Abstract
Tetraspanins are integral transmembrane proteins organized in microdomains displaying specific and direct interactions with other tetraspanins and molecular partners. Among them, CD81 has been implicated in a variety of physiological and pathological processes. CD81 also plays a crucial role in pathogen entry into host cells, including hepatitis C virus (HCV) entry into hepatocytes. HCV is a major cause of liver cirrhosis and hepatocellular carcinoma. HCV entry into hepatocytes is a complex process that requires the coordinated interaction of viral and host factors for the initiation of infection, including CD81, scavenger receptor BI, claudin-1, occludin, membrane-bound host cell kinases, Niemann-Pick C1 Like 1, Harvey rat sarcoma viral oncogene homolog (HRas), CD63 and transferrin receptor 1. Furthermore, recent data in HCV model systems have demonstrated that targeting critical components of tetraspanins and associated cell membrane proteins open new avenues to prevent and treat viral infection.
1. Introduction
1.1. The Tetraspanin Family of Proteins
Tetraspanins are part of a family of transmembrane proteins with significant sequence homology that has been conserved during evolution. In mammals, there are thirty two tetraspanins and only a minority has been extensively studied. The tetraspanin family is composed of type IV glycoproteins, discovered in the late 1980s with the subsequent cloning of CD81 as a protein of 26 kDa [1]. Tetraspanins are relatively small proteins (200–300 amino acids) composed of a small extracellular loop (SEL), a large extracellular loop (LEL), four transmembrane domains and intracellular N- and C−terminal domains [2,3]. Tetraspanins are characterized by the presence of four cysteines and a CCG motif in the LEL. The N-terminal domain forms an alpha helix containing positively charged residues important for protein interactions, as well as palmitoylation sites causing anchorage of the protein to the inner leaflet of the plasma membrane. The C-terminal domain also contains palmitoylation sites on intracellular cysteine residues [4], which are required for tetraspanin association with cholesterol complexes [5]. Palmitoylated tetraspanins are important for the assembly of the tetraspanin web by linking tetraspanins and their associated proteins in cholesterol rich regions of the plasma membrane and by their association with proteins of the cytoskeleton and signaling molecules [6,7].
While all cells, except sperm cells, express tetraspanins, the expression level of the individual proteins in different tissues is variable [3]. Some tetraspanins may be considered to be cell-specific while others are characterized by a very broad expression, without being ubiquitous. For example, the tetraspanin CD81 is expressed on hepatocytes, epithelial cells, fibroblasts, endothelial cells and on most of the blood cells, excluding erythrocytes, platelets and neutrophils. Tetraspanins form large complexes with other membrane proteins and given the heterogeneity in the composition as well as the dynamic nature of these complexes, tetraspanins are implicated in various biological processes, such as adhesion, migration, proliferation, signal transduction, intracellular trafficking and differentiation.
1.2. Interaction of CD81 with Other Host Factors
Several tetraspanins interact with other tetraspanins and partner transmembrane proteins like integrins, molecules of the immunoglobulin superfamily, cellular enzymes, signaling molecules and precursors of growth factors [5,8,9,10,11,12,13]. These interactions are direct and highly specific. Specialized regions on the surface of the cell membrane where tetraspanin interactions take place are termed as “tetraspanin-enriched microdomains” (TEMs) [3]. The composition of TEMs is cell- and tissue-specific. Thus, in each cell type, these networks consist of different tetraspanin-associated partners, which define their function. TEMs are highly regulated structures governed by cholesterol and lipid composition, by physiological stage of the cell and by palmitoylation of putative sites in juxtamembrane domains [6,7,14].
CD81 is a key tetraspanin protein expressed in numerous cell types, either alone or as a part of TEM. It is involved in myriad of physiological functions through association with other tetraspanins and membrane proteins. Among the known interaction partners of CD81 are integrin α4β1 [15] and members of the immunoglobulin family EWI-F and EWI-2 [16,17] that link CD81 to the actin cytoskeleton, thereby regulating cell motility and polarity [18,19]. On the surface of a B-cell, CD81 participates in forming CD19-signaling complex, which in conjunction with the B-cell antigen receptor (BCR) lowers the activation threshold of BCR, leading to antibody production in response to antigenic stimulation. While CD81 does not affect B-cell and T-cell development in CD81-knock-out mice, it regulates lymphocyte proliferation through multiple ways. Thus, CD81 deficiency results in enhanced antibody response to type II T-independent antigens but impaired antibody response to T-dependent antigens in CD81-null mice [20,21,22]. In line with this, a case study reported absence of CD19 expression in a patient with normal CD19 gene but possessing a rare homozygous CD81 gene defect as a cause of profound hypogammaglobulinemia [23].
Interestingly, CD81 is also able to activate intracellular signaling pathways, such as the mitogen−activated protein kinase (MAPK) pathway. Indeed, CD81 recruits Src homology 2 domain containing transforming protein (Shc) to the plasma membrane via its phosphotyrosine-binding (PTB) domain and induces activation of extracellular signal-regulated kinases (Erk) leading to tumor cell proliferation [24]. In addition, activated protein kinase C (PKC) migrates to the plasma membrane and associates with tetraspanins CD9, CD53, CD81, CD82 and CD151 [25]. PKC is required for integrin−mediated cell adhesion, but the formation of tetraspanin-PKC complexes is not integrin−dependent. Tetraspanins function rather as linker molecules that recruit PKC to a close proximity of integrin β1 (ITGB1) by associating ITGB1 to the extracellular domain of tetraspanins and PKC to their cytoplasmic domain [11].
2. Co-Receptor Association(s) and HCV Entry
2.1. CD81-HCV Interactions
Tetraspanins are not only essential for cell biology; they are also involved in various steps of pathogen infection including parasites, bacteria and viruses. The tetraspanin CD81 plays a role in Plasmodium sporozoite infection [26,27] and in Listeria monocytogenes entry [28]. Regarding viral pathogens, it is established that CD81 is an entry factor for hepatitis C virus (HCV) [29,30,31]. Contrarily, CD81 has been shown to negatively regulate human immunodeficiency virus-1 (HIV-1) infection by modulating envelope-mediated membrane fusion [32].
