Distinctive Temporal Profiles of Interferon-Stimulated Genes in Natural Infection, Viral Challenge, and Vaccination
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptome Datasets on Natural Infection
2.2. Transcriptome Dataset on Viral Challenge
2.3. Transcriptome Datasets on Vaccination
2.4. Data Analysis of Transcriptional Response to Infection and Vaccination
3. Results
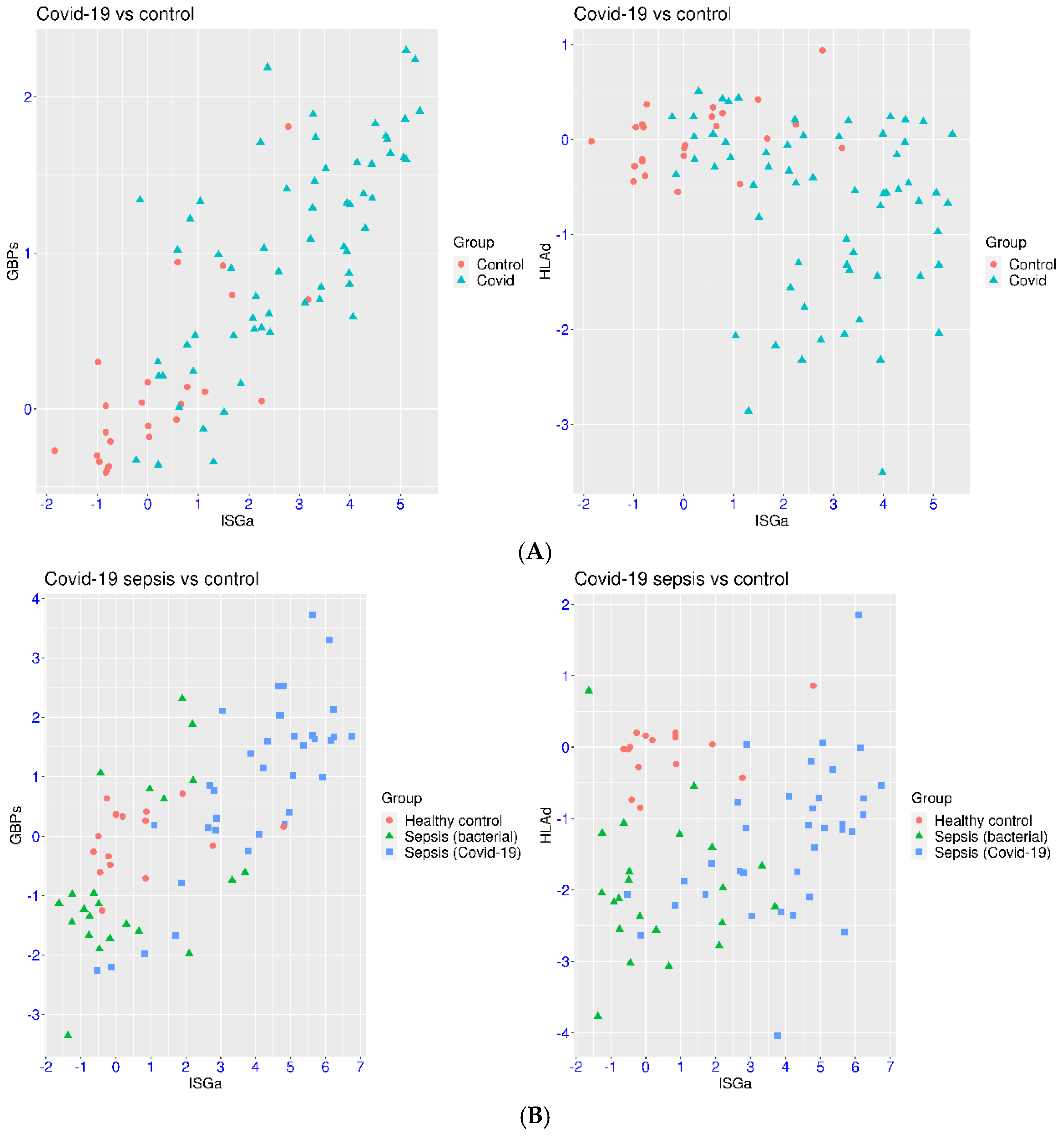
3.1. Selection of Marker Genes for IFN Signaling
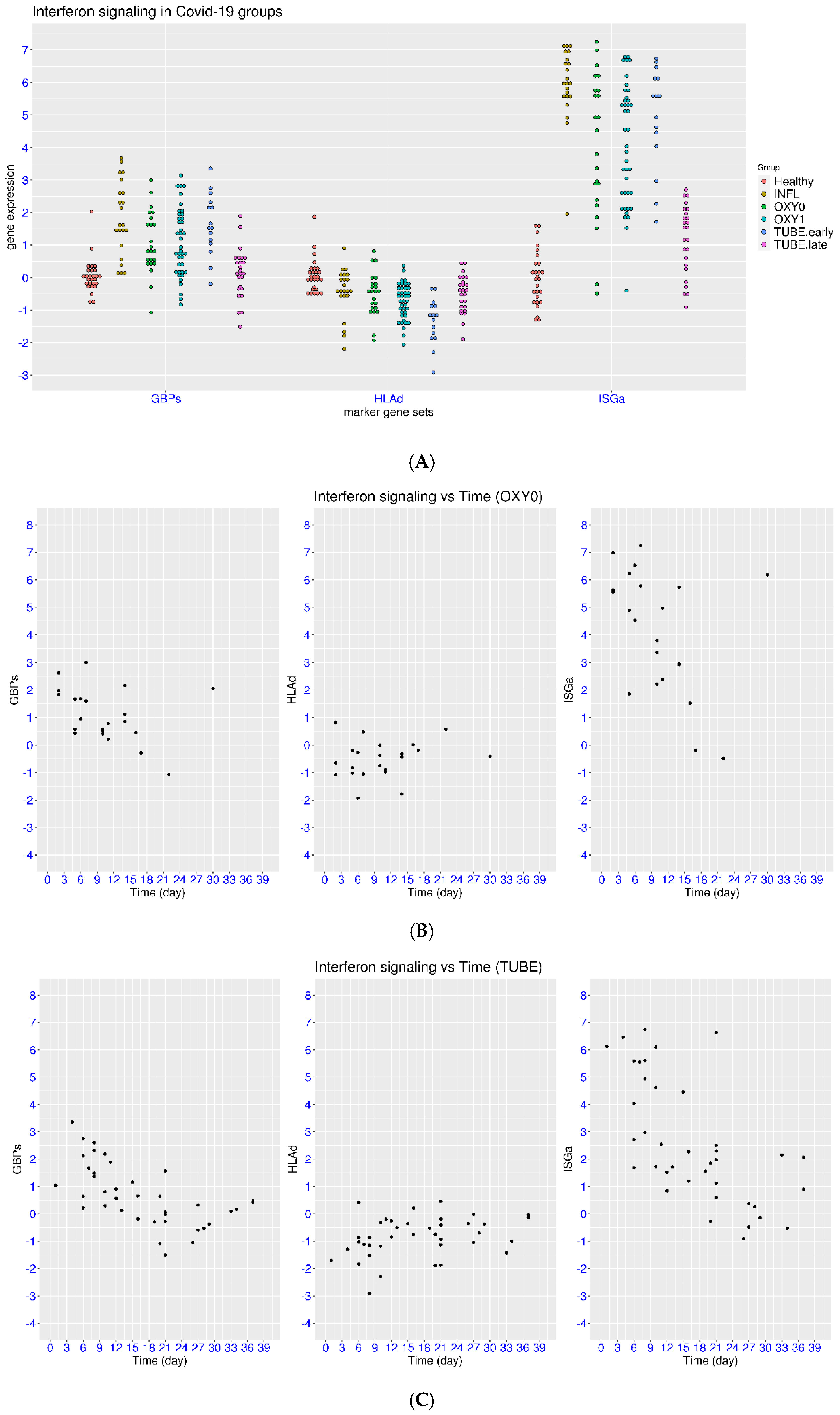
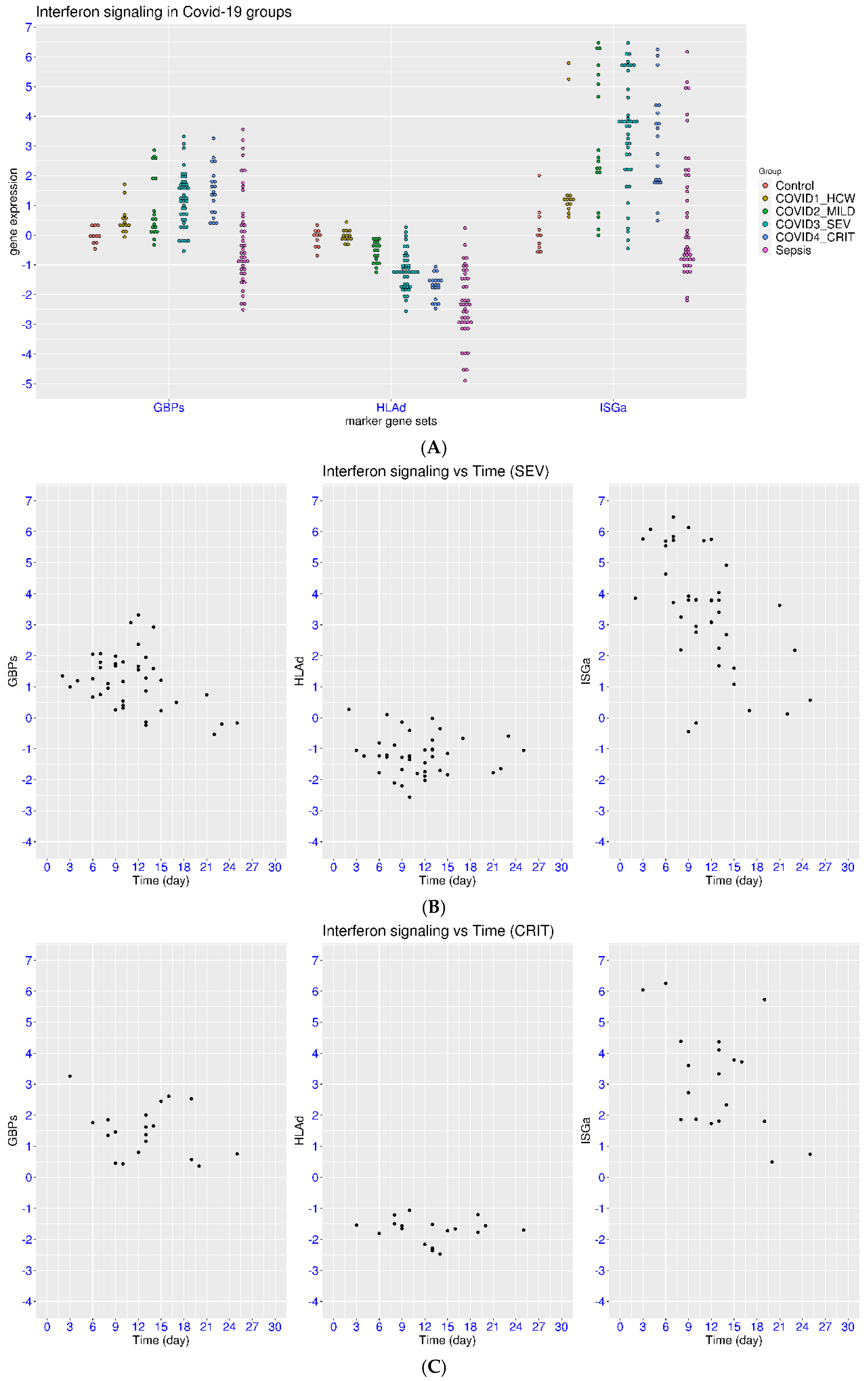
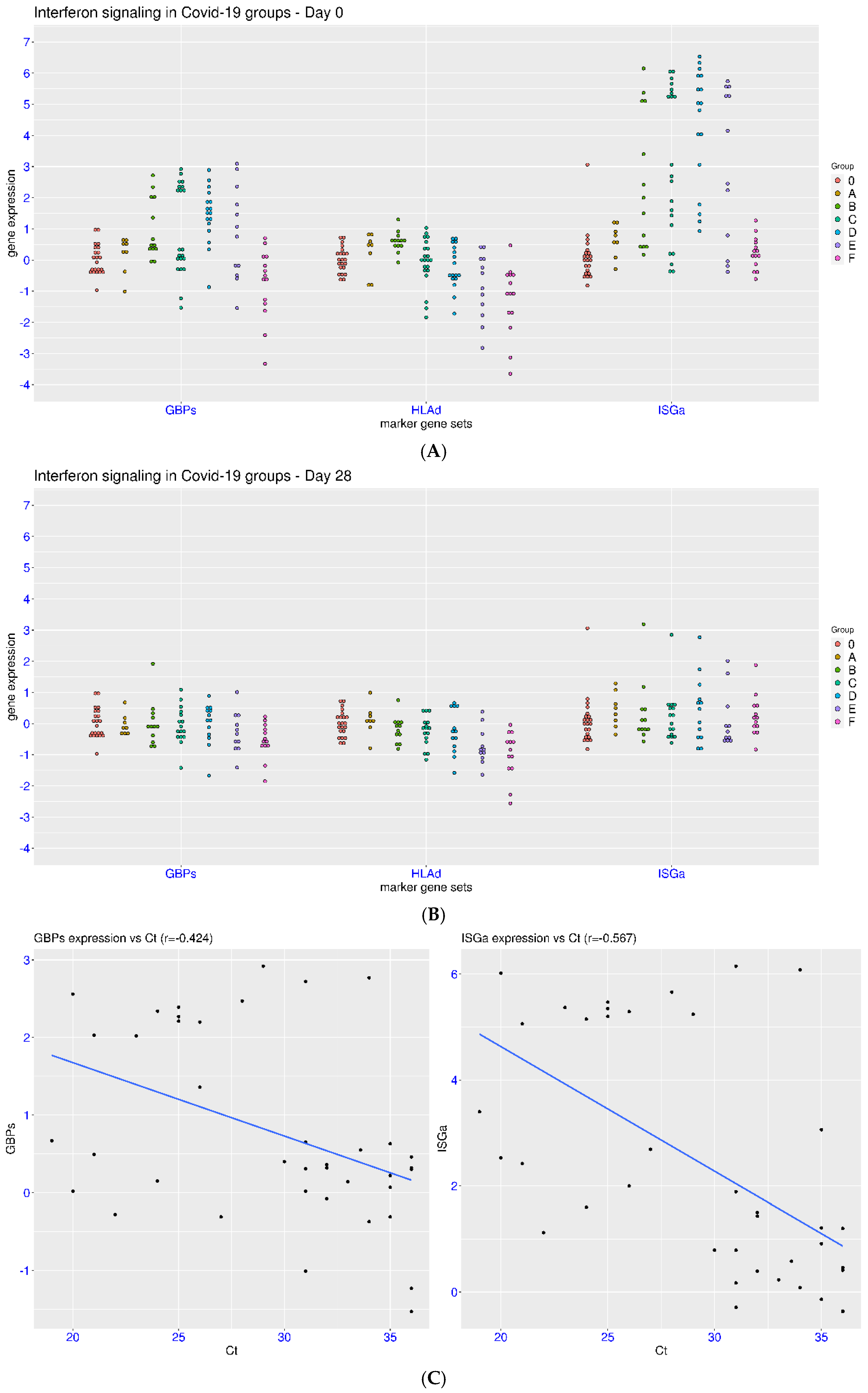
3.2. IFN Signaling in Hospitalized Patients with COVID-19
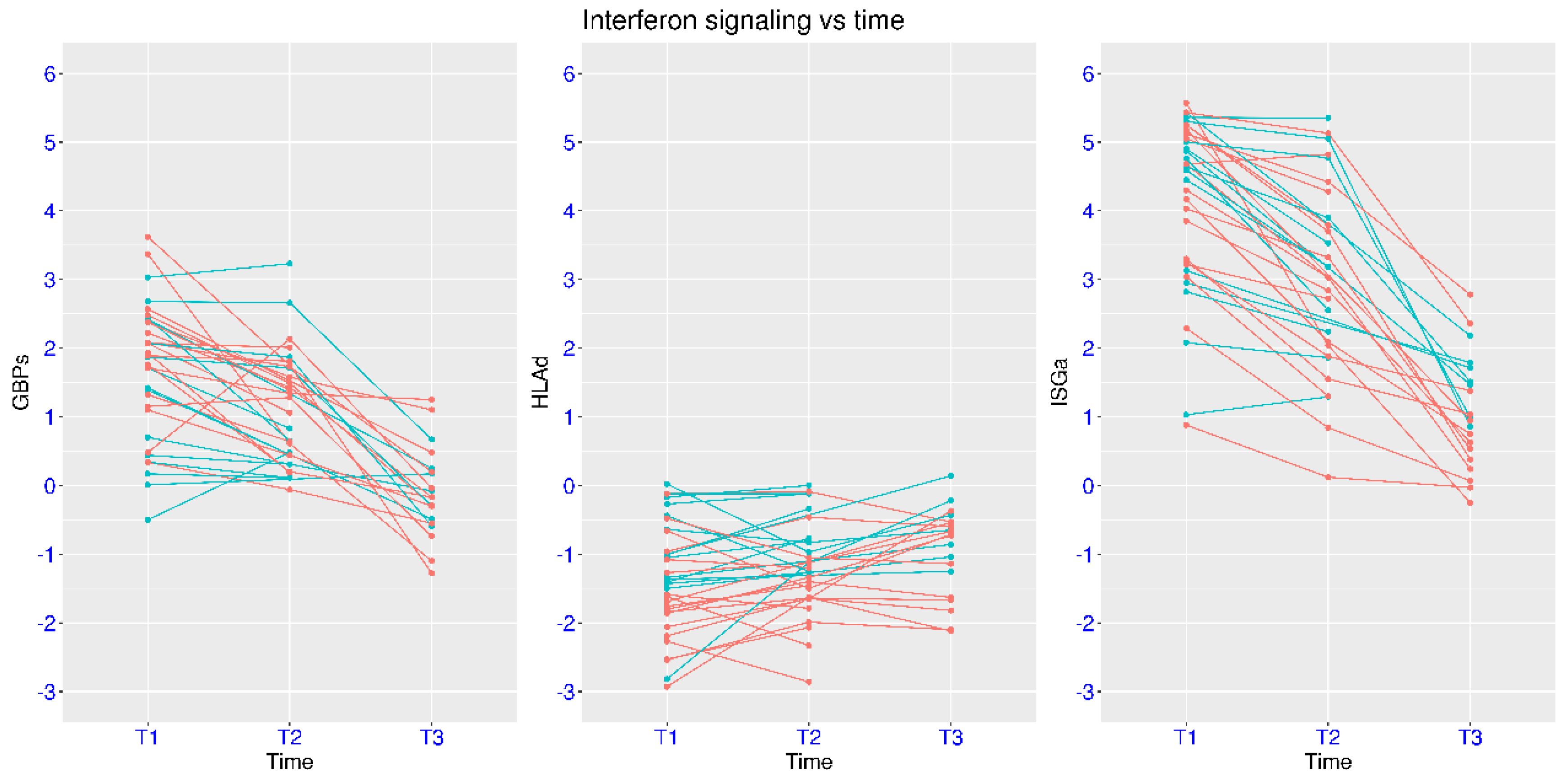
3.3. Disease Severity, Time, and IFN Signaling in Natural Infection
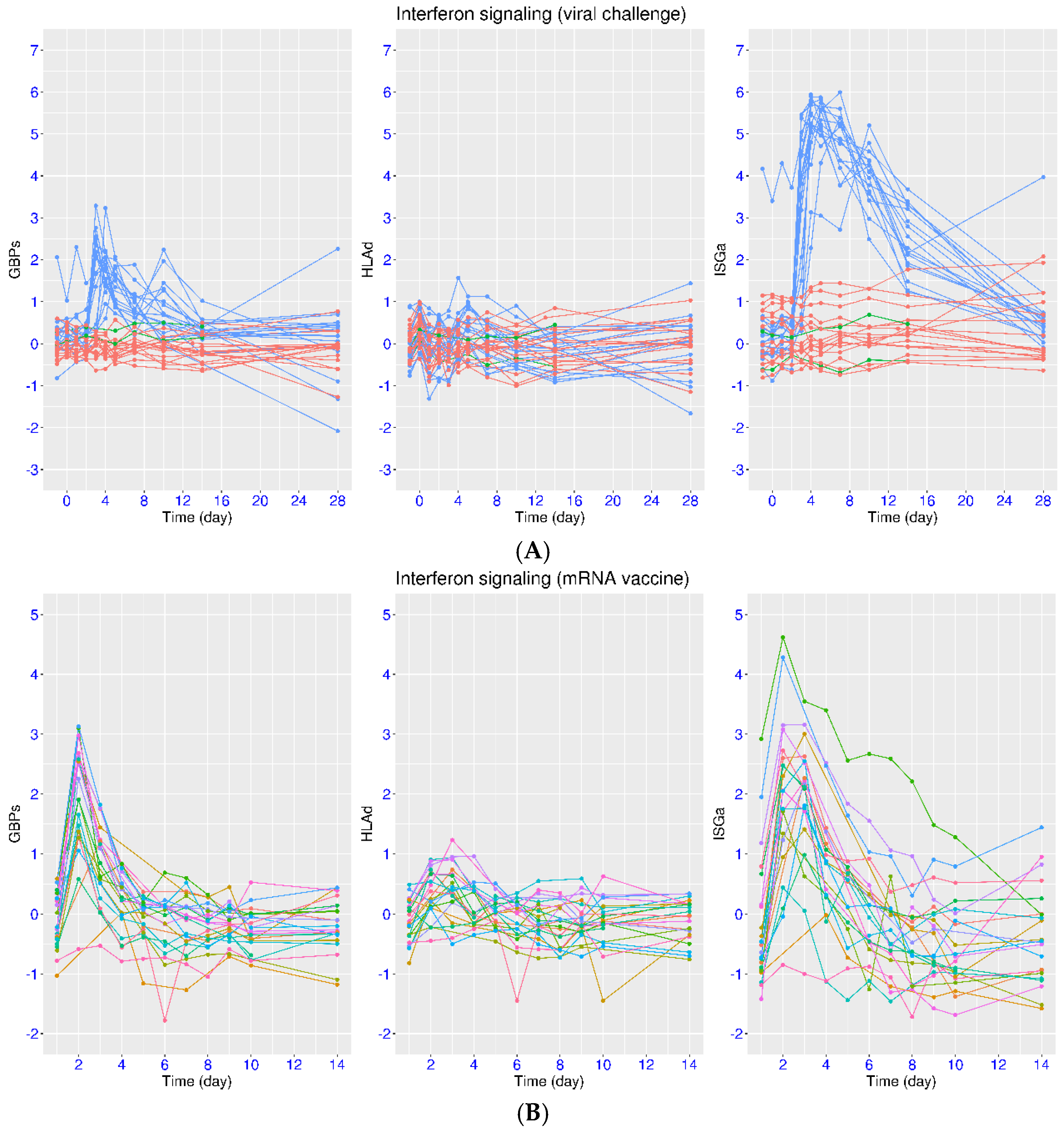
3.4. Regulation of IFN Signaling upon SARS-CoV-2 Challenge
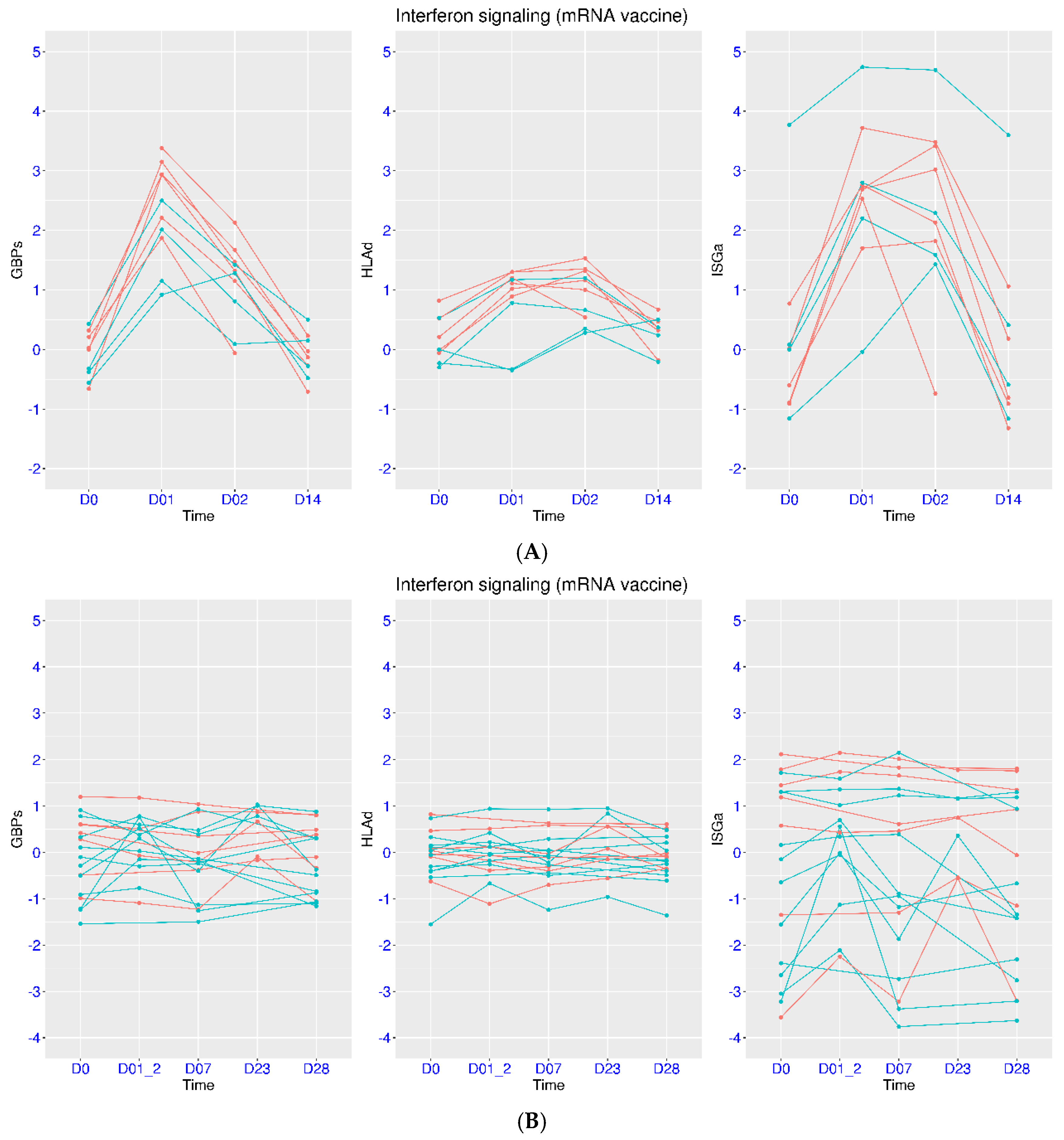
3.5. Regulation of IFN Signaling upon Vaccination
3.6. Differential Host Transcriptional Response upon Vaccination
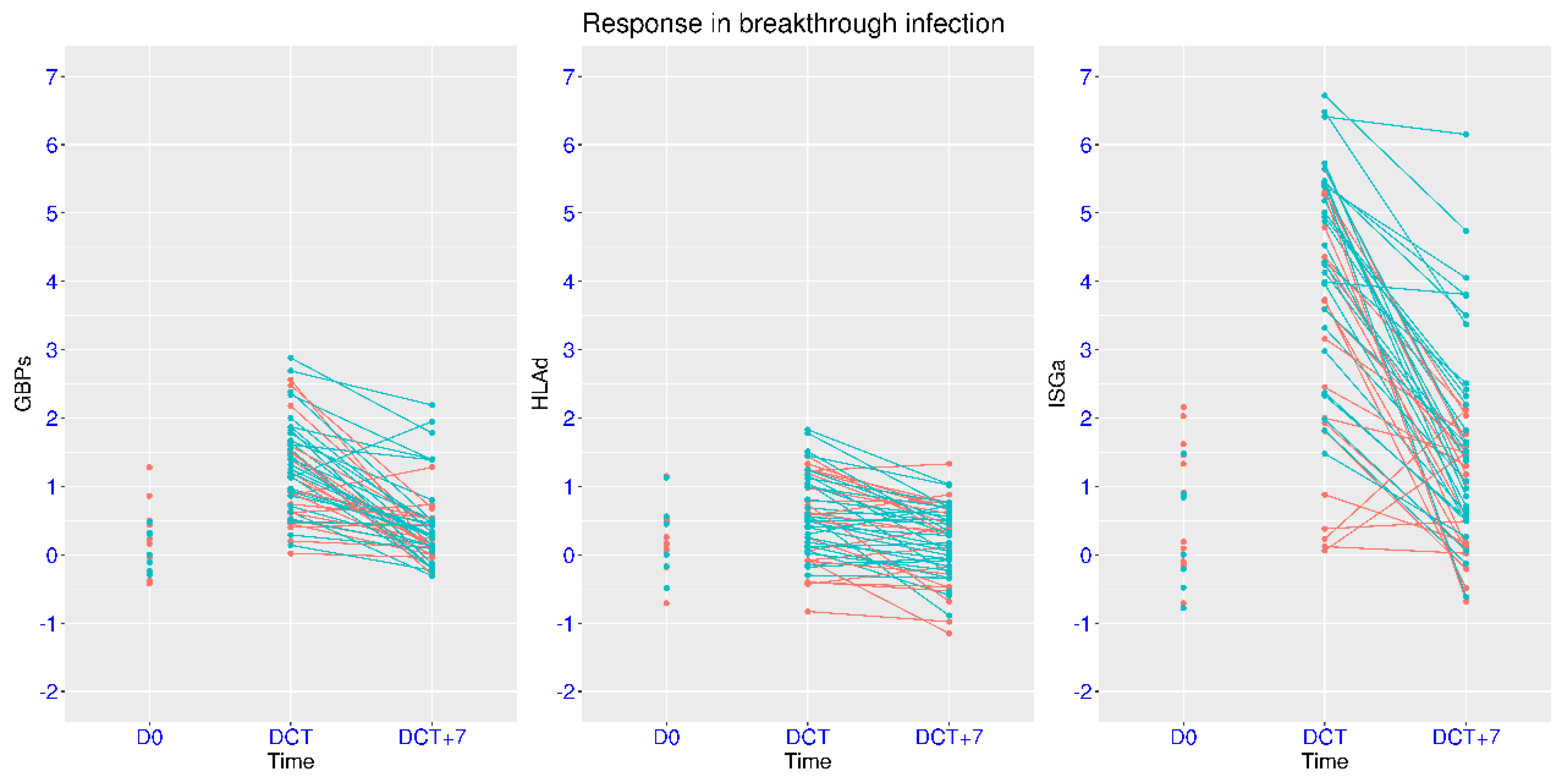
3.7. Modulation of IFN Signaling in Mild Breakthrough Infection
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons-Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Todorovic-Rakovic, N.; Whitfield, J.R. Between immunomodulation and immunotolerance: The role of IFNγ in SARS-CoV-2 disease. Cytokine 2021, 146, 155637. [Google Scholar] [CrossRef]
- Lin, Q.X.X.; Rajagopalan, D.; Gamage, A.M.; Tan, L.M.; Venkatesh, P.N.; Chan, W.O.Y.; Kumar, D.; Agrawal, R.; Chen, Y.; Fong, S.W.; et al. Longitudinal single cell atlas identifies complex temporal relationship between type I interferon response and COVID-19 severity. Nat. Commun. 2024, 15, 567. [Google Scholar] [CrossRef]
- Huang, B.; Huang, J.; Chiang, N.H.; Chen, Z.; Lui, G.; Ling, L.; Kwan, M.Y.W.; Wong, J.S.C.; Mak, P.Q.; Ling, J.W.H.; et al. Interferon response and profiling of interferon response genes in peripheral blood of vaccine-naive COVID-19 patients. Front. Immunol. 2023, 14, 1315602. [Google Scholar] [CrossRef]
