Functional RNA Elements in the Dengue Virus Genome
Abstract
:1. Introduction: DENV Life Cycle
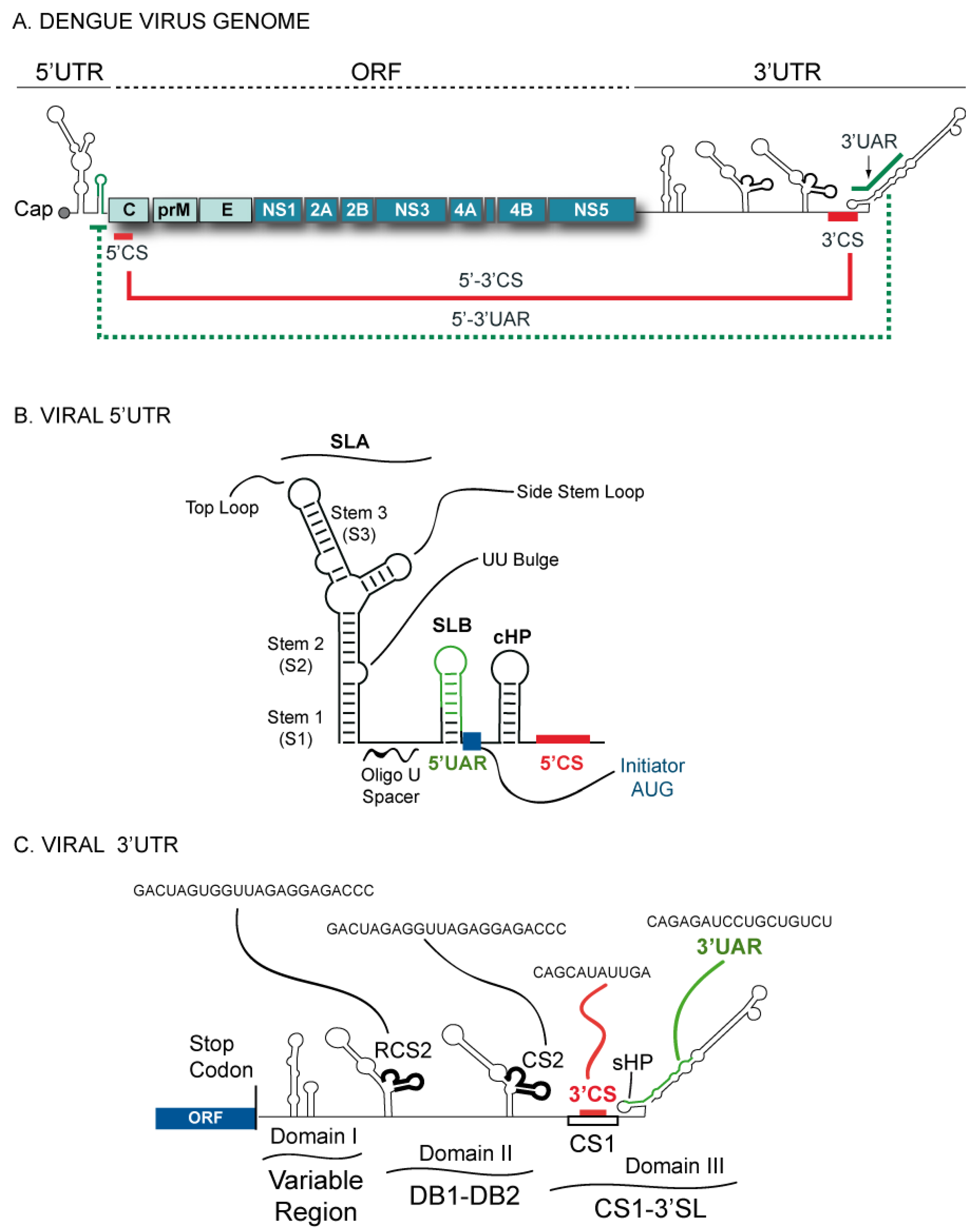
2. RNA Structures in the DENV Genome
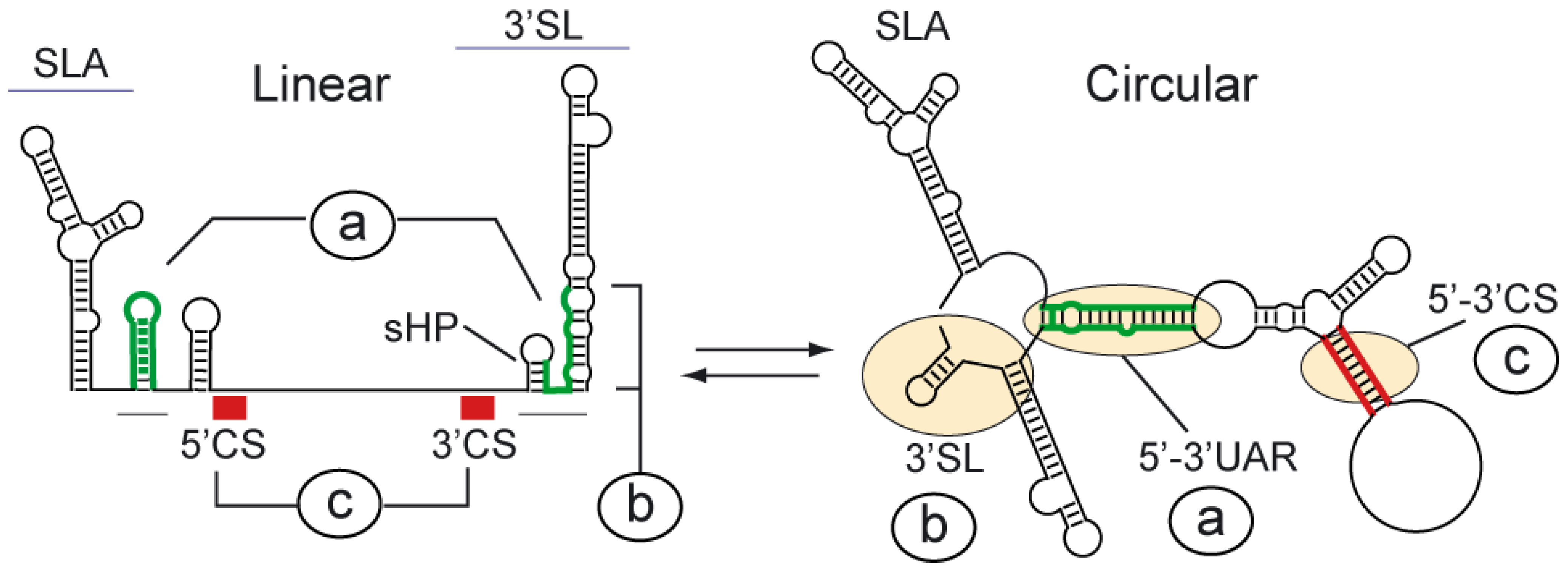
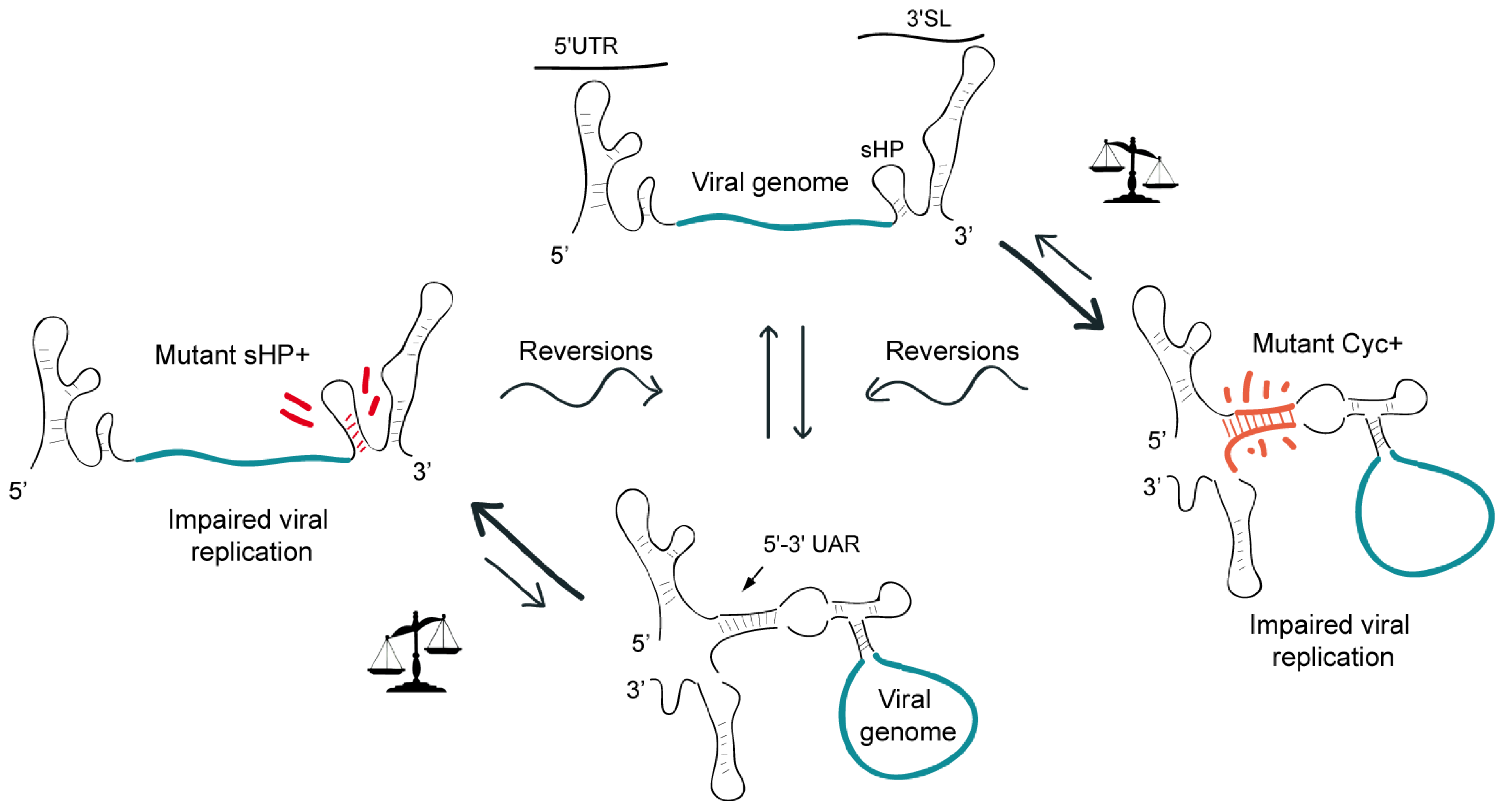
3. Functional Significance of DENV Genome Cyclization
4. Elements of the SLA Promoter for NS5 Binding and Polymerase Activity
5. RNA Synthesis Silencing by the 3’SL Structure
6. Elements Downstream of the SLA that Modulate RNA Synthesis
7. Dynamic Conformations of the DENV Genome Are Necessary for RNA Synthesis
8. Future Prospects
Acknowledgments
Conflict of Interest
References and Notes
- Lindenbach, B.D.; Rice, C.M. Flaviviridae: The viruses and their replication. In Fields Virology; Lippincott-Raven: Philadelphia, PA, USA, 2007; Volume 1, pp. 1101–1152. [Google Scholar]
- Rice, C.M.; Lenches, E.M.; Eddy, S.R.; Shin, S.J.; Sheets, R.L.; Strauss, J.H. Nucleotide sequence of yellow fever virus: Implications for flavivirus gene expression and evolution. Science 1985, 229, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Westaway, E.G.; Mackenzie, J.M.; Khromykh, A.A. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 2003, 59, 99–140. [Google Scholar]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [PubMed]
- Stadler, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997, 71, 8475–8481. [Google Scholar] [CrossRef]
- Allison, S.L.; Tao, Y.J.; O’Riordain, G.; Mandl, C.W.; Harrison, S.C.; Heinz, F.X. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J. Virol. 2003, 77, 11357–11366. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Sachidanandam, R.; Garcia-Sastre, A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 16303–16308. [Google Scholar] [CrossRef] [PubMed]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2’-o methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef]
- Gritsun, T.S.; Gould, E.A. Origin and evolution of flavivirus 5’UTRs and panhandles: Trans-terminal duplications? Virology 2007, 366, 8–15. [Google Scholar] [CrossRef]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef]
- Brinton, M.A.; Dispoto, J.H. Sequence and secondary structure analysis of the 5’-terminal region of flavivirus genome RNA. Virology 1988, 162, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Leyssen, P.; Charlier, N.; Lemey, P.; Billoir, F.; Vandamme, A.M.; De Clercq, E.; de Lamballerie, X.; Neyts, J. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology 2002, 293, 125–140. [Google Scholar] [CrossRef]