HCV infection is a leading cause of liver cirrhosis and hepatocellular carcinoma (HCC) world-wide [33,34,35]. HCV is a small enveloped virus possessing a single-stranded positive sense RNA genome. It belongs to the hepacivirus genus within the Flaviviridae family. De novo infection of hepatocytes by HCV is facilitated by two mechanisms, namely cell-free and cell-cell transmission [36,37]. Both modes of transmission rely on the viral envelope glycoproteins E1 and E2 and several host cell entry factors including CD81, scavenger receptor class B type 1 (SR-BI), claudin-1 (CLDN1), occludin (OCLN), epidermal growth factor receptor (EGFR) and its signal transducer Harvey rat sarcoma viral oncogene homolog (HRas) [37,38,39,40,41,42]. Within the past years, the molecular mechanisms of cell-free entry and the subsequent steps of the viral life cycle have been intensively characterized. Upon interaction with specific cellular receptors via its envelope glycoproteins, HCV particles are endocytosed. In the endocytic vesicle, low pH triggers fusion of the viral and the host membranes releasing the ~9.6 Kb viral genome into the cytoplasm of the newly infected cell [43]. The highly conserved un-translated regions (UTR) at the 5' and 3' ends mediate replication of the viral genome and translation of viral proteins. Internal ribosomal entry site (IRES)-dependent translation of HCV genome results in a ~3,010 amino acid polyprotein that is cleaved by host and viral proteases to yield 10 mature viral proteins consisting of three structural proteins (the core and glycoproteins E1 and E2), six non-structural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) and a small integral transmembrane protein (p7). Viral replication and assembly occurs at the endoplasmic reticulum (ER) membrane and in close association with lipid droplets (LDs) [44,45,46,47]. Assembly and release of HCV particles appear to be closely linked with very low density lipoprotein (VLDL) secretory pathway [48,49,50]. The released viral particles can then infect neighboring hepatocytes via cell-free infection. Of note, these viral particles are sensitive to neutralizing antibodies targeting the viral envelope glycoproteins. In addition, assembled viral particles can also be directly transmitted from an infected cell to an adjacent cell in a process that is resistant to most of the neutralizing antibodies uncovered to date, but the underlying molecular mechanisms have not been fully characterized. While cell-free infection plays an important role during initiation of infection, cell−cell transmission is thought to play a major role during maintenance of infection and viral dissemination.
The tetraspanin CD81was the first reported host factor interacting with a soluble form of the HCV glycoprotein E2 [29]. It was subsequently shown that CD81 is required for HCV infection of hepatocytes. Indeed, HCV entry and infectivity is inhibited in a pan-genotypic manner by CD81−specific antibodies [38,43,51,52,53,54,55], by a soluble recombinant form of the CD81 LEL [43,56], and by silencing CD81 expression [31]. In contrast, CD81 expression confers susceptibility to HCV infection in hepatoma cell lines lacking CD81, such as HepG2 cells [31,57,58]. Furthermore, CD81 expression levels have been shown to affect the efficiency of HCV entry [59,60]. Interestingly, a recent study demonstrated modulation of HCV RNA replication depending on CD81 expression [61]. These results suggest multiple and diverse roles of CD81 in the HCV life-cycle.
Various studies identified regions and residues of CD81 involved in the interaction with E2 and the viral particle (Figure 1). Indeed, E2 interacts with the LEL of CD81. E2-CD81 interaction is specific, since E2 does not bind other tetraspanins such as CD9 or CD151 [29,30,31,62,63,64]. Moreover, whereas CD81 LEL plays a direct role in HCV infection by mediating E2 binding, CD81 SEL plays an indirect role by regulating the optimal cell surface expression of LEL [65]. Several other regions of CD81, such as the C-terminal region, transmembrane residues and post-translational modification (e.g., palmitoylation of cysteines in the juxtamembrane domain) have been shown to be important for HCV entry via indirect mechanisms e.g., by mediating oligomerization of CD81, by facilitating interaction with other proteins and by cholesterol partitioning [66]. It is worth noting that residues in transmembrane domains and/or cysteine-mediated palmitoylation seem to exert only moderate inhibitory effects on HCV entry. This indicates that CD81 LEL is the key determinant of viral entry and that additional regions of CD81 only enhance viral entry.
CD81 expression on the cell surface is regulated by the membrane lipid composition. Sphingolipids associate with cholesterol to form lipid rafts and thus are important for plasma membrane organization. It has been shown that enrichment of ceramide in the plasma membrane induces internalization of CD81, thereby inhibiting HCV entry [67]. The cholesterol content of the plasma membrane is also important for HCV entry. It has been demonstrated that depletion of cholesterol—that is required for maintaining membrane fluidity—from cellular membranes inhibits HCV infection. This correlates with decreased amounts of CD81 at the cell surface since CD81 physically interacts with cholesterol [5,68]. Furthermore, it has been shown that the dynamic nature of CD81 and its lateral diffusion is dependent on cell polarization and correlate with HCV infection [69]. In addition, recent data suggest a role of CD81 trafficking in the HCV entry process [70]. Indeed, CD81 engagement with HCV or a CD81−specific antibody promotes clathrin-dependent internalization of CD81. Interestingly, the CD81−specific antibody also appears to neutralize HCV after its internalization, suggesting that intracellular CD81 plays a role in HCV infection [70].
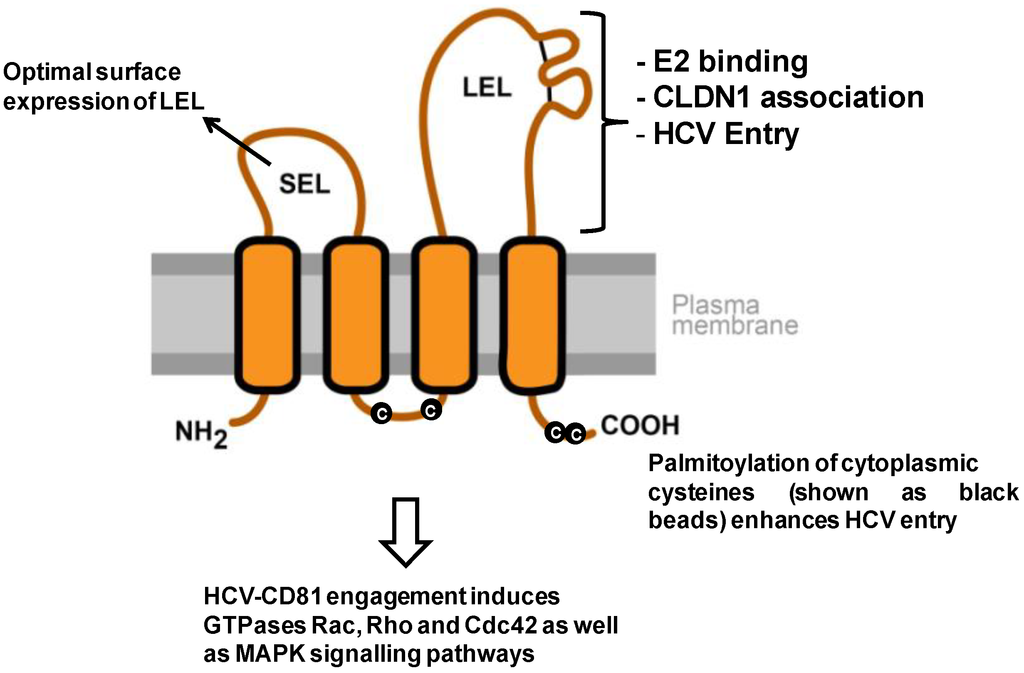
Figure 1.