- Islam, M.M.; Islam, S.; Ahmed, R.; Majumder, M.; Sarkar, B.; Himu, M.E.R.; Kawser, M.; Hossain, A.; Mia, M.J.; Parag, R.R.; et al. Reduced IFN-y levels along with changes in hematologic and immunologic parameters are key to COVID-19 severity in Bangladeshi patients. Exp. Hematol. 2023, 118, 53–64.e1. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Butt, A.R.; Bibi, A.; Mushtaq, S.; Ullah, I.; Alshahrani, F.; Khan, A.; Mansoor, A. Expression of IFN-Gamma is significantly reduced during severity of covid-19 infection in hospitalized patients. PLoS ONE 2023, 18, e0291332. [Google Scholar] [CrossRef]
- Kim, M.H.; Salloum, S.; Wang, J.Y.; Wong, L.P.; Regan, J.; Lefteri, K.; Manickas-Hill, Z.; Gao, C.; Li, J.Z.; Sadreyev, R.; et al. Type I, II, and III Interferon Signatures Correspond to Coronavirus Disease 2019 Severity. J. Infect. Dis. 2021, 224, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Maurya, R.; Swaminathan, A.; Shamim, U.; Arora, S.; Mishra, P.; Raina, A.; Ravi, V.; Tarai, B.; Budhiraja, S.; Pandey, R. Co-evolution of SARS-CoV-2 variants and host immune response trajectories underlie COVID-19 pandemic to epidemic transition. iScience 2023, 26, 108336. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, M.; Cheng, L.L.; Wang, Z.R.; Wu, Z.Y.; Feng, F.T.; Feng, X.X.; Wang, S.Y.; Guo, Y.; Li, Y.Z. Identification of aberrant interferon-stimulated gene associated host responses potentially linked to poor prognosis in COVID-19 during the Omicron wave. Virol. J. 2025, 22, 89. [Google Scholar] [CrossRef]
- Cha, H.; Lee, C.M.; Kim, S.; Kang, C.K.; Choe, P.G.; Jeon, Y.K.; Jo, H.J.; Kim, N.J.; Park, W.B.; Kim, H.J. Innate immune signatures in the nasopharynx after SARS-CoV-2 infection and links with the clinical outcome of COVID-19 in Omicron-dominant period. Cell. Mol. Life Sci. 2024, 81, 364. [Google Scholar] [CrossRef] [PubMed]
- Viox, E.G.; Hoang, T.N.; Upadhyay, A.A.; Nchioua, R.; Hirschenberger, M.; Strongin, Z.; Tharp, G.K.; Pino, M.; Nguyen, K.; Harper, J.L.; et al. Modulation of type I interferon responses potently inhibits SARS-CoV-2 replication and inflammation in rhesus macaques. Sci. Immunol. 2023, 8, eadg0033. [Google Scholar] [CrossRef]
- Lukaszewicz, A.C.; Venet, F.; Faure, A.; Vignot, E.; Monneret, G. Immunostimulation with interferon-γ in protracted SARS-CoV-2 pneumonia. J. Med. Virol. 2021, 93, 5710–5711. [Google Scholar] [CrossRef]
