatment of Viral Infections
Abstract
:1. Introduction
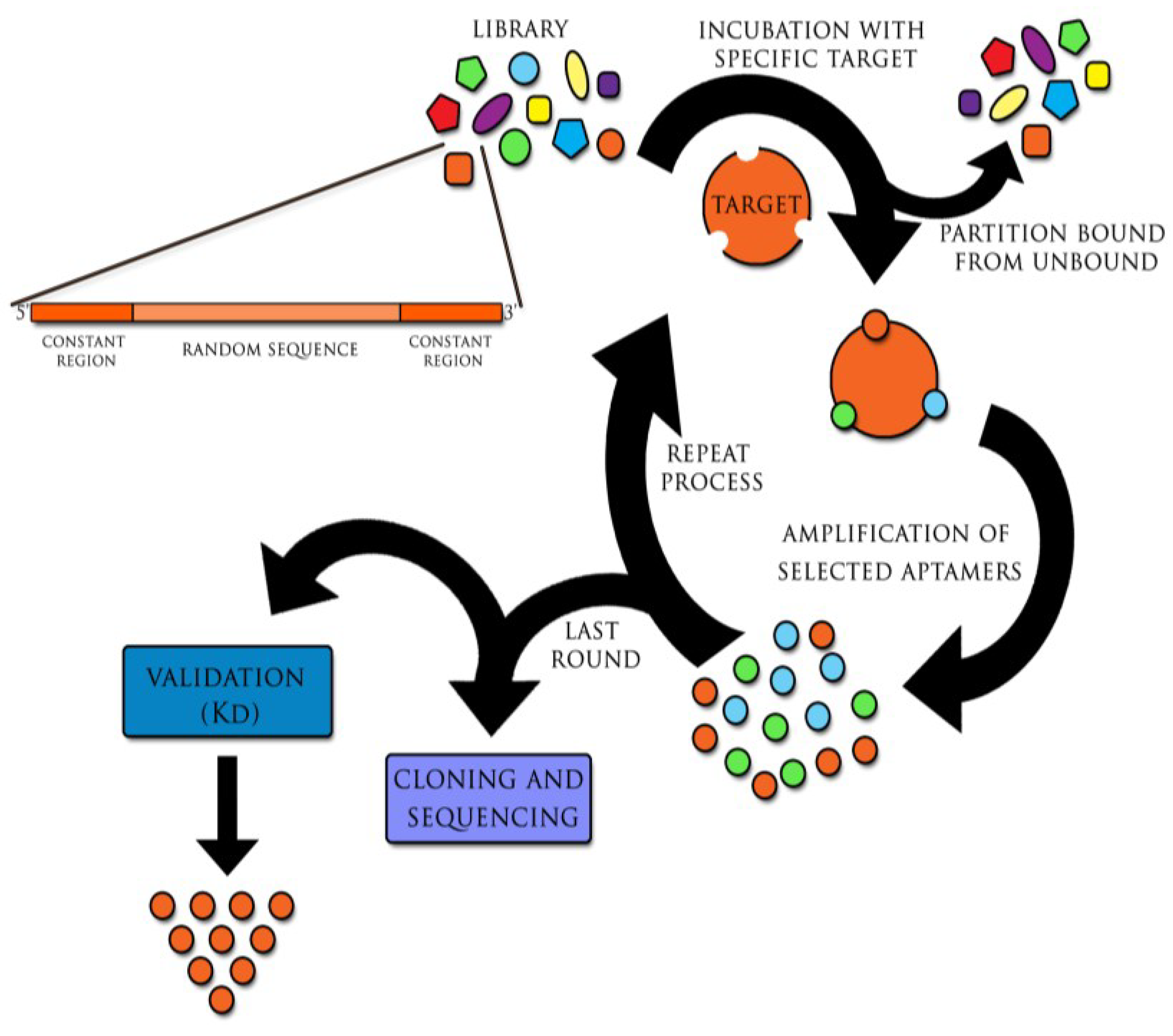
2. Aptamer in the Diagnostics of Viral Infections
2.1. Experimental Diagnostic Studies with Aptamers and Aptamer-Based Biosensors Conducted on Laboratory Model Samples
| Virus | Aptamer Name | Type | Target | Binding Affinity (Kd) | Detection Technique | Limit of Detection | Refs. |
|---|---|---|---|---|---|---|---|
| Influenza H5N1 | RHA0006 RHA0385 | DNA | Hemagglutinin | 15.3 nM 24.7 nM | sandwich enzyme linked aptamer assay (ELASA) | 0.1 µg/well | [37] |
| n/d | surface protein | 4.65 nM | QCM-based biosensor coated with the hydrogel | 0.0128 HAU | [36] | ||
| HIV-1 | n/d | RNA | Tat protein | 1nM | FET-based biosensor | 1.2 × 109 molecules | [23] |
| n/d | QCM-based biosensor SPR-based biosensor | 0.25ppm | [24] | ||||
| HCV | E2-B E2-D | DNA | E2 glycoprotein | 4 nM 0.8 nM | enzyme linked apto-sorbent assay (ELASA) | 3.13–6.25 × 102 FFU/mL, 16 ng/mL of glycoprotein E2 | [38] |
| Vaccinia | n/d | DNA | vaccinia particles | 25 nM | AptaVISens-V aptamer-based viability impedimetric sensor | 330 PFU | [39] |
| PP3 | Hemagglutinin | 3.24 nM | fluorescence microscope using Alexa Fluor 594-labeled aptamer PP3 | n/d | [41] | ||
| TV01 | surface protein | 7.3 nM | flow cytometry assay using Cy5-labeled aptamer TV01 | [40] | |||
| HPV | 13 14 20 28 | DNA | epitopes on cell surface proteins of non-infected cells | 2.5 nM 7.1 nM 1.6 nM 6.9 nM | confocal microscope | n/d | [42] |
| G5α3N.4 | RNA | oncoprotein E7 | 1.9 µM | EMSA assay | [27] | ||
| Chikungunya, Dengue, West Nile | spectrum of selected aptamers | DNA | viral envelope proteins | spectrum of data | lateral flow chromatographic test strip fluorescent aptamer-magnetic bead sandwich assay | n/d | [29] |
| Dengue | apt_EcoRI | n/d | EcoRI enzyme—one of biosensor modules | n/d | modular biosensor detecting the genetic sequences of Dengue genome | n/d | [21] |
2.2. Experimental Diagnostic Studies with Aptamer-Based Biosensors Conducted on Natural or Clinical-Based Samples
| Virus | Aptamer Name | Type | Target | Binding Affinity (Kd) | Detection Technique | Limit of Detection | Sample Type | Refs. |
|---|---|---|---|---|---|---|---|---|
| Influenza H5N1 | n/d | DNA | hemagglutinin | 4.65 nM | Spreeta SPR sensing chip | 0.128 HAU | poultry swab samples | [19] |
| HCV | ZE2 | DNA | glycoprotein E2 | 1.05 nM | sandwich ELISA | n/d | HCV infected patients’ sera | [26] |
| 9-14 9-15 | RNA | core antigen | 142 nM 224 nM | sol-gel chip-based fluorescence assay | [43] |
2.3. Advantages and Disadvantages of Aptamer-Based Tests in Comparison to Other Diagnostics Methods
| Virus | Method | Detection limit | Advantages | Disadvantages | Refs. |
|---|---|---|---|---|---|
| Influenza | isolation and identification of the virus | 1 EID50/mL | sensitivity | time consuming | [47] |
| ELISA | 1.0 ng | rapid | high rate of false positive results | [48] | |