Model of CD81 topology and its relevance for HCV entry. The depicted CD81 regions comprise the cytoplasmic N- and C-terminus, a small extracellular loop (SEL), a large extracellular loop (LEL), four transmembrane domains and a cytoplasmic loop. The two disulfide bonds in the LEL are shown as black lines. The LEL mediates multiple functions that are important for HCV entry. It binds HCV envelope glycoprotein E2 (L162, K171, I181, I182, N184, F186, D196) and mediates viral entry (K171, I181, I182, F186 [64,66,71,72]). Additionally, CD81 LEL is implicated in CLDN1 association (T149, E152, T153 [73]). The SEL plays an indirect role in HCV entry by facilitating optimal expression of LEL on the cell surface by proper translocation of CD81 during its synthesis [65,74]. The post-translational palmitoylation of the cysteines (shown as black beads) and the transmembrane domains of CD81 have also been shown to enhance HCV entry [66,74]. Finally, CD81 engagement by HCV/soluble E2/CD81-specific antibodies has been shown to activate GTPases Rac, Rho and Cdc42, as well as the MAPK signaling pathways [75,76].
2.2. The Tetraspanin Complex Formation during HCV Entry and Downstream Signaling Pathways
HCV relies on multiple host factors to gain entry into hepatocytes (reviewed in [77]). This includes two molecules of the tetraspanin superfamily, namely CD81 and CD63. Interestingly, both tetraspanins have been shown to interact with the viral envelope glycoprotein E2, and may thus directly interact with HCV during the viral entry process [29,78]. While CD81 does not bind any known endogenous ligand nor possess an internalization motif, binding of HCV glycoprotein E2 or CD81-specific antibodies to CD81 has been shown to activate GTPases Rac, Rho and Cdc42 as well as the MAPK signaling pathways [75,76]. These cellular events could regulate HCV interactions with its co−receptors and establish viral entry into target cell. In addition, CD63 is involved in clathrin−dependent endocytosis and vesicle trafficking to lysosomes, suggesting that CD63 may facilitate HCV uptake. Furthermore, while CD81 and CD63 have been involved in TEM formation [79], it is not yet known whether they interact with each other in hepatocytes.
Besides the potential direct effect in HCV envelope glycoprotein binding and subsequent downstream events described above, CD81 has also been shown to contribute to HCV entry by forming a co-receptor complex through its interaction with other proteins. A major breakthrough was the identification of the CD81-CLDN1 co-receptor complex [80,81]. Noteworthy, while there is no physiological role for this complex known to date, the association of CD81 to CLDN1 is a mandatory step of the HCV entry process. CLDN1 is an integral transmembrane protein of 25 kDa with membrane topology similar to CD81 [82]. However, it is not classified as a classical tetraspanin because of the lack of four cysteines and a CCG motif in the extracellular loop two, one of the defining features of tetraspanins. It has been demonstrated that CLDN1 is an essential host cell factor for HCV entry as cells lacking CLDN1 are resistant to HCV [83]. However, in contrast to CD81, CLDN1 is not seen as a classical HCV receptor as it does not directly bind to soluble HCV glycoprotein E2, a key viral protein involved in HCV entry. Neither CD81 nor CLDN1 appear to bind infectious viral particles [83,84], probably due to the masking of the viral envelope by host-derived lipoproteins, however, a recent study reported that E1E2 complexes are able to interact with CLDN1 [85]. These data suggest that CLDN1, like CD81, may contribute to HCV envelope glycoprotein binding, but that both proteins recognize distinct parts of the viral envelope. While predominantly expressed at the tight junction (TJ), CLDN1 also localizes on the basolateral membrane of hepatocytes. Noteworthy, this pool of CLDN1 outside of the TJ co-localizes with CD81 and allows the formation of the CD81−CLDN1 co-receptor complex that is essential for HCV entry [80,81,84]. Interestingly, CLDN1 co-localizes not only with CD81 at the plasma membrane, but also with SR-BI, another important HCV entry factor, suggesting that these HCV entry factors may be part of a larger membrane complex important for viral entry [42,86,87].
A genome-wide host kinase RNAi screen demonstrated an important regulatory role of kinases for HCV entry and infection [41]. Several kinases have been implicated in CLDN1 cellular localization, relocation and CD81-CLDN1 co-receptor complex formation including protein kinase A (PKA) [88] and receptor tyrosine kinases (RTKs) [41]. Inhibition of EGFR kinase activity by erlotinib or silencing EGFR expression reduces CD81 association to CLDN1 at the cell surface [41]. This highlights a role of EGFR signaling in the formation of the CD81-CLDN1 co-receptor complex and subsequent HCV entry. While EGFR is known to activate many downstream signals in various cell types, it has been demonstrated that EGFR predominantly activates MAPK signaling in hepatocytes [42]. Indeed, HRas GTPase, a molecular switch for the activation of the MAPK pathway, was identified as a cellular transducer of RTK signals required for HCV entry [42]. A differential proteomic approach allowed to identify HRas as well as CLDN1, SR-BI, integrin beta 1 (ITGB1) and Rap2B as specifically CD81 TEM-associated proteins [42]. Given that all these host factors play a role in HCV entry, these data indicate the existence of a functional membrane network of proteins involved in viral entry [42]. As HRas signaling has been demonstrated to modulate lateral membrane diffusion of CD81 which allows assembly of the tetraspanin receptor complex subsequently mediating HCV entry [42], HRas appears at the crossroad of interplay between EGFR signaling and the CD81 receptor complex (Figure 2). Taken together, these findings suggest that HCV may manipulate RTK signaling to promote its propagation. Indeed, it has been shown that virus engagement to CD81 activates phosphatidylinositol-3-kinase (PI3K)/Akt pathway [89] and EGFR that could contribute to virus internalization [90]. This indicates that HCV actively influences the composition of CD81 TEMs via signaling events in order to promote its own entry into target cells. As CD81 has also been shown to be important for influenza entry and HRas has been shown to play a role in influenza [91] and measles virus [42] entry, perturbation of TEMs may represent a novel concept for the development of antiviral strategies that may be effective against several pathogens using the same machinery.