- Ryoo, S.; Koh, D.H.; Yu, S.Y.; Choi, M.; Huh, K.; Yeom, J.S.; Heo, J.Y. Clinical efficacy and safety of interferon (Type I and Type III) therapy in patients with COVID-19: A systematic review and meta-analysis of randomized controlled trials. PLoS ONE 2023, 18, e0272826. [Google Scholar]
- Buchynskyi, M.; Kamyshna, I.; Lyubomirskaya, K.; Moshynets, O.; Kobyliak, N.; Oksenych, V.; Kamyshnyi, A. Efficacy of interferon alpha for the treatment of hospitalized patients with COVID-19: A meta-analysis. Front. Immunol. 2023, 14, 1069894. [Google Scholar] [CrossRef]
- Tat, V.Y.; Huang, P.; Khanipov, K.; Tat, N.Y.; Tseng, C.K.; Golovko, G. Evaluation of Type I Interferon Treatment in Hospitalized COVID-19 Patients: A Retrospective Cohort Study. Pathogens 2024, 13, 539. [Google Scholar] [CrossRef]
- Jackson, H.; Rivero Calle, I.; Broderick, C.; Habgood-Coote, D.; D’Souza, G.; Nichols, S.; Vito, O.; Gomez-Rial, J.; Rivero-Velasco, C.; Rodriguez-Nunez, N.; et al. Characterisation of the blood RNA host response underpinning severity in COVID-19 patients. Sci. Rep. 2022, 12, 12216. [Google Scholar] [CrossRef]
- Uzun, O.; Akpolat, T.; Varol, A.; Turan, S.; Bektas, S.G.; Cetinkaya, P.D.; Dursun, M.; Bakan, N.; Ketencioglu, B.B.; Bayrak, M.; et al. COVID-19: Vaccination vs. hospitalization. Infection 2022, 50, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Panahi, Y.; Einollahi, B.; Beiraghdar, F.; Darvishi, M.; Fathi, S.; Javanbakht, M.; Shafiee, S.; Akhavan-Sigari, R. Fully understanding the efficacy profile of the COVID-19 vaccination and its associated factors in multiple real-world settings. Front. Immunol. 2022, 13, 947602. [Google Scholar] [CrossRef]
- Mohamed, K.; Rzymski, P.; Islam, M.S.; Makuku, R.; Mushtaq, A.; Khan, A.; Ivanovska, M.; Makka, S.A.; Hashem, F.; Marquez, L.; et al. COVID-19 vaccinations: The unknowns, challenges, and hopes. J. Med. Virol. 2022, 94, 1336–1349. [Google Scholar] [CrossRef]
- Hall, J.C.; Casciola-Rosen, L.; Berger, A.E.; Kapsogeorgou, E.K.; Cheadle, C.; Tzioufas, A.G.; Baer, A.N.; Rosen, A. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. Proc. Natl. Acad. Sci. USA 2012, 109, 17609–17614. [Google Scholar] [CrossRef]
- Lei, H.; Xu, X.; Wang, C.; Xue, D.; Wang, C.; Chen, J. A host-based two-gene model for the identification of bacterial infection in general clinical settings. Int. J. Infect. Dis. 2021, 105, 662–667. [Google Scholar] [CrossRef]