| RT-PCR | 0.0256 HAU | specificity sensitivity | expensive complicated, highly skilled stuff | [49] | |
| qRT-PCR | 10 copies /reaction | [50] | |||
| HBV | ELISA | 0.5 pg/mL | as presented above | [51] | |
| HIV | ELISA | 0.9–1.2 IU/mL | [53] | ||
| Method | Virus Isolation | ELISA | RT-PCR | qRT-PCR | SPR Aptasensor |
|---|---|---|---|---|---|
| detection time | 120–170 h | 3 h | 5 h | 3 h | 1.5 h |
2.4. Future Perspectives of Aptamers in Diagnostic Procedures
3. Aptamers in the Treatment of Viral Infections
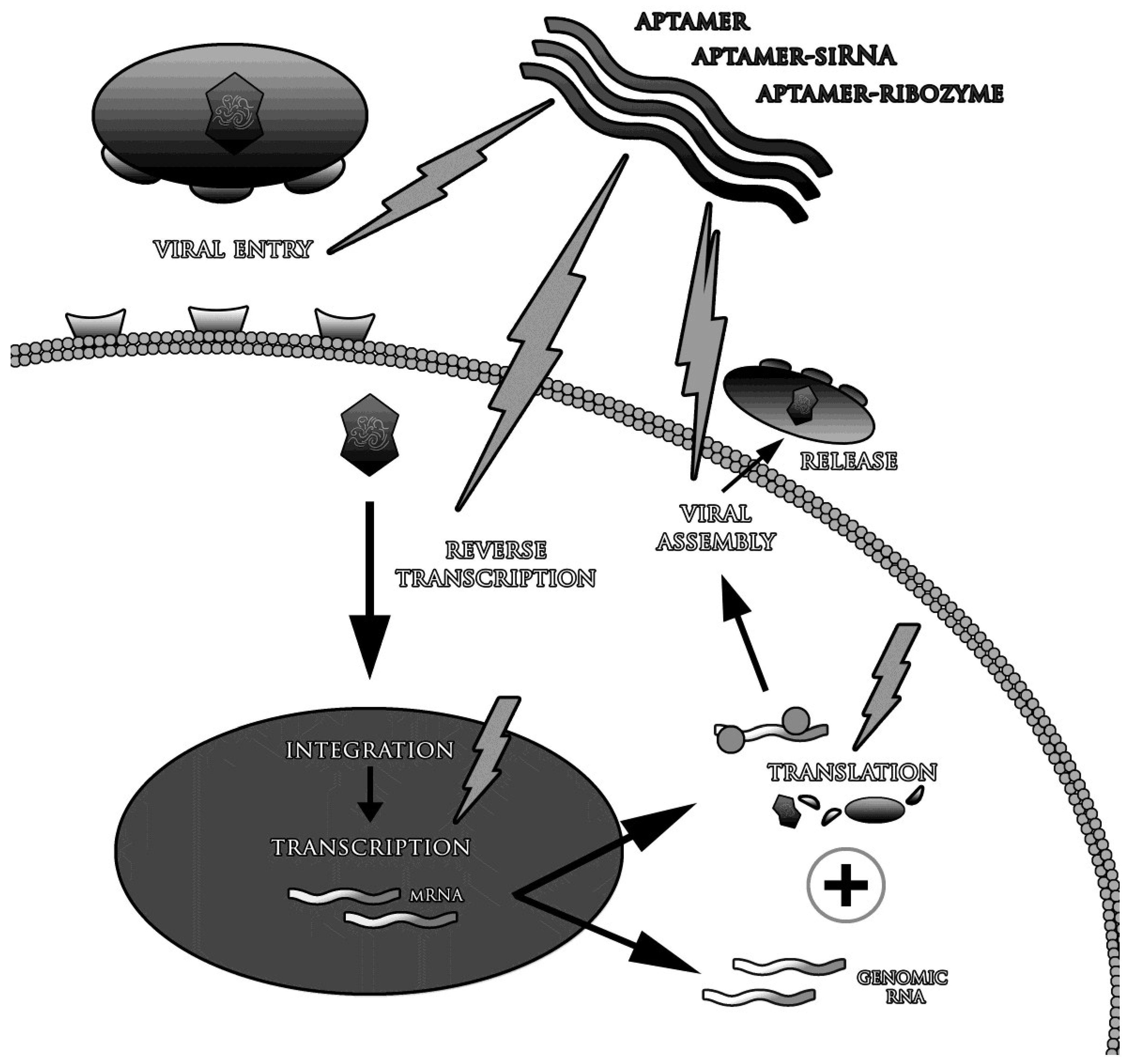
| Virus | Aptamer Name | Type | Target | Aptamer Application Method | Modification Enhancing Biostability | Inhibitory Effect | Kd/IC50 | Refs. |
|---|---|---|---|---|---|---|---|---|
| Influenza H5N1 | A22 | DNA | HA | BALB/c mice were intranasally inoculated with the A22 solution | --- | >90% decrease in viral loads in mice lungs | n/d | [80] |
| Influenza H9N2 | C7-35M | DNA | HA | MDCK-infected culture cells incubated with aptamer | --- | inhibition of viral infection in an aptamer-dose dependent manner (1000 pmole inhibits the viral infection by 55%) | n/d | [81] |
| Influenza H3N2 | HA12-16 | RNA | gHA1 | MDCK-infected culture cells incubated with aptamer | none | efficient suppression of viral infection of the cells | n/d | [82] |
| HIV-1 | B40, B40t77 | RNA | gp120-CCR5 | PBMC culture cells incubated with aptamer before infection | 2'-fluoro modification | inhibition of viral infectivity (50% at 2 nM) | Kd B40 = 21 ± 2 nM Kd B40t77 = 31 ± 2 nM IC50 = 2 nM | [79] |
| B40t77 iii_4 | gp120-CCR5 | PBMCs and blood monocyte-derived macrophages (BDMs)- infected cultures incubated with aptamer | inverted thymidine at the 3'-end; dimethoxyltrityloxy-(CH2)6-SS-(CH2)6-phospho linker at the 5'-end | inhibition of viral infectivity by 85% | n/d | [83] | ||
| 37 NT | HIV-RT | aptamer added to HIV-RT in vitro reaction | three 5'-nt and three loop-nt replaced by phosphothionucleosides | reaction rate decreased (100% by 50 nM of aptamer) | Kd = 0.66 nM IC50 = 2.5 nM | [84] | ||
| DP6-12 | Gag protein | 293T cells transfected with plasmid encoding aptamer | --- | 20-fold inhibition of virus production | Kd = 130 ± 9 nM | [85] | ||
| Ch A-1 (anti-gp120 aptamer-siRNA chimera) | gp120 (aptamer) tat/rev (siRNA) | RAG-Hu mice were injected with the chimera solution | 2'-fluoro modification | reduction in tat/rev mRNA transcript level in mice T lymphocytes between 75% and 90% | n/d | [86] | ||
| anti-gp120 aptamer- siRNA chmiera | gp120 (aptamer) tat/rev, CD4, transportin-3 (siRNA) | RAG-Hu mice were injected intravenously with chimera solution | 2'-fluoro modification | significant decrease in viral loads level; stable level of CD4 T lymphocytes | n/d | [87] | ||
| CD4-AsiCs | CD4 (aptamer) gag/vif CCR5 (siRNA) | NSG-BLT mice were administrated intravaginaly with aptamer | none | protection against HIV vaginal transmission | n/d | [88] | ||
