Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis
Abstract
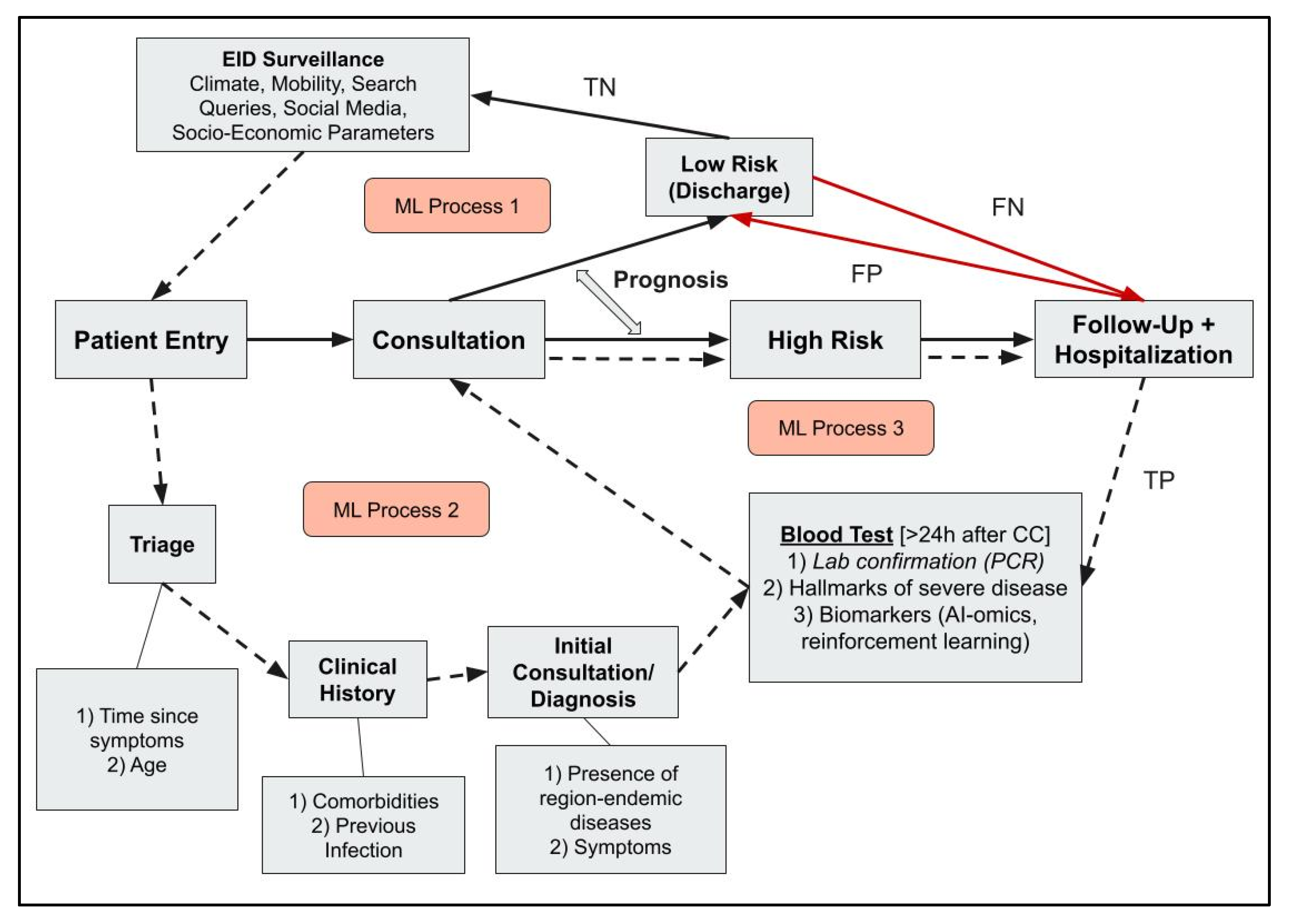
1. Introduction
2. Methods
2.1. Information Sources and Search Strategy
2.2. Selection and Data Collection Process
3. Results
3.1. Systematic Search for AI and ML Models Used in Infectious Disease Management
3.2. Supervised, Unsupervised, and Reinforcement ML Models Used in Infectious Disease Management
Methods Used to Evaluate AI and ML Model Performance
| Metric | Calculation | Description |
|---|---|---|
| Positive Predictive Value (PPV)/ Precision | Probability of the presence of disease given a positive test result [35] | |
| Negative Predictive Value (NPV) | Probability of the absence of disease given a negative test result [35] | |
| Accuracy | Measurement of how well a model predicts the correct class or the fraction of predictions that the model correctly identified out of all the cases [36] | |
| Sensitivity/ Recall | Probability of a positive test result given the presence of disease [35,36] | |
| Specificity | Probability of all negative samples that are correctly predicted by the model [36] | |
| AUROC | The area under the graph of sensitivity against 1-specificity | |
| F1-Score | Weighted harmonic mean between precision and recall [36] | |
| Matthew’s Correlation Coefficient (MCC) | Weighted classifier score factoring all four confusion matrix categories and imbalanced class data into account [36,37] |
3.3. Applications of AI and ML in Infectious Disease Management
3.4. Roles of ML in Infectious Disease Public Health and Surveillance
3.4.1. Climate
3.4.2. Mobility
3.4.3. Search Engine Queries (SEQs) and Social Media
3.4.4. SocioEconomic Factors
3.4.5. Web-Based Surveillance
3.5. Roles of AI and ML Models in Diagnosis
3.5.1. Imaging
| Image Category | Architecture | Accuracy | Precision/PPV | Recall/ Sensitivity | F1-Score | AUROC | Specificity | MCC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| X-ray | DenseNet201 | 0.99 | 0.97 | 0.97 | 0.97 | - | 0.9895 | - | [101] |
| Custom CNN | 0.9819 | 0.9767 | 0.9833 | 0.9733 | - | - | - | [98] | |
| VGG19 | 0.9888 | 0.9870 | 0.9904 | 0.9987 | 0.9939 | - | - | [99] | |
| Texture Extraction and SVM | 0.9547 | 0.9471 | 0.9618 | 0.9544 | - | 0.9624 | - | [96] | |
| EfficientNetB4 and ResNet50 | 0.92 | 0.97 | 0.92 | 0.94 | 0.90 | - | - | [97] | |
| Stacking NN and SVM | 0.9962 | 0.9966 | 0.9962 | 0.9962 | - | - | - | [97] | |
| EfficientNet and ResNet | - | - | - | - | 0.89 | 0.79 | - | [100] | |
| Custom CNN | 0.9872 | 0.9989 | 0.9966 | 0.9977 | - | - | - | [102] | |
| GoogleNet and ResNet50 | 0.98 | 0.9471 | 0.9402 | 0.9389 | - | 0.9633 | - | [103] | |
| X-ray and CT | Custom CNN | 0.9940 | 0.9886 | 0.9941 | 0.9846 | - | - | - | [104] |