- Lodeiro, M.F.; Filomatori, C.V.; Gamarnik, A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009, 83, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5’ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nomaguchi, M.; Padmanabhan, R.; Markoff, L. Specific requirements for elements of the 5’ and 3’ terminal regions in flavivirus RNA synthesis and viral replication. Virology 2008, 374, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Lodeiro, M.F.; Luduena, S.J.; Pietrasanta, L.I.; Gamarnik, A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005, 79, 6631–6643. [Google Scholar] [CrossRef]
- Polacek, C.; Foley, J.E.; Harris, E. Conformational changes in the solution structure of the dengue virus 5’ end in the presence and absence of the 3’ untranslated region. J. Virol. 2009, 83, 1161–1166. [Google Scholar] [CrossRef]
- Gamarnik, A.V. Role of the dengue virus 5’ and 3’ untranslated regions in viral replicación. In Frontiers in Dengue Virus Reserach; Hanley, K.A., Weaver, S.C., Eds.; Caister Academic Press: Norfolk, UK, 2010; pp. 55–78. [Google Scholar]
- Gritsun, T.S.; Venugopal, K.; Zanotto, P.M.; Mikhailov, M.V.; Sall, A.A.; Holmes, E.C.; Polkinghorne, I.; Frolova, T.V.; Pogodina, V.V.; Lashkevich, V.A.; et al. Complete sequence of two tick-borne flaviviruses isolated from siberia and the uk: Analysis and significance of the 5’ and 3’-UTRs. Virus Res. 1997, 49, 27–39. [Google Scholar] [CrossRef]
- Mandl, C.W.; Holzmann, H.; Kunz, C.; Heinz, F.X. Complete genomic sequence of powassan virus: Evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 1993, 194, 173–184. [Google Scholar] [CrossRef]
- Clyde, K.; Barrera, J.; Harris, E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and west nile virus RNA synthesis. Virology 2008, 379, 314–323. [Google Scholar] [CrossRef]
- Alvarez, D.E.; De Lella Ezcurra, A.L.; Fucito, S.; Gamarnik, A.V. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005, 339, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Mammen, M.P., Jr.; Klungthong, C.; Chinnawirotpisan, P.; Vaughn, D.W.; Nimmannitya, S.; Kalayanarooj, S.; Holmes, E.C.; Zhang, C. Comparative analysis reveals no consistent association between the secondary structure of the 3’-untranslated region of dengue viruses and disease syndrome. J. Gen. Virol. 2006, 87, 2595–2603. [Google Scholar] [CrossRef]