| HCV | ODN 27v | DNA | NSB5 | Huh7- JHF1 strain infected cells incubated with aptamer; aptamer enter cells without transfection reagent | none | reduction in virus mRNA levels (90% reduction at aptamer concentrations of 5 µM) | Kd = 132.2 ± 20 nM IC50 = 196 ± 16 nM | [74] |
| B.2 | RNA | aptamer added to HCV-NS5B in vitro reaction | --- | inhibition of NS5B polymerase activity | Kd = 1.5 ± 0.2 nM IC50 = 10 ± 0.5 nM; | [89] | ||
| NEO-35-s41 G925-s50 | NS3 | aptamer added to HCV-NS3 protease cleavage and helicase unwinding in vitro reactions | --- | Inhibition of NS3 helicase and protease activity | protease/helicase NEO-35-s41 IC50 = 0.2 µM/20 nM G925-s50 IC50 = 0.2 µM/15 nM | [90] | ||
| NEO-III-14U | HeLa-NS3-expressing cells were transfected with aptamer | --- | protease activity inhibited in 60% | Kd = 4 nM | [91] | |||
| AP30 | (-)IRES domain I | aptamer preincubated with template and added to NS5B in vitro reaction | --- | genetic material replication inhibited by 50% | Kd = 36 nM | [92] | ||
| HCMV | L13 | RNA | glycoprotein B | virus particles preincubated with aptamer used to infect HFF cells | 2'-amino-modified pyrimidines | infectivity reduction | IC50 = 125 ± 20 nM | [93] |
| L19 | glycoprotein H | 100-fold reduction in viral yield blockade of viral entry | IC50 = 35 ± 7 nM | |||||
| HSV | Aptamer-1 | RNA | glycoprotein D | virus particles preincubated with aptamer used to infect VERO cells | 2'-fluoro modification | blockade of viral entry | Kd = 109 nM IC50 = 0.8 µM | [75] |
| HBV | S9 | RNA | P protein | HepG2.2.15 cells trasfected with plasmid encoding aptamer | --- | reduction of replicative intermediates by about 80%–85% | n/d | [94] |
| SCV | ES15 | RNA | NsP10 | aptamer added to SCV helicase unwinding in vitro reaction | --- | helicase unwinding activity inhibited in 85% | IC50 = 1.2 nM | [95] |
| Ebola | 1G8-14 2F11-14 | RNA | eVP35 IID | n/d | --- | inhibition of EBOV polymerase activity and VP36-nucleoprotein interaction | Kd = 30-50 nM | [96] |
3.1. Blocking of Viral Fusion with the Target Cell
3.2. Inhibition of Proteins and Enzymes of Viral Replication Cycle
3.2.1. Blocking of Viral Enzymes with Polymerase Activity
3.2.2. Blocking the Activity of Other Enzymes Involved in Viral Replication
3.2.3. Blocking the Nucleocapsid Protein of HIV-1
3.3. Inhibition of Nucleic Acid Sequences Essential for Virus Replication Cycle
3.4. Delivery of Therapeutic Molecules to Cells Infected with Viruses
3.5. Other Strategies
4. Aptamers against Ebola Infection
5. Aptamers—“For” and “against”
5.1. The Target Site of Action—Does it Matter? Aptamer vs. siRNA
5.2. Aptamer’s Stability under Physiological Conditions
5.3. Renal Clearance
5.4. Toxicity
6. Conclusions
7. Executive Summary
7.1. Introduction
- Aptamers are single strand nucleic acid molecules, consisted of DNA or RNA, which bind to organic or nonorganic molecules with high specificity and affinity.