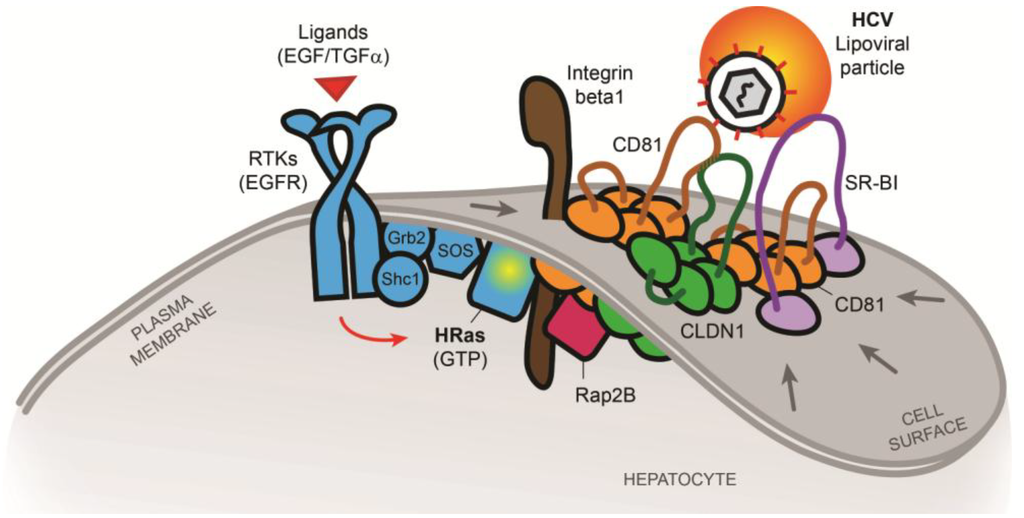
Figure 2.
Model of tetraspanin co-receptor formation(s) and HCV entry according to [42]. RTK signaling mediated by e.g., EGFR is relevant for viral entry including HCV [41]. HRas, recruited and activated by EGFR via the scaffolding proteins Shc1 and Grb2, act as a key host signaling transducer for viral entry. HRas signaling modulates lateral membrane diffusion of CD81 and promotes CD81-CLDN1 co-receptor complex formation that is essential for HCV entry. Moreover, HRas is associated with TEMs containing host receptors CD81, CLDN1, SR-BI and the previously unknown HCV entry factors integrin beta 1 (ITGB1) and Rap2B. Viruses may thus exploit HRas signaling for cellular entry by compartmentalization of entry factors and receptor trafficking. This highlights a new mechanism to regulate CD81-dependent pathogen invasion of the liver [42].
4. Conclusions and Perspectives
TEMs play a role in membrane compartmentalization leading to coupling and/or regulating molecular machinery and signaling pathways in a tissue specific manner. The tetraspanin CD81 is not only a key HCV entry factor, but also a molecular organizer of plasma membrane microdomains that contain the molecular machinery used by HCV. Signal transduction through associated tetraspanins and partner proteins likely induce actin remodeling allowing lateral movement of CD81, which appears to be required for HCV entry. This suggests a cooperative action of HCV entry factors and molecules required for vesicle formation and trafficking, leading to compartmentalization of entry factors in TEMs. Moreover, recent studies have also highlighted a central role of CD81 TEMs and virus-induced host cell signaling for entry of HCV [42,90]. Taken together, these data support a model where CD81 complexes, activated either by the virus itself or by RTK signaling, provide a functional link between CD81 trafficking and CD81-CLDN1 association that are prerequisites of HCV entry and highlight a crucial role of TEM “platforms” in the HCV entry process. These findings suggest that CD81 TEMs are highly relevant for pathogen entry such as HCV. Additionally, as CD81 mediated TEMs are required by other viruses, they present potential targets for novel broad spectrum antivirals.
Acknowledgments
We thank Olga Koutsopoulos (INSERM U1110) for critical reading of the manuscript. The authors work is supported by the European Union (ERC-2008-AdG-233130-HEPCENT, Interreg IV FEDER-Hepato-Regio-Net 2012 and FP7 HEPAMAB GAN 305600), the Agence Nationale de Recherches sur le SIDA et les hépatites virales (ANRS) (2012/239, 2013/081, 2013/249), the Direction Générale de l'Offre de Soins (A12027MS), Inserm, University of Strasbourg and ARC (TheraHCC). This work has been published under the framework of the LABEX ANR-10-LABX-0028_HEPSYS and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program.
Author Contributions
LZ, RGT, MBZ and TFB prepared the manuscript. FX and JL designed illustrations. LZ, RGT, MBZ, FX, CS, JL and TFB edited the manuscript. MBZ and TFB supervised the work. LZ and RGT contributed equally.
Conflicts of Interest
The authors declare no conflict of interest. Inserm, the University of Strasbourg and Aldevron/Genovac have filed patent applications on monoclonal antibodies targeting host factors and inhibiting HCV infection and kinases as antiviral targets.