- Lei, H. A single transcript for the prognosis of disease severity in COVID-19 patients. Sci. Rep. 2021, 11, 12174. [Google Scholar] [CrossRef]
- Lei, H. A two-gene marker for the two-tiered innate immune response in COVID-19 patients. PLoS ONE 2023, 18, e0280392. [Google Scholar] [CrossRef]
- Lei, H.X. Hypoxia and Activation of Neutrophil Degranulation-Related Genes in the Peripheral Blood of COVID-19 Patients. Viruses 2024, 16, 201. [Google Scholar] [CrossRef]
- Lei, H. Quantitative and Longitudinal Assessment of Systemic Innate Immunity in Health and Disease Using a 2D Gene Model. Biomedicines 2024, 12, 969. [Google Scholar] [CrossRef]
- Thair, S.A.; He, Y.D.; Hasin-Brumshtein, Y.; Sakaram, S.; Pandya, R.; Toh, J.; Rawling, D.; Remmel, M.; Coyle, S.; Dalekos, G.N.; et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 2021, 24, 101947. [Google Scholar] [CrossRef]
- Muratsu, A.; Oda, S.; Onishi, S.; Yoshimura, J.; Matsumoto, H.; Togami, Y.; Mitsuyama, Y.; Ito, H.; Okuzaki, D.; Ogura, H.; et al. Bacterial sepsis causes more dramatic pathogenetic changes in the Th1 pathway than does viral (COVID-19) sepsis: A prospective observational study of whole blood transcriptomes. Virol. J. 2024, 21, 190. [Google Scholar] [CrossRef]
- Bibert, S.; Guex, N.; Lourenco, J.; Brahier, T.; Papadimitriou-Olivgeris, M.; Damonti, L.; Manuel, O.; Liechti, R.; Götz, L.; Tschopp, J.; et al. Transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients. Front. Immunol. 2021, 12, 666163. [Google Scholar] [CrossRef]
- Ahern, D.J.; Ai, Z.C.; Ainsworth, M.; Allan, C.; Allcock, A.; Angus, B.; Ansari, M.A.; Arancibia-Carcamo, C.V.; Aschenbrenner, D.; Attar, M.; et al. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell 2022, 185, 916–938.e58. [Google Scholar] [CrossRef]
- Prebensen, C.; Lefol, Y.; Myhre, P.; Lüders, T.; Jonassen, C.; Blomfeldt, A.; Omland, T.; Nilsen, H.; Berdal, J.E. Longitudinal whole blood transcriptomic analysis characterizes neutrophil activation and interferon signaling in moderate and severe COVID-19. Sci. Rep. 2023, 13, 10368. [Google Scholar]
- Bergamaschi, L.; Mescia, F.; Turner, L.; Hanson, A.L.; Kotagiri, P.; Dunmore, B.J.; Ruffieux, H.; De Sa, A.; Huhn, O.; Morgan, M.D.; et al. Longitudinal analysis reveals that delayed bystander CD8 T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity 2021, 54, 1257–1275.e8. [Google Scholar] [CrossRef]