| VGG19 | 0.9167 | 0.86 | 1 | 0.92 | 0.92 | - | - | [8] | |
| CT | VGG16 | 0.98 | 0.9799 | 0.9799 | 0.9799 | 0.9790 | - | - | [109] |
| AlexNet | 0.9310 | - | 0.9180 | - | 0.9870 | 0.9460 | - | [106] | |
| Inception-ResNetV2 and ResNet18 and Multi-Layer Perceptron | 0.994 | - | 0.843 | - | 0.92 | 0.828 | - | [107] | |
| ResNet34 | 0.9547 | 0.9947 | 0.9216 | 0.9567 | 0.9974 | 0.9942 | - | [110] | |
| ResNet34 | 0.90 | 0.95 | 0.87 | - | 0.83 | 0.94 | - | [105] | |
| Custom CNN and Ensemble | 0.9973 | 0.9946 | 1 | 0.9973 | 0.9973 | - | - | [108] | |
| Photograph | ResNet50 | 0.8417 | - | - | - | - | - | 0.7715 | [112] |
| MonkeyNet and Grad-CAM | 0.9891 | 0.9892 | 0.9891 | 0.9891 | 0.9997 | - | - | [111] | |
| InceptionV3 | 0.94 | - | 0.88 | - | - | 1 | - | [113] | |
| Microscopy | YOLOv2 and ResNet50 | - | 0.7120 | 0.9190 | - | - | 0.8970 | - | [117] |
| Patch-U-Net | - | 0.9380 | 0.8170 | - | 0.9740 | - | - | [115] | |
| MobileNetV3Large | 0.9920 | 0.9840 | 1 | 0.9920 | 0.993 | 0.9850 | - | [116] |
3.5.2. Clinical Signs and Symptoms
| Category | Architecture | Accuracy | Precision/PPV | Recall/ Sensitivity | F1-Score | AUROC | Specificity | MCC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Clinical Signs | GBM Ensemble + SHAP | 0.96 | 0.94 | 0.95 | 0.94 | 0.98 | - | - | [120] |
| XGBoost | - | - | 0.819 | - | 0.97 | 0.979 | - | [121] | |
| DNN Multi-Layer Perceptron | 0.86 | - | 0.93 | - | 0.95 | 0.81 | - | [125] | |
| XGBoost | 0.822 | - | 0.797 | - | 0.905 | 0.845 | - | [122] | |
| DNN Multi-Layer Perceptron | - | 0.94 | 0.91 | 0.92 | - | - | - | [127] | |
| RF | 0.827 | 0.575 | 0.339 | 0.427 | 0.785 | 0.941 | - | [126] | |
| XGBoost | - | 0.73 | 0.56 | - | 0.86 | 0.92 | - | [124] | |
| RF, LR, SVM, Multi-Layered Perceptron, XGBoost, AdaBoost Ensemble | - | 0.29 | 0.93 | - | 0.91 | 0.64 | - | [123] | |
| Symptoms | Boosted LR | 0.57 | 0.64 | 0.35 | 0.43 | - | 0.80 | 0.15 | [129] |
| LR and Minority Data Upsampling | 0.73 | 0.25 | 0.60 | 0.35 | 0.68 | 0.75 | 0.25 | [128] |
3.5.3. Unstructured Text Classification
3.6. Roles of AI and ML Models in Clinical Prognosis
| Category | Architecture | Accuracy | Precision/PPV | Recall/ Sensitivity | F1-Score | AUROC | Specificity | MCC | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Clinical Biomarkers | SVM | 0.903 | - | - | - | - | - | - | [137] |
| Ensemble (Bagging) | - | 0.86 | 0.98 | 0.91 | 0.79 | - | - | [138] | |
| XGBoost | 0.73 | - | 0.66 | - | 0.79 | 0.85 | - | [139] | |
| XGBoost + SHAP | - | 0.29 | 0.64 | - | 0.85 | 0.91 | - | [140] | |
| XGBoost | 0.9602 | 0.9533 | 0.9613 | 0.9573 | 0.9603 | 0.9591 | 0.8520 | [141] | |