- Aptamers are generated in the method referred to as Systematic Evolution of Ligands by Exponential Enrichment (SELEX).
- Properties of aptamers make them competitive to monoclonal antibodies used in conventional laboratory practice.
- The first pharmaceutical aptamer, Macugen (pegaptanib sodium) has been admitted by US Agency for Food and Drug Administration (FDA) for the treatment of Age-Related Macular Degeneration (AMD) in 2004.
7.2. Aptamers in the Diagnostics of Viral Infections
- The success of treatment in viral diseases depends on the early detection of the infective agent.
- Aptamers allow for detection of both early (viral genes and proteins), and late (antibodies produced by the host) infection markers.
- There are strategies enabling differentiation between infected host cells and uninfected ones.
- Aptamers can differentiate active and inactive virus forms.
7.3. Aptamers in the Viral Infections Treatment
- Aptamers are promising solution in viral diseases, if presently used drugs and vaccines are not effective enough. Aptamers can target any element of the virus-infected host cell complex.
- Possible strategies of aptamer application in the treatment of viral diseases include:
- o
- blockade of the virion penetration into the cells;
- o
- inhibition of enzymes responsible for viral replication and other crucial processes;
- o
- conjugation and delivery of therapeutic molecules to virus-infected cells;
- o
- prevention of infection; and
- o
- selective activation of the immune system.
Author Contributions
Conflicts of Interest
References
- Zhou, J.; Bobbin, M.L.; Burnett, J.C.; Rossi, J.J. Current progress of RNA aptamer-based therapeutics. Front. Genet. 2012, 3, e234. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Pagratis, N.C.; Bell, C.; Chang, Y.F.; Jennings, S.; Fitzwater, T.; Jellinek, D.; Dang, C. Potent 2'-amino-, and 2'-fluoro-2'-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997, 15, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Fennewald, S.; Luxon, B.A.; Aronson, J.; Herzog, N.K.; Gorenstein, D.G. Aptamers containing thymidine 3'-Ophosphorodithioates: Synthesis and binding to nuclear factor-kappaB. Bioorg. Med. Chem. Lett. 1999, 9, 3357–3362. [Google Scholar] [CrossRef] [PubMed]
- Kusser, W. Chemically modified nucleic acid aptamers for in vitro selections: Evolving evolution. J. Biotechnol. 2000, 74, 27–38. [Google Scholar] [PubMed]
- Eulberg, D.; Klussmann, S. Spiegelmers: Biostable aptamers. ChemBioChem 2003, 4, 979–983. [Google Scholar] [CrossRef] [PubMed]
- Mendosa, S.D.; Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004, 126, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.; Krylov, S.N. Using nonequilibrium capillary electrophoresis of equilibrium mixtures for the determination of temperature in capillary electrophoresis. Anal. Chem. 2004, 6, 7114–7117. [Google Scholar] [CrossRef]
- Misono, T.S.; Kumar, P.K. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal. Biochem. 2005, 342, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, P.E.; Lewis, S.D.; Silva, R.F.; Preiss, J.R.; Horwitz, L.R.; Pendergrast, P.S.; McCauley, T.G.; Kurz, J.C.; Epstein, D.M.; Wilson, C. Direct in vitro selection of a 2'-O-methyl aptamer to VEGF. Chem. Biol. 2005, 12, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Somasunderam, A.; Ferguson, M.R.; Rojo, D.R.; Thiviyanathan, V.; Li, X.; O’Brien, W.A.; Gorenstein, D.G. Combinatorial selection, inhibition, and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 2005, 44, 10388–10395. [Google Scholar] [CrossRef] [PubMed]
- Szpechciński, A.; Grzanka, A. Aptamers in clinical diagnostics. Postepy Biochem. 2006, 52, 260–270. [Google Scholar] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Kurniawan, H.; Byrne, M.E.; Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.C.; DeFeo-Fraulini, T.; Hutabarat, R.M.; Horvath, C.J.; Merlino, P.G.; Marsh, H.N.; Healy, J.M.; Boufakhreddine, S.; Holohan, T.V.; Schaub, R.G. First-in human evaluation of anti-von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 2007, 116, 2678–2686. [Google Scholar] [CrossRef] [PubMed]
- Jilma, B.; Paulinska, P.; Jilma-Stohlawetz, P.; Gilbert, J.C.; Hutabarat, R.; Knöbl, P. A randomized pilot trial of the anti von Willebrand factor aptamer ARC1779 in ptients with type 2b von Willebrand disease. Thromb Haemost. 2010, 104, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Laber, D.A.; Taft, B.S.; Kloecker, G.H.; Bates, P.J.; Trent, J.O.; Miller, D.M. Extended phase I study of AS1411 in renal and non-small cell lung cancers. In Proceedings of the American Society of Clinical Oncology Annual Meeting, Atlanta, GA, USA, 2–6 June 2006; p. 13098.