- Lindeboom, R.G.H.; Worlock, K.B.; Dratva, L.M.; Yoshida, M.; Scobie, D.; Wagstaffe, H.R.; Richardson, L.; Wilbrey-Clark, A.; Barnes, J.L.; Kretschmer, L.; et al. Human SARS-CoV-2 challenge uncovers local and systemic response dynamics. Nature 2024, 631, 189–198. [Google Scholar] [CrossRef]
- Rinchai, D.; Deola, S.; Zoppoli, G.; Kabeer, B.S.A.; Taleb, S.; Pavlovski, I.; Maacha, S.; Gentilcore, G.; Toufiq, M.; Mathew, L.; et al. High-temporal resolution profiling reveals distinct immune trajectories following the first and second doses of COVID-19 mRNA vaccines. Sci. Adv. 2022, 8, eabp9961. [Google Scholar] [CrossRef]
- Odak, I.; Riemann, L.; Sandrock, I.; Cossmann, A.; Ramos, G.M.; Hammerschmidt, S.I.; Ritter, C.; Friedrichsen, M.; Hassan, A.; Dopfer-Jablonka, A.; et al. Systems biology analysis reveals distinct molecular signatures associated with immune responsiveness to the BNT162b COVID-19 vaccine. EBioMedicine 2024, 99, 104947. [Google Scholar] [CrossRef]
- Ryan, F.J.; Norton, T.S.; McCafferty, C.; Blake, S.J.; Stevens, N.E.; James, J.; Eden, G.L.; Tee, Y.C.; Benson, S.C.; Masavuli, M.G.; et al. A systems immunology study comparing innate and adaptive immune responses in adults to COVID-19 mRNA and adenovirus vectored vaccines. Cell Rep. Med. 2023, 4, 100971. [Google Scholar] [CrossRef]
- Sarin, K.Y.; Zheng, H.; Chaichian, Y.; Arunachalam, P.S.; Swaminathan, G.; Eschholz, A.; Gao, F.; Wirz, O.F.; Lam, B.; Yang, E.; et al. Impaired innate and adaptive immune responses to BNT162b2 SARS-CoV-2 vaccination in systemic lupus erythematosus. JCI Insight 2024, 9, e176556. [Google Scholar] [CrossRef]
- Drury, R.E.; Camara, S.; Chelysheva, I.; Bibi, S.; Sanders, K.; Felle, S.; Emary, K.; Phillips, D.; Voysey, M.; Ferreira, D.M.; et al. Multi-omics analysis reveals COVID-19 vaccine induced attenuation of inflammatory responses during breakthrough disease. Nat. Commun. 2024, 15, 3402. [Google Scholar] [CrossRef]
- Song, F.; Qian, Y.; Peng, X.; Li, X.; Xing, P.; Ye, D.; Lei, H. The frontline of immune response in peripheral blood. PLoS ONE 2017, 12, e0182294. [Google Scholar] [CrossRef]