References and Notes
- Oren, R.; Takahashi, S.; Doss, C.; Levy, R.; Levy, S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell. Biol. 1990, 10, 4007–4015. [Google Scholar]
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005, 5, 136–148. [Google Scholar] [CrossRef]
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205. [Google Scholar] [CrossRef]
- Charrin, S.; Manie, S.; Oualid, M.; Billard, M.; Boucheix, C.; Rubinstein, E. Differential stability of tetraspanin/tetraspanin interactions: Role of palmitoylation. FEBS Lett. 2002, 516, 139–144. [Google Scholar] [CrossRef]
- Charrin, S.; Manie, S.; Thiele, C.; Billard, M.; Gerlier, D.; Boucheix, C.; Rubinstein, E. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 2003, 33, 2479–2489. [Google Scholar] [CrossRef]
- Berditchevski, F.; Odintsova, E.; Sawada, S.; Gilbert, E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002, 277, 36991–37000. [Google Scholar] [CrossRef]
- Yang, X.; Claas, C.; Kraeft, S.K.; Chen, L.B.; Wang, Z.; Kreidberg, J.A.; Hemler, M.E. Palmitoylation of tetraspanin proteins: Modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell 2002, 13, 767–781. [Google Scholar] [CrossRef]
- Claas, C.; Stipp, C.S.; Hemler, M.E. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 2001, 276, 7974–7984. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef]
- Le Naour, F.; Andre, M.; Boucheix, C.; Rubinstein, E. Membrane microdomains and proteomics: Lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 2006, 6, 6447–6454. [Google Scholar]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar]
- Martin, F.; Roth, D.M.; Jans, D.A.; Pouton, C.W.; Partridge, L.J.; Monk, P.N.; Moseley, G.W. Tetraspanins in viral infections: A fundamental role in viral biology? J. Virol. 2005, 79, 10839–10851. [Google Scholar] [CrossRef]
- Perrault, M.; Pecheur, E.I. The hepatitis C virus and its hepatic environment: A toxic but finely tuned partnership. Biochem. J. 2009, 423, 303–314. [Google Scholar] [CrossRef]
- Kovalenko, O.V.; Yang, X.; Kolesnikova, T.V.; Hemler, M.E. Evidence for specific tetraspanin homodimers: Inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 2004, 377, 407–417. [Google Scholar] [CrossRef]
- Serru, V.; Le Naour, F.; Billard, M.; Azorsa, D.O.; Lanza, F.; Boucheix, C.; Rubinstein, E. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem. J. 1999, 340, 103–111. [Google Scholar]
- Charrin, S.; Le Naour, F.; Labas, V.; Billard, M.; Le Caer, J.P.; Emile, J.F.; Petit, M.A.; Boucheix, C.; Rubinstein, E. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 2003, 373, 409–421. [Google Scholar] [CrossRef]
- Zhang, X.A.; Lane, W.S.; Charrin, S.; Rubinstein, E.; Liu, L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Canc. Res. 2003, 63, 2665–2674. [Google Scholar]
- Sala-Valdes, M.; Ursa, A.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sanchez-Madrid, F.; Yanez-Mo, M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 2006, 281, 19665–19675. [Google Scholar] [CrossRef]
- Berditchevski, F. Complexes of tetraspanins with integrins: More than meets the eye. J. Cell Sci. 2001, 114, 4143–4151. [Google Scholar]
- Miyazaki, T.; Muller, U.; Campbell, K.S. Normal development but differentially altered proliferative responses of lymphocytes in mice lacking CD81. EMBO J. 1997, 16, 4217–4225. [Google Scholar] [CrossRef]
- Tsitsikov, E.N.; Gutierrez-Ramos, J.C.; Geha, R.S. Impaired CD19 expression and signaling, enhanced antibody response to type II T independent antigen and reduction of B-1 cells in CD81-deficient mice. Proc. Natl. Acad. Sci. USA 1997, 94, 10844–10849. [Google Scholar] [CrossRef]
- Maecker, H.T.; Levy, S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 1997, 185, 1505–1510. [Google Scholar] [CrossRef]
- van Zelm, M.C.; Smet, J.; Adams, B.; Mascart, F.; Schandene, L.; Janssen, F.; Ferster, A.; Kuo, C.C.; Levy, S.; van Dongen, J.J.; et al. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest. 2010, 120, 1265–1274. [Google Scholar] [CrossRef]
- Carloni, V.; Mazzocca, A.; Ravichandran, K.S. Tetraspanin CD81 is linked to ERK/MAPKinase signaling by Shc in liver tumor cells. Oncogene 2004, 23, 1566–1574. [Google Scholar] [CrossRef]
- Zhang, X.A.; Bontrager, A.L.; Hemler, M.E. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem. 2001, 276, 25005–25013. [Google Scholar]
- Silvie, O.; Rubinstein, E.; Franetich, J.F.; Prenant, M.; Belnoue, E.; Renia, L.; Hannoun, L.; Eling, W.; Levy, S.; Boucheix, C.; et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 2003, 9, 93–96. [Google Scholar]
- Yalaoui, S.; Zougbede, S.; Charrin, S.; Silvie, O.; Arduise, C.; Farhati, K.; Boucheix, C.; Mazier, D.; Rubinstein, E.; Froissard, P. Hepatocyte permissiveness to Plasmodium infection is conveyed by a short and structurally conserved region of the CD81 large extracellular domain. PLoS Pathog. 2008, 4, e1000010. [Google Scholar] [CrossRef]
- Tham, T.N.; Gouin, E.; Rubinstein, E.; Boucheix, C.; Cossart, P.; Pizarro-Cerda, J. Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect. Immun. 2010, 78, 204–209. [Google Scholar] [CrossRef]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef]
- Cormier, E.G.; Tsamis, F.; Kajumo, F.; Durso, R.J.; Gardner, J.P.; Dragic, T. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 2004, 101, 7270–7274. [Google Scholar] [CrossRef]