- Shurtleff, A.C.; Beasley, D.W.; Chen, J.J.; Ni, H.; Suderman, M.T.; Wang, H.; Xu, R.; Wang, E.; Weaver, S.C.; Watts, D.M.; et al. Genetic variation in the 3’ non-coding region of dengue viruses. Virology 2001, 281, 75–87. [Google Scholar] [CrossRef]
- Silva, R.L.; de Silva, A.M.; Harris, E.; MacDonald, G.H. Genetic analysis of dengue 3 virus subtype III 5’ and 3’ non-coding regions. Virus Res. 2008, 135, 320–325. [Google Scholar] [CrossRef]
- Roche, C.; Cassar, O.; Laille, M.; Murgue, B. Dengue-3 virus genomic differences that correlate with in vitro phenotype on a human cell line but not with disease severity. Microbes Infect. 2007, 9, 63–69. [Google Scholar] [CrossRef]
- Aquino, V.H.; Anatriello, E.; Goncalves, P.F.; EV, D.A.S.; Vasconcelos, P.F.; Vieira, D.S.; Batista, W.C.; Bobadilla, M.L.; Vazquez, C.; Moran, M.; et al. Molecular epidemiology of dengue type 3 virus in brazil and paraguay, 2002–2004. Am. J. Trop. Med. Hyg. 2006, 75, 710–715. [Google Scholar] [CrossRef]
- Vasilakis, N.; Fokam, E.B.; Hanson, C.T.; Weinberg, E.; Sall, A.A.; Whitehead, S.S.; Hanley, K.A.; Weaver, S.C. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology 2008, 377, 296–307. [Google Scholar] [CrossRef]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3’ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987, 198, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Gritsun, T.S.; Gould, E.A. Direct repeats in the 3’ untranslated regions of mosquito-borne flaviviruses: Possible implications for virus transmission. J. Gen. Virol. 2006, 87, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Olsthoorn, R.C.; Bol, J.F. Sequence comparison and secondary structure analysis of the 3’ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 2001, 7, 1370–1377. [Google Scholar]
- Romero, T.A.; Tumban, E.; Jun, J.; Lott, W.B.; Hanley, K.A. Secondary structure of dengue virus type 4 3’ untranslated region: Impact of deletion and substitution mutations. J. Gen. Virol. 2006, 87, 3291–3296. [Google Scholar] [CrossRef]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of cis-acting elements in the 3’-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef] [PubMed]