- Wei, L.; Li, X.; Wang, L.J.; Song, Y.Y.; Dong, H.M. Comprehensive Analysis of RNA Expression Profile Identifies Hub miRNA-circRNA Interaction Networks in the Hypoxic Ischemic Encephalopathy. Comput. Math. Methods Med. 2021, 2021, 6015473. [Google Scholar] [CrossRef]
- Yang, J.; Wang, F.; Chen, B.A. HLA-DPA1 gene is a potential predictor with prognostic values in multiple myeloma. BMC Cancer 2020, 20, 915. [Google Scholar] [CrossRef]
- Dickel, S.; Grimm, C.; Amschler, K.; Schnitzler, S.U.; Schanz, J.; Moerer, O.; Payen, D.; Tampe, B.; Winkler, M.S. Case Report: Interferon-γ Restores Monocytic Human Leukocyte Antigen Receptor (mHLA-DR) in Severe COVID-19 With Acquired Immunosuppression Syndrome. Front. Immunol. 2021, 12, 645124. [Google Scholar] [CrossRef]
- Nabilah, M.H.; Wardhani, P.; Aryati, A.; Hernaningsih, Y.; Semedi, B.P. The Comparison of Monocyte Human Leukocyte Antigen-D-Related (mHLA-DR) Expression Levels Between Corona Virus Disease 2019 (COVID-19) Patients and Healthy Subjects. Acta Med. Indones 2023, 55, 52–60. [Google Scholar]
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A.G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, 1401–1418.e18. [Google Scholar] [CrossRef]
- Monneret, G.; Voirin, N.; Richard, J.C.; Cour, M.; Rimmele, T.; Garnier, L.; Yonis, H.; Coudereau, R.; Gossez, M.; Malcus, C.; et al. Monitoring monocyte HLA-DR expression and CD4 + T lymphocyte count in dexamethasone-treated severe COVID-19 patients. Ann. Intensiv. Care 2024, 14, 76. [Google Scholar] [CrossRef]
- Henao-Agudelo, J.S.; Ayala, S.; Badiel, M.; Zea-Vera, A.F.; Matta Cortes, L. Classical monocytes-low expressing HLA-DR is associated with higher mortality rate in SARS-CoV-2+ young patients with severe pneumonia. Heliyon 2024, 10, e24099. [Google Scholar] [CrossRef]
- Dhindsa, S.; Zhang, N.; McPhaul, M.J.; Wu, Z.R.; Ghoshal, A.K.; Erlich, E.C.; Mani, K.; Randolph, G.J.; Edwards, J.R.; Mudd, P.A.; et al. Association of Circulating Sex Hormones With Inflammation and Disease Severity in Patients with COVID-19. JAMA Netw. Open 2021, 4, e2111398. [Google Scholar] [CrossRef]
- Aschenbrenner, A.C.; Mouktaroudi, M.; Krämer, B.; Oestreich, M.; Antonakos, N.; Nuesch-Germano, M.; Gkizeli, K.; Bonaguro, L.; Reusch, N.; Basser, K.; et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021, 13, 7. [Google Scholar] [CrossRef]