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.; Li, Y. A SPR Aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, H.; Li, S.; Zaia, J.; Rossi, J.J. Novel dual inhibitory function aptamer–siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008, 16, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.J.; Phillips, L.W.; Milligan, A.S.; Rodda, S.J. Toward specific detection of Dengue virus serotypes using a novel modular biosensor. Biosens. Bioelectron. 2010, 26, 1696–1700. [Google Scholar] [CrossRef] [PubMed]
- Negri, P.; Chen, G.; Kage, A.; Nitsche, A.; Naumann, D.; Xu, B.; Dluhy, R.A. Direct optical detection of viral nucleoprotein binding to an anti-Influenza aptamer. Anal. Chem. 2012, 84, 5501–5508. [Google Scholar] [CrossRef] [PubMed]
- Ruslinda, R.A.; Tanabe, K.; Ibori, S.; Wang, X.; Kawarada, H. Effects of diamond-FET-based RNA aptamer sensing for detection of real sample of HIV-1 Tat protein. Biosens. Bioelectron. 2013, 40, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Luzi, E.; Mascicni, M. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 2005, 67, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, Y.; Hu, B.; Ma, Z.; Huang, H.; Yu, Y.; Liu, S.; Lu, M.; Yang, D. Development of HBsAg-binding aptamers that bind HepG2.2.15 cells via HBV surface antigen. Virol. Sin. 2010, 25, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Li, D.; Chen, H.; Zhang, X. CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for Hepatitis C Virus envelope glycoprotein E2. PLOS ONE 2009, 4, e8142. [Google Scholar] [CrossRef] [PubMed]
- Toscano-Garibay, J.D.; Benitez-Hess, M.L.; Alvarez-Salas, L.M. Isolation and characterization of an RNA aptamer for the HPV-16 E7 oncoprotein. Arch. Med. Res. 2011, 42, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Woo, H.M.; Kim, K.S.; Oh, J.W.; Jeong, Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011, 112, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P.; Richarte, A.M.; Phillips, T.; Anfrews, C.; Lee, J.S. Development, screening, and analysis of DNA aptamer libraries potentially useful for diagnosis and passive immunity of arboviruses. BMC Res. Notes 2012, 5, e633. [Google Scholar] [CrossRef]
- Ellenbecker, M.; Sears, L.; Li, P.; Lanchy, J.M.; Lodmell, S.J. Characterization of RNA aptamers directed against the nucleocapsid protein of Rift Valley fever virus. Antiviral Res. 2012, 93, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.M. The impact of avian influenza viruses on public health. Avian Dis. 2003, 47, 914–920. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO, 2003–2015. Available online: http://www.who.int/influenza/human_animal_interface/EN_GIP_20150106CumulaiveNumberH5N1cases.pdf?ua=1 (accessed on 20 January 2015).
- Capua, I.; Cattoli, G. Prevention and control of highly pathogenic avian influenza with particular reference to H5N1. Virus Res. 2013, 178, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Kawasaki, K.; Kumar, P.K. Selection of RNA-aptamer against human influenza B virus. Nucleic Acids Symp. Ser. 2005, 49, 85–86. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Misono, T.S.; Kawasaki, K.; Mizuno, T.; Imai, M.; Odagiri, T.; Kumar, P.K. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006, 87, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens. Bioelectron. 2013, 42, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jee, M.H.; Kwon, O.S.; Keum, S.J.; Jang, S.K. Infectivity of hepatitis C virus correlates with the amount of envelope protein E2: Development of a new aptamer-based assay system suitable for measuring the infectious titer of HCV. Virology 2013, 439, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Zamay, A.S.; Muharemagic, D.; Chechik, A.V.; Bell, J.C.; Berezovski, M.V. Aptamer-based viability impedimetric sensor for viruses. Anal. Chem. 2012, 84, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Parekh, P.; Tang, Z.; Turner, P.C.; Moyer, R.W.; Tan, W. Aptamers recognize glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 2010, 82, 8642–8649. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.C.; Zarbl, H. Use of Cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLOS ONE 2012, 7, e36103. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, Y.S.; Jo, M.; Jin, M.; Lee, D.; Kim, S. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 2007, 358, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Leon, R.; Medina, M.; Schiff, E.R. Diagnostic tools in the evaluation of patients with viral hepatitis undergoing liver transplantation. Liver Transpl. Surg. 1998, 4, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Ocadiz-Delgado, R.; Albino-Sanchez, M.E.; Garcia-Villa, E.; Aguilar-Gonzalez, M.G.; Cabello, C.; Rosete, D.; Mejia, F.; Manjarrez-Zavala, M.E.; Ondarza-Aguilera, C.; Rivera-Rosales, R.M.; et al. In situ molecular identification of the Influenza A (H1N1) 2009 Neuraminidase in patients with severe and fatal infections during a pandemic in Mexico City. BMC Infect. Dis. 2013, 13, e20. [Google Scholar] [CrossRef]
- Caliendo, A.M. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin. Infect. Dis. 2011, 52, 326–330. [Google Scholar] [CrossRef]