- Men, R.; Bray, M.; Clark, D.; Chanock, R.M.; Lai, C.J. Dengue type 4 virus mutants containing deletions in the 3’ noncoding region of the RNA genome: Analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 1996, 70, 3930–3937. [Google Scholar] [CrossRef]
- Mandl, C.W.; Holzmann, H.; Meixner, T.; Rauscher, S.; Stadler, P.F.; Allison, S.L.; Heinz, F.X. Spontaneous and engineered deletions in the 3’ noncoding region of tick-borne encephalitis virus: Construction of highly attenuated mutants of a flavivirus. J. Virol. 1998, 72, 2132–2140. [Google Scholar] [CrossRef]
- Bredenbeek, P.J.; Kooi, E.A.; Lindenbach, B.; Huijkman, N.; Rice, C.M.; Spaan, W.J. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 2003, 84, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Tilgner, M.; Bernard, K.A.; Shi, P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3’ untranslated region of west nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003, 77, 10004–10014. [Google Scholar] [CrossRef] [PubMed]
- Takegami, T.; Washizu, M.; Yasui, K. Nucleotide sequence at the 3’ end of Japanese encephalitis virus genomic RNA. Virology 1986, 152, 483–486. [Google Scholar] [CrossRef]
- Proutski, V.; Gould, E.A.; Holmes, E.C. Secondary structure of the 3’ untranslated region of flaviviruses: Similarities and differences. Nucleic Acids Res. 1997, 25, 1194–1202. [Google Scholar] [CrossRef]
- Grange, T.; Bouloy, M.; Girard, M. Stable secondary structures at the 3’-end of the genome of yellow fever virus (17 D vaccine strain). FEBS Lett. 1985, 188, 159–163. [Google Scholar] [CrossRef]
- Deng, R.; Brock, K.V. 5’ and 3’ untranslated regions of pestivirus genome: Primary and secondary structure analyses. Nucleic Acids Res. 1993, 21, 1949–1957. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A.; Fernandez, A.V.; Dispoto, J.H. The 3’-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 1986, 153, 113–121. [Google Scholar] [CrossRef]
- Blight, K.J.; Rice, C.M. Secondary structure determination of the conserved 98-base sequence at the 3’ terminus of hepatitis c virus genome RNA. J. Virol. 1997, 71, 7345–7352. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Falgout, B.; Markoff, L. Identification of specific nucleotide sequences within the conserved 3’-SL in the dengue type 2 virus genome required for replication. J. Virol. 1998, 72, 7510–7522. [Google Scholar] [CrossRef] [PubMed]