- Zhang, J.; Randall, G.; Higginbottom, A.; Monk, P.; Rice, C.M.; McKeating, J.A. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004, 78, 1448–1455. [Google Scholar] [CrossRef]
- Gordon-Alonso, M.; Yanez-Mo, M.; Barreiro, O.; Alvarez, S.; Munoz-Fernandez, M.A.; Valenzuela-Fernandez, A.; Sanchez-Madrid, F. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 2006, 177, 5129–5137. [Google Scholar]
- Alter, M.J. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 2007, 13, 2436–2441. [Google Scholar]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef]
- Arzumanyan, A.; Reis, H.M.; Feitelson, M.A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Canc. 2013, 13, 123–135. [Google Scholar] [CrossRef]
- Marsh, M.; Helenius, A. Virus entry: Open sesame. Cell 2006, 124, 729–740. [Google Scholar] [CrossRef]
- Timpe, J.M.; Stamataki, Z.; Jennings, A.; Hu, K.; Farquhar, M.J.; Harris, H.J.; Schwarz, A.; Desombere, I.; Roels, G.L.; Balfe, P.; et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 2008, 47, 17–24. [Google Scholar]
- Brimacombe, C.L.; Grove, J.; Meredith, L.W.; Hu, K.; Syder, A.J.; Flores, M.V.; Timpe, J.M.; Krieger, S.E.; Baumert, T.F.; Tellinghuisen, T.L.; et al. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J. Virol. 2011, 85, 596–605. [Google Scholar] [CrossRef]
- Schwarz, A.K.; Grove, J.; Hu, K.; Mee, C.J.; Balfe, P.; McKeating, J.A. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J. Virol. 2009, 83, 12407–12414. [Google Scholar] [CrossRef]
- Fofana, I.; Fafi-Kremer, S.; Carolla, P.; Fauvelle, C.; Zahid, M.N.; Turek, M.; Heydmann, L.; Cury, K.; Hayer, J.; Combet, C.; et al. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology 2012, 143, 223–233 e229. [Google Scholar]
- Lupberger, J.; Zeisel, M.B.; Xiao, F.; Thumann, C.; Fofana, I.; Zona, L.; Davis, C.; Mee, C.J.; Turek, M.; Gorke, S.; et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011, 17, 589–595. [Google Scholar]
- Zona, L.; Lupberger, J.; Sidahmed-Adrar, N.; Thumann, C.; Harris, H.J.; Barnes, A.; Florentin, J.; Tawar, R.G.; Xiao, F.; Turek, M.; et al. HRas Signal Transduction Promotes Hepatitis C Virus Cell Entry by Triggering Assembly of the Host Tetraspanin Receptor Complex. Cell Host Microbe 2013, 13, 302–313. [Google Scholar] [CrossRef]
- Hsu, M.; Zhang, J.; Flint, M.; Logvinoff, C.; Cheng-Mayer, C.; Rice, C.M.; McKeating, J.A. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 2003, 100, 7271–7276. [Google Scholar]
- Steinmann, E.; Penin, F.; Kallis, S.; Patel, A.H.; Bartenschlager, R.; Pietschmann, T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007, 3, e103. [Google Scholar] [CrossRef]
- Jirasko, V.; Montserret, R.; Appel, N.; Janvier, A.; Eustachi, L.; Brohm, C.; Steinmann, E.; Pietschmann, T.; Penin, F.; Bartenschlager, R. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 2008, 283, 28546–28562. [Google Scholar] [CrossRef]
- Jones, D.M.; Patel, A.H.; Targett-Adams, P.; McLauchlan, J. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 2009, 83, 2163–2177. [Google Scholar]
- Ma, Y.; Yates, J.; Liang, Y.; Lemon, S.M.; Yi, M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 2008, 82, 7624–7639. [Google Scholar] [CrossRef]
- Gastaminza, P.; Cheng, G.; Wieland, S.; Zhong, J.; Liao, W.; Chisari, F.V. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 2008, 82, 2120–2129. [Google Scholar]
- Huang, H.; Sun, F.; Owen, D.M.; Li, W.; Chen, Y.; Gale, M., Jr.; Ye, J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 2007, 104, 5848–5853. [Google Scholar]
- Benga, W.J.; Krieger, S.E.; Dimitrova, M.; Zeisel, M.B.; Parnot, M.; Lupberger, J.; Hildt, E.; Luo, G.; McLauchlan, J.; Baumert, T.F.; et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 2010, 51, 43–53. [Google Scholar] [CrossRef]
- Bartosch, B.; Dubuisson, J.; Cosset, F.L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 2003, 197, 633–642. [Google Scholar] [CrossRef]
- Bartosch, B.; Vitelli, A.; Granier, C.; Goujon, C.; Dubuisson, J.; Pascale, S.; Scarselli, E.; Cortese, R.; Nicosia, A.; Cosset, F.L. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 2003, 278, 41624–41630. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Zhong, J.; Gastaminza, P.; Cheng, G.; Kapadia, S.; Kato, T.; Burton, D.R.; Wieland, S.F.; Uprichard, S.L.; Wakita, T.; Chisari, F.V. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 9294–9299. [Google Scholar] [CrossRef]
- Lavillette, D.; Tarr, A.W.; Voisset, C.; Donot, P.; Bartosch, B.; Bain, C.; Patel, A.H.; Dubuisson, J.; Ball, J.K.; Cosset, F.L. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 2005, 41, 265–274. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Evans, M.J.; Syder, A.J.; Wolk, B.; Tellinghuisen, T.L.; Liu, C.C.; Maruyama, T.; Hynes, R.O.; Burton, D.R.; McKeating, J.A.; et al. Complete replication of hepatitis C virus in cell culture. Science 2005, 309, 623–626. [Google Scholar] [CrossRef]
- McKeating, J.A.; Zhang, L.Q.; Logvinoff, C.; Flint, M.; Zhang, J.; Yu, J.; Butera, D.; Ho, D.D.; Dustin, L.B.; Rice, C.M.; Balfe, P. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 2004, 78, 8496–8505. [Google Scholar] [CrossRef]
- Mee, C.J.; Harris, H.J.; Farquhar, M.J.; Wilson, G.; Reynolds, G.; Davis, C.; van, I.S.C.; Balfe, P.; McKeating, J.A. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J. Virol. 2009, 83, 6211–6221. [Google Scholar] [CrossRef]
- Koutsoudakis, G.; Herrmann, E.; Kallis, S.; Bartenschlager, R.; Pietschmann, T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 2007, 81, 588–598. [Google Scholar] [CrossRef]