- Charlton, B.; Crossley, B.; Hietala, S.; et al. Conventional and future diagnostics for avian influenza. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Huang, H.; Zou, W.; Dan, H.; Guo, X.; Zhang, A.; Yu, Z.; Chen, H.; Jin, M. An indirect sandwich ELISA for the detection of avian influenza H5 subtype viruses using anti-hemagglutinin protein monoclonal antibody. Vet. Microbiol. 2009, 137, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Dhumpa, R.; Handberg, K.J.; Jørgensen, P.H.; Yi, S.; Wolff, A.; Bang, D.D. Rapid detection of avian influenza virus in chicken fecal samples by immunomagnetic capture reverse transcriptasepoly-merase chain reaction assay. Diagn. Microbiol. Infect. Dis. 2011, 69, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Sidoti, F.; Rizzo, F.; Costa, C.; Astegiano, S.; Curtoni, A.; Mandola, M.L.; Cavallo, R.; Bergallo, M. Development of real time RT-PCR assays for detection of type A influenza virus and for subtyping of avian H5 and H7 hemagglutinin subtypes. Mol. Biotechnol. 2010, 44, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kim, J.H.; Kim, Y. Comparison of nine different qualitative HBsAg assay kits. Korean J. Lab. Med. 2010, 30, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Shin, S.; Oh, E.J.; Kahng, J.; Kim, Y.; Lee, H.K.; Kwon, H.J. Comparison of the AdvanSure HBV real-time PCR test with three other HBV DNA quantification assays. Ann. Clin. Lab. Sci. 2013, 43, 230–237. [Google Scholar] [PubMed]
- Muhlbacher, A.; Schennach, H.; Helden, J.; Hebell, T.; Pantaleo, G.; Bürgisser, P.; Cellerai, C.; Perm-pikul, P.; Rodriguez, M.I.; Eiras, A.; et al. Performance evaluation of a new fourth-generation HIV combination. Med. Microbiol. Immunol. 2013, 202, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Saune, K.; Delaugerre, C.; Raymond, S.; Nicot, F.; Boineau, J.; Pasquier, C.; Izopet, J. Analytical sensitivity of three real-time PCR assays for measuring subtype B HIV-1 RNA. J. Clin. Virol. 2013, 57, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Dunning, J.; Baillie, J.K.; Cao, B.; Hayden, F.G. Antiviral combinations for severe influenza. Lancet Infect. Dis. 2014, 14, Jeo9–1270. [Google Scholar] [CrossRef]
- Webster, R.G.; Govorkova, E.A. Continuing challenges in influenza. Ann. N.Y. Acad. Sci. 2014, 1323, 115–139. [Google Scholar] [CrossRef] [PubMed]
- Sahu, G.K. Potential implication of residual viremia in patients on effective antiretroviral therapy. AIDS Res. Hum. Retrovir. 2015, 31, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Memish, Z.A.; Zumla, A. Severe acute respiratory syndrome vs. the Middle East respiratory syndrome. Curr. Opin. Pulm. Med. 2014, 20, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Ferir, G.; Gordts, S.C.; Schols, D. HIV-1 and its resistance to peptidic Carbohydrate-Binding Agents (CBAs): An overview. Molecules 2014, 19, 21085–21112. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Memish, Z.A. What are our pharmacotherapeutic options for MERS-CoV? Expert Rev. Clin. Pharmacol. 2014, 7, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Cortez, K.J.; Kottilil, S. Beyond interferon: Rationale and prospects for newer treatment paradigms for chronic hepatitis C. Ther. Adv. Chronic Dis. 2015, 6, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K.G.; Wood, J.M.; Zambon, M. Influenza. Lancet 2003, 362, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Marascio, N.; Torti, C.; Liberto, M.; Foca, A. Update on different aspects of HCV variability: Focus on NS5B polymerase. BMC Infect. Dis. 2014, 14, e1. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, I. Clinical implications of hepatitis B virus mutations: Recent advances. World J. Gastroenterol. 2014, 20, 7653–7664. [Google Scholar] [CrossRef] [PubMed]
- Fall-Malick, F.Z.; Tchiakpé, E.; Ould Soufiane, S.; Diop-Ndiaye, H.; Mouhamedoune Baye, A.; Ould Horma Babana, A.; Touré Kane, C.; Lo, B.; Mboup, S. Drug resistance mutations and genetic diversity in adults treated for HIV type 1 infection in Mauritania. J. Med. Virol. 2014, 86, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.; Spindler, K.; Horodyski, F.; Grabau, E.; Nichol, S.; VandePol, S. Rapid evolution of RNA genomes. Science 1982, 215, 1577–1585. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; McFadden, G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Soriano, V.; Labarga, P.; Barreiro, P.; Fernandez-Montero, J.V.; Mendoza, C.; Esposito, I.; Benítez-Gutiérrez, L.; Peña, J.M. Drug interactions with new hepatitis C oral drugs. Expert Opin. Drug Metab. Toxicol. 2015, 2, 1–9. [Google Scholar]
- Zornitzki, T.; Malnick, S.; Lysyy, L.; Knobler, H. Interferon therapy in hepatitis C leading to chronic type 1 diabetes. World J. Gastroenterol. 2015, 21, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; McHutchison, J.G.; Gordon, S.C.; Rustgi, V.K.; Shiffman, M.; Reindollar, R.; Goodman, Z.D.; Koury, K.; Ling, M.; Albrecht, J.K. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet 2001, 358, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Naghavi, M.; Foreman, K.; Lim, S.; Shibuya, K.; Aboyans, V.; Abraham, J.; Adair, T.; Aggarwal, R.; Ahn, S.Y.; et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095–2128. [Google Scholar] [CrossRef] [PubMed]
- Mufhandu, H.T.; Gray, E.S.; Madiga, M.C.; Tumba, N.; Alexandre, K.B.; Khoza, T.; Wibmer, C.K.; Moore, P.L.; Morris, L.; Khati, M. UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits entry of Human Immunodeficiency Virus type 1 Subtype C. J. Virol. 2012, 86, 4989–4999. [Google Scholar] [CrossRef] [PubMed]