- Tilgner, M.; Deas, T.S.; Shi, P.Y. The flavivirus-conserved penta-nucleotide in the 3’ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 2005, 331, 375–386. [Google Scholar] [CrossRef]
- Yu, L.; Markoff, L. The topology of bulges in the long stem of the flavivirus 3’ stem-loop is a major determinant of RNA replication competence. J. Virol. 2005, 79, 2309–2324. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef]
- You, S.; Padmanabhan, R. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3’-end of exogenous viral RNA templates requires 5’- and 3’-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999, 274, 33714–33722. [Google Scholar] [CrossRef]
- Markoff, L. 5’ and 3’ NCRs in Flavivirus RNA. In The Flaviviruses; Elsevier Academic Press: San Diego, CA, USA, 2003; Volume 59, pp. 177–223. [Google Scholar]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef]
- Friebe, P.; Shi, P.Y.; Harris, E. The 5’ and 3’ downstream aug region elements are required for mosquito-borne flavivirus RNA replication. J. Virol. 2011, 85, 1900–1905. [Google Scholar] [CrossRef]
- Corver, J.; Lenches, E.; Smith, K.; Robison, R.A.; Sando, T.; Strauss, E.G.; Strauss, J.H. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 2003, 77, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Filomatori, C.V.; Gamarnik, A.V. Functional analysis of dengue virus cyclization sequences located at the 5’ and 3’UTRs. Virology 2008, 375, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Lin, W.; Zhang, J.; Simon, A.E.; Kushner, D.B. Structural plasticity and rapid evolution in a viral RNA revealed by in vivo genetic selection. J. Virol. 2009, 83, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.M.; Hoenninger, V.M.; Thurner, C.; Mandl, C.W. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 2006, 80, 4099–4113. [Google Scholar] [CrossRef]
- Alvarez, D.E.; Lodeiro, M.F.; Filomatori, C.V.; Fucito, S.; Mondotte, J.A.; Gamarnik, A.V. Structural and functional analysis of dengue virus RNA. Novartis Found. Symp. 2006, 277, 120–132; discussion 132–125, 251–123. [Google Scholar] [PubMed]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef]
- Yap, T.L.; Xu, T.; Chen, Y.L.; Malet, H.; Egloff, M.P.; Canard, B.; Vasudevan, S.G.; Lescar, J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007, 81, 4753–4765. [Google Scholar] [CrossRef]