- Akazawa, D.; Date, T.; Morikawa, K.; Murayama, A.; Miyamoto, M.; Kaga, M.; Barth, H.; Baumert, T.F.; Dubuisson, J.; Wakita, T. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 2007, 81, 5036–5045. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Zhang, B.H.; Ishii, K.; Liang, T.J. Novel function of CD81 in controlling hepatitis C virus replication. J. Virol. 2010, 84, 3396–3407. [Google Scholar] [CrossRef]
- Flint, M.; Maidens, C.; Loomis-Price, L.D.; Shotton, C.; Dubuisson, J.; Monk, P.; Higginbottom, A.; Levy, S.; McKeating, J.A. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 1999, 73, 6235–6244. [Google Scholar]
- Petracca, R.; Falugi, F.; Galli, G.; Norais, N.; Rosa, D.; Campagnoli, S.; Burgio, V.; Di Stasio, E.; Giardina, B.; Houghton, M.; et al. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 2000, 74, 4824–4830. [Google Scholar] [CrossRef]
- Drummer, H.E.; Wilson, K.A.; Poumbourios, P. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 2002, 76, 11143–11147. [Google Scholar]
- Masciopinto, F.; Campagnoli, S.; Abrignani, S.; Uematsu, Y.; Pileri, P. The small extracellular loop of CD81 is necessary for optimal surface expression of the large loop, a putative HCV receptor. Virus Res. 2001, 80, 1–10. [Google Scholar] [CrossRef]
- Bertaux, C.; Dragic, T. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 2006, 80, 4940–4948. [Google Scholar] [CrossRef]
- Voisset, C.; Lavie, M.; Helle, F.; Op De Beeck, A.; Bilheu, A.; Bertrand-Michel, J.; Terce, F.; Cocquerel, L.; Wychowski, C.; Vu-Dac, N.; et al. Ceramide enrichment of the plasma membrane induces CD81 internalization and inhibits hepatitis C virus entry. Cell. Microbiol. 2008, 10, 606–617. [Google Scholar] [CrossRef]
- Kapadia, S.B.; Barth, H.; Baumert, T.; McKeating, J.A.; Chisari, F.V. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 2007, 81, 374–383. [Google Scholar]
- Harris, H.J.; Clerte, C.; Farquhar, M.J.; Goodall, M.; Hu, K.; Rassam, P.; Dosset, P.; Wilson, G.K.; Balfe, P.; Ijzendoorn, S.C.; et al. Hepatoma polarization limits CD81 and hepatitis C virus dynamics. Cell. Microbiol. 2013, 15, 430–445. [Google Scholar] [CrossRef]
- Farquhar, M.J.; Hu, K.; Harris, H.J.; Davis, C.; Brimacombe, C.L.; Fletcher, S.J.; Baumert, T.F.; Rappoport, J.Z.; Balfe, P.; McKeating, J.A. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J. Virol. 2012, 86, 4305–4316. [Google Scholar] [CrossRef]
- Flint, M.; von Hahn, T.; Zhang, J.; Farquhar, M.; Jones, C.T.; Balfe, P.; Rice, C.M.; McKeating, J.A. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 2006, 80, 11331–11342. [Google Scholar] [CrossRef]
- Higginbottom, A.; Quinn, E.R.; Kuo, C.C.; Flint, M.; Wilson, L.H.; Bianchi, E.; Nicosia, A.; Monk, P.N.; McKeating, J.A.; Levy, S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 2000, 74, 3642–3649. [Google Scholar] [CrossRef]
- Davis, C.; Harris, H.J.; Hu, K.; Drummer, H.E.; McKeating, J.A.; Mullins, J.G.; Balfe, P. In silico directed mutagenesis identifies the CD81/claudin-1 hepatitis C virus receptor interface. Cell. Microbiol. 2012, 14, 1892–1903. [Google Scholar] [CrossRef]
- Montpellier, C.; Tews, B.A.; Poitrimole, J.; Rocha-Perugini, V.; D'Arienzo, V.; Potel, J.; Zhang, X.A.; Rubinstein, E.; Dubuisson, J.; Cocquerel, L. Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 2011, 286, 13954–13965. [Google Scholar] [CrossRef]
- Brazzoli, M.; Bianchi, A.; Filippini, S.; Weiner, A.; Zhu, Q.; Pizza, M.; Crotta, S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 2008, 82, 8316–8329. [Google Scholar] [CrossRef]
- Mazzocca, A.; Sciammetta, S.C.; Carloni, V.; Cosmi, L.; Annunziato, F.; Harada, T.; Abrignani, S.; Pinzani, M. Binding of hepatitis C virus envelope protein E2 to CD81 up-regulates matrix metalloproteinase-2 in human hepatic stellate cells. J. Biol. Chem. 2005, 280, 11329–11339. [Google Scholar]
- Zeisel, M.B.; Felmlee, D.J.; Baumert, T.F. Hepatitis C virus entry. Curr. Top. Microbiol. Immunol. 2013, 369, 87–112. [Google Scholar]
- Park, J.H.; Park, S.; Yang, J.S.; Kwon, O.S.; Kim, S.; Jang, S.K. Discovery of cellular proteins required for the early steps of HCV infection using integrative genomics. PLoS One 2013, 8, e60333. [Google Scholar]
- Levy, S.; Shoham, T. Protein-protein interactions in the tetraspanin web. Physiology 2005, 20, 218–224. [Google Scholar] [CrossRef]
- Harris, H.J.; Davis, C.; Mullins, J.G.; Hu, K.; Goodall, M.; Farquhar, M.J.; Mee, C.J.; McCaffrey, K.; Young, S.; Drummer, H.; et al. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 2010, 285, 21092–21102. [Google Scholar] [CrossRef]
- Harris, H.J.; Farquhar, M.J.; Mee, C.J.; Davis, C.; Reynolds, G.M.; Jennings, A.; Hu, K.; Yuan, F.; Deng, H.; Hubscher, S.G.; et al. CD81 and claudin 1 coreceptor association: Role in hepatitis C virus entry. J. Virol. 2008, 82, 5007–5020. [Google Scholar] [CrossRef]
- Furuse, M.; Hata, M.; Furuse, K.; Yoshida, Y.; Haratake, A.; Sugitani, Y.; Noda, T.; Kubo, A.; Tsukita, S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 2002, 156, 1099–1111. [Google Scholar] [CrossRef]
- Evans, M.J.; von Hahn, T.; Tscherne, D.M.; Syder, A.J.; Panis, M.; Wolk, B.; Hatziioannou, T.; McKeating, J.A.; Bieniasz, P.D.; Rice, C.M. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 2007, 446, 801–805. [Google Scholar] [CrossRef]