- Bellecave, P.; Cazenave, C.; Rumi, J.; Staedel, C.; Cosnefroy, O.; Andreola, M.L.; Ventura, M.; Tarrago-Litvak, L.; Astier-Gin, T. Inhibition of Hepatitis C Virus (HCV) RNA polymerase by DNA aptamers: Mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob. Agents Chemother. 2008, 52, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.; Hayashi, K.; Kumar, P.K. Aptamer that binds to the gD protein of Herpes Simplex Virus 1 and efficiently inhibits viral entry. J. Virol. 2012, 86, 6732–6744. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.Y.; Sun, H.Y.; Lee, K.H.; Oh, B.H.; Cha, Y.J.; Kim, B.H.; Yoo, J.Y. 5'-triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012, 40, 2724–2733. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Berzal-Herranz, B.; Gomez, J.; Berzal-Herranz, A. An engineered inhibitor RNA that efficiently interferes with hepatitis C virus translation and replication. Antivir. Res. 2012, 94, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Shibata, T.; Kabashima, T.; Kai, M. Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur. J. Med. Chem. 2012, 56, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.H.; Kayhan, B.; Ben-Yedidia, T.; Arnon, R. A DNA aptamer prevents Influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 2004, 279, 48410–48419. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.K.; Lee, C.; Lee, K.S.; Choe, S.Y.; Mo, I.P.; Seong, R.H.; Hong, S.; Jeon, S.H. DNA aptamers against the receptor binding region of hemagglutinin prevent Avian Influenza viral infection. Mol. Cells 2011, 32, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.M.; Lee, K.H.; Han, B.W.; Han, M.R.; Kim, D.H.; Kim, D.E. An RNA aptamer that specifically binds to the glycosylated hemagglutinin of Avian Influenza virus and suppresses viral infection in cells. PLOS ONE 2014, 9, e97574. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.; Forzan, M.; Sproat, B.; Pantophlet, R.; McGowan, I.; Burton, D.; James, W. An aptamer that neutralizes R5 strains of HIV-1 binds to core residues of gp120 in the CCR5 binding site. Virology 2008, 381, 46–54. [Google Scholar] [CrossRef] [PubMed]
- DeStefano, J.J.; Nair, G.R. Novel aptamer inhibitors of Human Immunodeficiency Virus reverse transcriptase. Oligonucleotides 2008, 18, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, D.; Duclair, S.; Datta, S.A.; Ellington, A.; Rein, A.; Prasad, V.R. RNA aptamers directed to Human Immunodeficiency Virus type Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J. Virol. 2011, 85, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci. Transl. Med. 2011, 3, e66ra6. [Google Scholar] [CrossRef]
- Zhou, J.; Neff, Ch.P.; Swiderski, P.; Li, H.; Smith, D.D.; Aboellail, T.; Remling-Mulder, L.; Akkina, R.; Rossi, J.J. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013, 21, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, L.A.; Trifonova, R.; Vrbanac, V.; Basar, R.; McKernan, S.; Xu, Z.; Seung, E.; Deruaz, M.; Dudek, T.; Einarsson, J.I.; et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Investig. 2011, 121, 2401–2412. [Google Scholar]
- Biroccio, A.; Hamm, J.; Incitti, I.; Francesco, R.; Tomei, L. Selection of RNA aptamers that are specific and high-affinity ligands of the Hepatitis C Virus RNA-dependent RNA polymerase. J. Virol. 2002, 76, 3688–3696. [Google Scholar] [CrossRef] [PubMed]
- Umehara, T.; Fukuda, K.; Nishikawa, F.; Sekiya, S.; Kohara, M.; Hasegawa, T.; Nishikawa, S. Designing and analysis of a potent bi-functional aptamers that inhibit protease and helicase activities of HCV NS3. Nucleic Acids Symp. Ser. 2004, 48, 195–196. [Google Scholar] [CrossRef]
- Fukuda, K.; Umehara, T.; Sekiya, S.; Kunio, K.; Hasegawa, T.; Nishikawa, S. An RNA ligand inhibits Hepatitis C Virus NS3 protease and helicase activities. Biochem. Biophys. Res. Commun. 2004, 325, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; Fujita, S.; Iizuka, M.; Nishikawa, S.; Hasegawa, T.; Fukuda, K. Isolation and characterization of RNA aptamers specific for the HCV minus-IRES domain I. Nucleic Acids Symp. Ser. 2008, 52, 493–494. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, H.; Liu, F. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA 2000, 6, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.H. A SELEX-screened aptamer of human Hepatitis B Virus RNA encapsidation signal suppresses viral replication. PLOS ONE 2011, 6, e27862. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Lee, N.R.; Yeo, W.S.; Jeong, Y.J.; Kim, D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008, 366, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Binning, J.M.; Wang, T.; Luthra, P.; Shabman, R.S.; Borek, D.M.; Liu, G.; Xu, W.; Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 2013, 52, 8406–8419. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Khati, M.; Tang, M.; Wyatt, R.; Lea, S.M.; James, W. An aptamer that neutralizes R5 strains of Human Immunodeficiency Virus type 1 blocks gp120-CCR5 interaction. J. Virol. 2005, 79, 13806–13810. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Dong, J.; Yao, L.; Chen, A.; Jia, R.; Huan, L.; Guo, J.; Shu, Y.; Zhang, Z. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008, 366, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, E.; Kumar, P.K. An aptamer that binds efficiently to the hemagglutinins of highly pathogenic avian influenza viruses (H5N1 and H7N7) and inhibits hemagglutinin-glycan interactions. Acta Biomater. 2014, 10, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Kanai, A.; Tanabe, K.; Kohara, M. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 1995, 376, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Gao, Y.; Xue, B.; Wang, X.; Yang, D.; Qin, Y.; Yu, R.; Liu, N.; Xu, L.; Fang, X.; Zhu, H. Inhibition of Hepatitis C Virus infection by NS5A-specific aptamer. Antivir. Res. 2014, 106, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yu, X.; Xue, B.; Zhou, F.; Wang, X.; Yang, D.; Liu, N.; Xu, L.; Fang, X.; Zhu, H. Inhibition of Hepatitis C Virus infection by DNA aptamer against NS2 protein. PLOS ONE 2014, 9, e90333. [Google Scholar] [CrossRef] [PubMed]