| LightGBM + SHAP | 0.754 | 0.792 | 0.816 | 0.802 | 0.847 | 0.764 | - | [142] | |
| Variational Autoencoders | - | 0.62 | 0.75 | - | - | 0.71 | - | [143] | |
| DNN + SHAP | - | 0.3765 | 0.869 | - | 0.937 | 0.867 | - | [144] | |
| GBM + SHAP | 0.79 | 0.21 | 0.85 | - | 0.89 | 0.79 | - | [145] | |
| ANN Backpropagation | - | - | - | - | 0.8768 | - | - | [146] | |
| Transformer + DNN | 0.918 | 0.914 | 0.916 | 0.913 | 0.96 | - | - | [147] | |
| Ensemble (RF, LightGBM) + SHAP | - | 0.79 | 0.53 | - | 0.86 | 0.93 | 0.53 | [148] | |
| LASSO + XGBoost + SHAP | - | 0.882 | 0.918 | 0.937 | 0.94 | - | - | [149] | |
| XGBoost | - | - | 0.929 | - | 0.80 | 0.385 | - | [150] | |
| DT | 0.98 | - | 1.0 | 0.93 | 0.99 | - | - | [151] | |
| ANN + SHAP | 0.7523 | - | - | - | 0.8324 | - | - | [152] | |
| LightGBM + SHAP | 0.882 | 0.271 | 0.861 | 0.629 | 0.934 | 0.883 | - | [153] | |
| DNN-Encoders + XGBoost | 0.8278 | - | - | - | - | - | - | [154] | |
| Ensemble (RF, LR, DT, KNN, AdaBoost, CatBoost, LightGBM, XGBoost) | 0.95 | 0.96 | - | 0.95 | 0.98 | - | 0.89 | [155] | |
| Gene and Pathway Identification | XGBoost | - | 0.209 | 0.864 | - | 0.94 | 0.797 | - | [156] |
| SVM, RF, LASSO | - | - | - | - | - | - | - | [157] | |
| RF | - | - | - | - | 0.889 | - | - | [158] | |
| LASSO | - | - | - | - | 0.98 | - | - | [159] |
4. Discussion
4.1. Implications
4.2. Limitations
4.3. Future Directions
4.4. Challenges in AI and ML Implementation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COVID-19 | Coronavirus Disease 2019 |
| LMICs | Low- and Middle-Income Countries |
| AI | Artificial Intelligence |
| ML | Machine Learning |
| XAI | Explainable AI |
| SHAP | Shapley Additive exPlanations |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| CNNs | Convolutional Neural Networks |
| CT | Computed Tomography |
| RF | Random Forest |
| AUROC | Area Under Receiver Operating Characteristic |
| MCC | Matthew’s Correlation Coefficient |
| CC | Chief Complaint |
| PCR | Polymerase Chain Reaction |
| FN | False Negative |
| TN | True Negative |
| FP | False Positive |
| TP | True Positive |
| EIDs | Emerging Infectious Diseases |
| HAIs | Hospital Acquired Infections |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome-Coronavirus 2 |
| RBD | Receptor Binding Domain |
| SEQs | Search Engine Queries |
| SVM | Support Vector Machine |
| RNNs | Recurrent Neural Networks |
| LSTM-ATT | Attention-Based Long-Short Term Memory |
| SEIR | Susceptible-Exposed-Infected-Recovered |
| LR | Logistic Regression |
| KNN | k-Nearest Neighbor |
| XGBoost | eXtreme Gradient Boost |
| NLP | Natural Language Processing |
| PCC | Pearson’s Correlation Coefficient |
| DNNs | Deep Neural Networks |