- Chhabra, S.; Kalia, M. Guanylate-binding proteins in virus infection. Biochem. Soc. Trans. 2023, 51, 1621–1633. [Google Scholar]
- Cillo, A.R.; Somasundaram, A.; Shan, F.; Cardello, C.; Workman, C.J.; Kitsios, G.D.; Ruffin, A.T.; Kunning, S.; Lampenfeld, C.; Onkar, S.; et al. People critically ill with COVID-19 exhibit peripheral immune profiles predictive of mortality and reflective of SARS-CoV-2 lung viral burden. Cell Rep. Med. 2021, 2, 100476. [Google Scholar] [CrossRef]
- Sommen, S.L.; Zhao, Z.; Segtnan, S.; Stiansen-Sonerud, T.; Selvakumar, J.; Beier Havdal, L.; Gjerstad, J.; Wyller, V.B.B.; Lund Berven, L. Bulk RNA sequencing for analysis of post COVID-19 condition in adolescents and young adults. J. Transl. Med. 2024, 22, 312. [Google Scholar] [CrossRef]
- Brunet-Ratnasingham, E.; Morin, S.; Randolph, H.E.; Labrecque, M.; Belair, J.; Lima-Barbosa, R.; Pagliuzza, A.; Marchitto, L.; Hultstrom, M.; Niessl, J.; et al. Sustained IFN signaling is associated with delayed development of SARS-CoV-2-specific immunity. Nat. Commun. 2024, 15, 4177. [Google Scholar] [CrossRef]
- Montenegro, A.F.L.; Clementino, M.A.F.; Yaochite, J.N.U. Type I interferon pathway genetic variants in severe COVID-19. Virus Res. 2024, 342, 199339. [Google Scholar] [CrossRef]
- Matuozzo, D.; Talouarn, E.; Marchal, A.; Zhang, P.; Manry, J.; Seeleuthner, Y.; Zhang, Y.; Bolze, A.; Chaldebas, M.; Milisavljevic, B.; et al. Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19. Genome Med. 2023, 15, 22. [Google Scholar] [CrossRef]
- Bastard, P.; Vazquez, S.E.; Liu, J.; Laurie, M.T.; Wang, C.Y.; Gervais, A.; Le Voyer, T.; Bizien, L.; Zamecnik, C.; Philippot, Q.; et al. Vaccine breakthrough hypoxemic COVID-19 pneumonia in patients with auto-Abs neutralizing type I IFNs. Sci. Immunol. 2023, 8, eabp8966. [Google Scholar] [CrossRef]
- Aoki, A.; Iwamura, C.; Kiuchi, M.; Tsuji, K.; Sasaki, A.; Hishiya, T.; Hirasawa, R.; Kokubo, K.; Kuriyama, S.; Onodera, A.; et al. Suppression of Type I Interferon Signaling in Myeloid Cells by Autoantibodies in Severe COVID-19 Patients. J. Clin. Immunol. 2024, 44, 104. [Google Scholar] [CrossRef]
- Kiyono, H.; Ernst, P.B. Nasal vaccines for respiratory infections. Nature 2025, 641, 321–330. [Google Scholar] [CrossRef]

| Accession ID | Condition | Sample Numbers | Ref. |