- Ackermann, M.; Padmanabhan, R. De novo synthesis of RNA by the dengue virus RNA- dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001, 276, 39926–39937. [Google Scholar] [CrossRef]
- Selisko, B.; Dutartre, H.; Guillemot, J.C.; Debarnot, C.; Benarroch, D.; Khromykh, A.; Despres, P.; Egloff, M.P.; Canard, B. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 2006, 351, 145–158. [Google Scholar] [CrossRef]
- Nomaguchi, M.; Ackermann, M.; Yon, C.; You, S.; Padmanabhan, R.; Padmanbhan, R. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J. Virol. 2003, 77, 8831–8842. [Google Scholar] [CrossRef]
- Filomatori, C.V.; Iglesias, N.G.; Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011, 286, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.G.; Filomatori, C.V.; Gamarnik, A.V. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J. Virol. 2011, 85, 5745–5756. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Falgout, B.; Markoff, L.; Padmanabhan, R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5’- and 3’-terminal regions that influence RNA structure. J. Biol. Chem. 2001, 276, 15581–15591. [Google Scholar] [CrossRef]
- Teramoto, T.; Kohno, Y.; Mattoo, P.; Markoff, L.; Falgout, B.; Padmanabhan, R. Genome 3’-end repair in dengue virus type 2. RNA 2008, 14, 2645–2656. [Google Scholar] [CrossRef]
- Simon, A.E.; Gehrke, L. RNA conformational changes in the life cycles of RNA viruses, viroids, and virus-associated RNAs. Biochim. Biophys. Acta 2009, 1789, 571–583. [Google Scholar] [CrossRef]
- Wu, B.; Pogany, J.; Na, H.; Nicholson, B.L.; Nagy, P.D.; White, K.A. A discontinuous RNA platform mediates RNA virus replication: Building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009, 5, e1000323. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Montalvo, B.M.; Medina, F.; del Angel, R.M. La protein binds to NS5 and NS3 and to the 5’ and 3’ ends of dengue 4 virus RNA. Virus Res. 2004, 102, 141–150. [Google Scholar] [CrossRef]
- Yocupicio-Monroy, M.; Padmanabhan, R.; Medina, F.; del Angel, R.M. Mosquito La protein binds to the 3’ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 2007, 357, 29–40. [Google Scholar] [CrossRef]
- Yocupicio-Monroy, R.M.; Medina, F.; Reyes-del Valle, J.; del Angel, R.M. Cellular proteins from human monocytes bind to dengue 4 virus minus-strand 3’ untranslated region RNA. J. Virol. 2003, 77, 3067–3076. [Google Scholar] [CrossRef]