| GBMs | Gradient Boosting Machines |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| ARI | Acute Respiratory Infection |
| DTs | Decision Trees |
| RMSE | Root Mean Squared Error |
| FDA | Food and Drug Administration |
| NS1 | Non-Structural Protein 1 |
| LLMs | Large Language Models |
| QSOFA | Quick Sepsis-related Organ Failure Assessment |
| PCT | Procalcitonin |
References
- Gray, A.; Sharara, F. Global and regional sepsis and infectious syndrome mortality in 2019: A systematic analysis. Lancet Glob. Health 2022, 10, S2. [Google Scholar] [CrossRef]
- Global Burden of Disease Study 2019 (GBD 2019) Data Resources. Institute for Health Metrics and Evaluation. 2019. Available online: https://ghdx.healthdata.org/gbd-2019 (accessed on 19 April 2024).
- Lewis, T.P.; McConnell, M.; Aryal, A.; Irimu, G.; Mehata, S.; Mrisho, M.; Kruk, M.E. Health service quality in 2929 facilities in six low-income and middle-income countries: A positive deviance analysis. Lancet Glob. Health 2023, 11, e862–e870. [Google Scholar] [CrossRef] [PubMed]
- Basto-Abreu, A.; Barrientos-Gutierrez, T.; Wade, A.N.; Oliveira de Melo, D.; Semeão de Souza, A.S.; Nunes, B.P.; Perianayagam, A.; Tian, M.; Yan, L.L.; Ghosh, A.; et al. Multimorbidity matters in low and middle-income countries. J. Multimorb. Comorbidity 2022, 12, 263355652211060. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, T.; Zhang, K.H. Data-driven decision-making for precision diagnosis of digestive diseases. Biomed. Eng. Online 2023, 22, 87. [Google Scholar] [CrossRef]
- Paszkiewicz, K.H.; van der Giezen, M. Omics, Bioinformatics, and infectious disease research. In Genetics and Evolution of Infectious Diseases; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 523–539. [Google Scholar]
- Edgar, R.C.; Taylor, B.; Lin, V.; Altman, T.; Barbera, P.; Meleshko, D.; Lohr, D.; Novakovsky, G.; Buchfink, B.; Al-Shayeb, B.; et al. Petabase-scale sequence alignment catalyses viral discovery. Nature 2022, 602, 142–147. [Google Scholar] [CrossRef]
- Mukhi, S.E.; Varshini, R.T.; Sherley, S.E.F. Diagnosis of COVID-19 from Multimodal Imaging Data Using Optimized Deep Learning Techniques. SN Comput. Sci. 2023, 4, 212. [Google Scholar] [CrossRef]
- Yang, I.S.; Ryu, C.; Cho, K.J.; Kim, J.K.; Ong, S.H.; Mitchell, W.P.; Kim, B.S.; Oh, H.B.; Kim, K.H. IDBD: Infectious disease biomarker database. Nucleic Acids Res. 2008, 36, D455–D460. [Google Scholar] [CrossRef]
- Milinovich, G.J.; Williams, G.M.; Clements, A.C.A.; Hu, W. Internet-based surveillance systems for monitoring emerging infectious diseases. Lancet Infect. Dis. 2014, 14, 160–168. [Google Scholar] [CrossRef]