|---|---|---|---|
| GSE152641 | COVID-19 infection Hospitalized patients | 62 patients + 24 controls, 23 patients intubated | [23] |
| GSE243217 | COVID-19 infection Sepsis patients | 57 sepsis + 15 controls 35 COVID-19, 22 bacterial | [24] |
| Mendeley 8wxhhykfnh.2 | COVID-19 infection Severity and time | 103 COVID-19 + 27 controls + 22 flu patients | [25] |
| Zenodo 6120249 | COVID-19 infection Severity and time | 91 COVID-19 + 10 controls + 42 sepsis patients | [26] |
| GSE213313 | COVID-19 infection Longitudinal and severity | 83 COVID-19, 11 controls, T1, T2, and T3 | [27] |
| EGAS00001005332 | COVID-19 infection Longitudinal and severity | 158 COVID-19 + 22 controls Day 0 and day 28 | [28] |
| E-MTAB-12993 | SARS-CoV-2 challenge Longitudinal | 374 blood samples, Day 0 to day 28 | [29] |
| GSE190001 | mRNA vaccination Longitudinal | 23 subjects, 11 time points 226 samples | [30] |
| GSE247401 | mRNA or mixed vaccination Second/third dose | 17 subjects, 4 time points 50 samples | [31] |
| GSE199750 | mRNA or mixed vaccination First/second/third dose | 86 subjects, 4 time points 260 samples | [32] |
| GSE246525 | mRNA vaccination High/low response | 10 subjects, 4 time points 38 samples | [31] |
| GSE250023 | mRNA vaccination SLE patients, Responder/non-responder | 18 subjects, 5 time points 75 samples | [33] |
| GSE228839 | Breakthrough infection w/o prior vaccination | 71 subjects, 3 time points 122 samples | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, H. Distinctive Temporal Profiles of Interferon-Stimulated Genes in Natural Infection, Viral Challenge, and Vaccination. Viruses 2025, 17, 1060. https://doi.org/10.3390/v17081060
Lei H. Distinctive Temporal Profiles of Interferon-Stimulated Genes in Natural Infection, Viral Challenge, and Vaccination. Viruses. 2025; 17(8):1060. https://doi.org/10.3390/v17081060
Chicago/Turabian StyleLei, Hongxing. 2025. "Distinctive Temporal Profiles of Interferon-Stimulated Genes in Natural Infection, Viral Challenge, and Vaccination" Viruses 17, no. 8: 1060. https://doi.org/10.3390/v17081060
APA StyleLei, H. (2025). Distinctive Temporal Profiles of Interferon-Stimulated Genes in Natural Infection, Viral Challenge, and Vaccination. Viruses, 17(8), 1060. https://doi.org/10.3390/v17081060