- Krieger, S.E.; Zeisel, M.B.; Davis, C.; Thumann, C.; Harris, H.J.; Schnober, E.K.; Mee, C.; Soulier, E.; Royer, C.; Lambotin, M.; et al. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 2010, 51, 1144–1157. [Google Scholar] [CrossRef]
- Douam, F.; Dao Thi, V.L.; Maurin, G.; Fresquet, J.; Mompelat, D.; Zeisel, M.B.; Baumert, T.F.; Cosset, F.L.; Lavillette, D. A critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 2013. [Google Scholar] [CrossRef]
- Reynolds, G.M.; Harris, H.J.; Jennings, A.; Hu, K.; Grove, J.; Lalor, P.F.; Adams, D.H.; Balfe, P.; Hubscher, S.G.; McKeating, J.A. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology 2008, 47, 418–427. [Google Scholar]
- Yang, W.; Qiu, C.; Biswas, N.; Jin, J.; Watkins, S.C.; Montelaro, R.C.; Coyne, C.B.; Wang, T. Correlation of the tight junction-like distribution of Claudin-1 to the cellular tropism of hepatitis C virus. J. Biol. Chem. 2008, 283, 8643–8653. [Google Scholar]
- Farquhar, M.J.; Harris, H.J.; Diskar, M.; Jones, S.; Mee, C.J.; Nielsen, S.U.; Brimacombe, C.L.; Molina, S.; Toms, G.L.; Maurel, P.; et al. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 2008, 82, 8797–8811. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, Y.; Machida, K.; Lai, M.M.; Luo, G.; Foung, S.K.; Ou, J.H. Transient Activation of the PI3K-AKT Pathway by Hepatitis C Virus to Enhance Viral Entry. J. Biol. Chem. 2012, 287, 41922–41930. [Google Scholar]
- Diao, J.; Pantua, H.; Ngu, H.; Komuves, L.; Diehl, L.; Schaefer, G.; Kapadia, S.B. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J. Virol. 2012, 86, 10935–10949. [Google Scholar] [CrossRef]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Lupberger, J.; Fofana, I.; Baumert, T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C - perspectives and challenges. J. Hepatol. 2013, 58, 375–384. [Google Scholar] [CrossRef]
- Fofana, I.; Xiao, F.; Thumann, C.; Turek, M.; Zona, L.; Tawar, R.G.; Grunert, F.; Thompson, J.; Zeisel, M.B.; Baumert, T.F. A novel monoclonal anti-CD81 antibody produced by genetic immunization efficiently inhibits Hepatitis C virus cell-cell transmission. PLoS One 2013, 8, e64221. [Google Scholar] [CrossRef]
- Morikawa, K.; Zhao, Z.; Date, T.; Miyamoto, M.; Murayama, A.; Akazawa, D.; Tanabe, J.; Sone, S.; Wakita, T. The roles of CD81 and glycosaminoglycans in the adsorption and uptake of infectious HCV particles. J. Med. Virol. 2007, 79, 714–723. [Google Scholar] [CrossRef]
- Meuleman, P.; Hesselgesser, J.; Paulson, M.; Vanwolleghem, T.; Desombere, I.; Reiser, H.; Leroux-Roels, G. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 2008, 48, 1761–1768. [Google Scholar] [CrossRef]
- Van Compernolle, S.E.; Wiznycia, A.V.; Rush, J.R.; Dhanasekaran, M.; Baures, P.W.; Todd, S.C. Small molecule inhibition of hepatitis C virus E2 binding to CD81. Virology 2003, 314, 371–380. [Google Scholar] [CrossRef]
- Fofana, I.; Krieger, S.E.; Grunert, F.; Glauben, S.; Xiao, F.; Fafi-Kremer, S.; Soulier, E.; Royer, C.; Thumann, C.; Mee, C.J.; et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 2010, 139, 953–964, 964, e951–e954. [Google Scholar] [CrossRef]
- Si, Y.; Liu, S.; Liu, X.; Jacobs, J.L.; Cheng, M.; Niu, Y.; Jin, Q.; Wang, T.; Yang, W. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology 2012, 56, 507–515. [Google Scholar]
- Hotzel, I.; Chiang, V.; Diao, J.; Pantua, H.; Maun, H.R.; Kapadia, S.B. Efficient production of antibodies against a mammalian integral membrane protein by phage display. Protein Eng. Des. Sel. 2011, 24, 679–689. [Google Scholar] [CrossRef]
- Pilot-Matias, T.; Lacy, S.; Ng, T.; Barbon, J.; Fung, E.; Pithawalla, R.; Barlow, E.; Kutskova, Y.; Hsieh, C.-M.; DiGiammarino, E.; et al. Evaluation of a panel of anti-CD81 antibodies using human liver-uPA/SCID mice. In Proceedings of the 17th International Meeting on Hepatitis C Virus and Related Viruses, Yokohama, Japan, 10–14 September 2010.
- Bardou-Jacquet, E.; Lorho, R.; Guyader, D. Kinase inhibitors in the treatment of chronic hepatitis C virus. Gut 2011, 60, 879–880. [Google Scholar] [CrossRef]
- Lupberger, J.; Duong, F.H.; Fofana, I.; Zona, L.; Xiao, F.; Thumann, C.; Durand, S.C.; Pessaux, P.; Zeisel, M.B.; Heim, M.H.; Baumert, T.F. EGFR signaling impairs the antiviral activity of interferon-alpha. Hepatology 2013, 58, 1225–1235. [Google Scholar] [CrossRef]
- Reuther, G.W.; Der, C.J. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr. Opin. Cell Biol. 2000, 12, 157–165. [Google Scholar] [CrossRef]
- Kohl, N.E.; Mosser, S.D.; deSolms, S.J.; Giuliani, E.A.; Pompliano, D.L.; Graham, S.L.; Smith, R.L.; Scolnick, E.M.; Oliff, A.; Gibbs, J.B. Selective inhibition of ras-dependent transformation by a farnesyltransferase inhibitor. Science 1993, 260, 1934–1937. [Google Scholar]
- James, G.L.; Goldstein, J.L.; Brown, M.S.; Rawson, T.E.; Somers, T.C.; McDowell, R.S.; Crowley, C.W.; Lucas, B.K.; Levinson, A.D.; Marsters, J.C., Jr. Benzodiazepine peptidomimetics: Potent inhibitors of Ras farnesylation in animal cells. Science 1993, 260, 1937–1942. [Google Scholar]
- Marom, M.; Haklai, R.; Ben-Baruch, G.; Marciano, D.; Egozi, Y.; Kloog, Y. Selective inhibition of Ras-dependent cell growth by farnesylthiosalisylic acid. J. Biol. Chem. 1995, 270, 22263–22270. [Google Scholar]
- Sawada, K.; Ohyagi-Hara, C.; Kimura, T.; Morishige, K. Integrin inhibitors as a therapeutic agent for ovarian cancer. J. Oncol. 2012, 2012, 915140. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