- Kao, R.Y.; Tsui, W.H.; Lee, T.S.; Tanner, J.A.; Watt, R.M.; Huang, J.D.; Hu, L.; Chen, G.; Chen, Z.; Zhang, L.; et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004, 11, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, M.Y.; Lee, J.H.; You, J.C.; Jeong, S. Selection and stabilization of the RNA aptamers against the Human Immunodeficiency Virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2002, 291, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Konno, K.; Iizuka, M.; Fujita, S.; Nishikawa, S.; Hasegawa, T.; Fukuda, K. An RNA aptamer containing two binding sites against the HCV minus-IRES domain I. Nucleosides Nucleotides Nucleic Acids 2011, 30, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Hwang, J.; Kuno, A.; Hasegawa, T.; Nishikawa, S. RNA aptamers targeted to domain II of Hepatitis C Virus IRES that bind to its apical loop region. J. Biochem. 2003, 133, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Umehara, T.; Fukuda, K.; Kuno, A.; Hasegawa, T.; Nishikawa, S. A Hepatitis C Virus (HCV) internal ribosome entry site (IRES) domain III-IV targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005, 33, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Umehara, T.; Nishikawa, F.; Fukuda, K.; Hasegawa, T.; Nishikawa, S. Increased inhibitory ability of conjugated RNA aptamers against the HCV IRES. Biochem. Biophys. Res. Commun. 2009, 386, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Srisawat, C.; Engelke, D.R. Selection of RNA aptamers that bind HIV-1 LTR DNA duplexes: Strand invaders. Nucleic Acids Res. 2010, 38, 8306–8315. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Swiderski, P.; Li, H.; Zhang, J.; Neff, C.P.; Akkina, R.; Rossi, J.J. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009, 37, 3094–3109. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, X.; Gao, G. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell 2010, 1, 752–729. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Banda, N.; Lee, N.S.; Rossi, J.; Akkina, R. RNA-based anti-HIV-1 gene therapeutic constructs in SCID-hu mouse model. Mol. Ther. 2002, 6, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Banerjea, A.; Li, M.J.; Remling, L.; Rossi, J.; Akkina, R. Lentiviral transduction of Tar Decoy and CCR5 ribozyme into CD34+ progenitor cells and derivation of HIV-1 resistant T cells and macrophages. AIDS Res. Ther. 2004, 1, e2. [Google Scholar] [CrossRef]
- Hacein-Bey-Abina, S.; Garrigue, A.; Wang, G.P.; Soulier, J.; Lim, A.; Morillon, E.; Clappier, E.; Caccavelli, L.; Delabesse, E.; Beldjord, K.; et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 2008, 118, 3132–3142. [Google Scholar]
- Zhou, J.; Li, H.; Zhang, J.; Swiderski, P.; Rossi, J. Development of cell-type specific anti-HIV gp120 aptamers for siRNA delivery. J. Vis. Exp. 2011, 52, e2954. [Google Scholar]
- Griffin, L.C.; Tidmarsh, G.F.; Bock, L.C.; Toole, J.J.; Leung, L.L. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracor-poreal circuits. Blood 1993, 81, 3271–3276. [Google Scholar] [PubMed]
- Beigelman, L.; Matulic-Adamic, J.; Haeberli, P.; Usman, N.; Dong, B.; Silverman, R.H.; Khamnei, S.; Torrence, P.F. Synthesis and biological activities of a phosphorodithioate analog of 2',5' oligoadenylate. Nucleic Acids Res. 1995, 23, 3989–3994. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Asakawa, H.; Tashiro, Y.; Juni, K.; Sueishi, T. Stability, specific binding activity, and plasma concentration in mice of an oligodeoxynucleotide modified at 5-terminal with poly(ethylene glycol). Biol. Pharm. Bull. 1995, 18, 474–476. [Google Scholar] [CrossRef] [PubMed]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef] [PubMed]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic silencing of an endogenous gene by systemic admin-istration of modified siRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, C.P.; Roberts, J.D.; Pitoc, G.A.; Nimjee, S.M.; White, R.R.; Quick, G.; Scardino, E.; Fay, W.P.; Sullenger, B.A. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat. Bio-Tech. 2004, 22, 1423–1428. [Google Scholar] [CrossRef]
- Verhoef, J.J.; Carpenter, J.F.; Anchordoquy, T.J.; Schellekens, H. Potential induction of anti-PEG anti-bodies and complement activation toward PEGylated therapeutics. Drug Discov. Today 2014, 19, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kiwada, H. Anti-polyethyleneglycol antibody response to PEGylated substances. Biol. Pharm. Bull. 2013, 36, 889–991. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.K.; Hempel, G.; Koling, S.; Chan, L.S.; Fisher, T.; Meiselman, H.J.; Garratty, G. Anti-body against poly(ethylene glycol) adversely affects PEG-asparaginase therapy in acute lymphoblastic leukemia patients. Cancer 2007, 110, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 2015, 7, 751-780. https://doi.org/10.3390/v7020751
Wandtke T, Woźniak J, Kopiński P. Aptamers in Diagnostics and Treatment of Viral Infections. Viruses. 2015; 7(2):751-780. https://doi.org/10.3390/v7020751
Chicago/Turabian StyleWandtke, Tomasz, Joanna Woźniak, and Piotr Kopiński. 2015. "Aptamers in Diagnostics and Treatment of Viral Infections" Viruses 7, no. 2: 751-780. https://doi.org/10.3390/v7020751
APA StyleWandtke, T., Woźniak, J., & Kopiński, P. (2015). Aptamers in Diagnostics and Treatment of Viral Infections. Viruses, 7(2), 751-780. https://doi.org/10.3390/v7020751