- Paranjape, S.M.; Harris, E. Y box-binding protein-1 binds to the dengue virus 3’-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007, 282, 30497–30508. [Google Scholar] [CrossRef]
- De Nova-Ocampo, M.; Villegas-Sepulveda, N.; del Angel, R.M. Translation elongation factor- 1alpha, La, and PTB interact with the 3’ untranslated region of dengue 4 virus RNA. Virology 2002, 295, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.L.; Brinton, M.A. Translation elongation factor-1 alpha interacts with the 3’ stem- loop region of West Nile virus genomic RNA. J. Virol. 1997, 71, 6433–6444. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Kedersha, N.; Anderson, P.; Emara, M.; Swiderek, K.M.; Moreno, G.T.; Brinton, M.A. Cell proteins TIA-1 and TIAR interact with the 3’ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002, 76, 11989–12000. [Google Scholar] [CrossRef]
- Ta, M.; Vrati, S. Mov34 protein from mouse brain interacts with the 3’ noncoding region of Japanese encephalitis virus. J. Virol. 2000, 74, 5108–5115. [Google Scholar] [CrossRef]
- Vashist, S.; Anantpadma, M.; Sharma, H.; Vrati, S. La protein binds the predicted loop structures in the 3’ non-coding region of Japanese encephalitis virus genome: Role in virus replication. J. Gen. Virol. 2009, 90, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Gomila, R.C.; Martin, G.W.; Gehrke, L. NF90 binds the dengue virus RNA 3’ terminus and is a positive regulator of dengue virus replication. PLoS ONE 2011, 6, e16687. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (https://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gebhard, L.G.; Filomatori, C.V.; Gamarnik, A.V. Functional RNA Elements in the Dengue Virus Genome. Viruses 2011, 3, 1739-1756. https://doi.org/10.3390/v3091739
Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA Elements in the Dengue Virus Genome. Viruses. 2011; 3(9):1739-1756. https://doi.org/10.3390/v3091739
Chicago/Turabian StyleGebhard, Leopoldo G., Claudia V. Filomatori, and Andrea V. Gamarnik. 2011. "Functional RNA Elements in the Dengue Virus Genome" Viruses 3, no. 9: 1739-1756. https://doi.org/10.3390/v3091739
APA StyleGebhard, L. G., Filomatori, C. V., & Gamarnik, A. V. (2011). Functional RNA Elements in the Dengue Virus Genome. Viruses, 3(9), 1739-1756. https://doi.org/10.3390/v3091739