- Beltrán-Silva, S.L.; Chacón-Hernández, S.S.; Moreno-Palacios, E.; Pereyra-Molina, J.Á. Clinical and differential diagnosis: Dengue, chikungunya and Zika. Revista Médica del Hospital General de México 2018, 81, 146–153. [Google Scholar] [CrossRef]
- Bi, Q.; Goodman, K.E.; Kaminsky, J.; Lessler, J. What is machine learning? A primer for the epidemiologist. Am. J. Epidemiol. 2019, 188, 2222–2239. [Google Scholar] [CrossRef]
- Cunningham, P.; Cord, M.; Delany, S.J. Supervised Learning. In Machine Learning Techniques for Multimedia, 1st ed.; Cord, M., Cunningham, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 21–49. [Google Scholar]
- Keshavamurthy, R.; Dixon, S.; Pazdernik, K.T.; Charles, L.E. Predicting infectious disease for biopreparedness and response: A systematic review of machine learning and deep learning approaches. One Health 2022, 15, 100439. [Google Scholar] [CrossRef] [PubMed]
- Alfred, R.; Obit, J.H. The roles of machine learning methods in limiting the spread of deadly diseases: A systematic review. Heliyon 2021, 7, e07371. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Bakhrebah, M.A.; Alotaibi, J.; Natto, Z.S.; Alkhaibari, R.S.; Alawad, E.; Alshammari, H.M.; Alwarthan, S.; Alhajri, M.; Almogbel, M.S.; et al. Unleashing the power of artificial intelligence for diagnosing and treating infectious diseases: A comprehensive review. J. Infect. Public Health 2023, 16, 1837–1847. [Google Scholar] [CrossRef]
- Alqaissi, E.Y.; Alotaibi, F.S.; Ramzan, M.S. Modern Machine-Learning Predictive Models for Diagnosing Infectious Diseases. Comput. Math. Methods Med. 2022, 2022, 6902321. [Google Scholar] [CrossRef]
- Tran, N.K.; Albahra, S.; May, L.; Waldman, S.; Crabtree, S.; Bainbridge, S.; Rashidi, H. Evolving Applications of Artificial Intelligence and Machine Learning in Infectious Diseases Testing. Clin. Chem. 2022, 68, 125–133. [Google Scholar] [CrossRef]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Pantelis, G.; Lescure, F.X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: A narrative review of current applications. Clin. Microbiol. Infect. 2020, 26, 584–595. [Google Scholar] [CrossRef]
- Theodosiou, A.A.; Read, R.C. Artificial intelligence, machine learning and deep learning: Potential resources for the infection clinician. J. Infect. 2023, 87, 287–294. [Google Scholar] [CrossRef]
- Al Meslamani, A.Z.; Sobrino, I.; de la Fuente, J. Machine learning in infectious diseases: Potential applications and limitations. Ann. Med. 2024, 56, 2362869. [Google Scholar] [CrossRef]
- Meira, D.D.; Zetum, A.S.S.; Casotti, M.C.; Campos da Silva, D.R.; de Araújo, B.C.; Vicente, C.R.; Duque, D.A.; Campanharo, B.P.; Garcia, F.M.; Campanharo, C.V.; et al. Bioinformatics and molecular biology tools for diagnosis, prevention, treatment and prognosis of COVID-19. Heliyon 2024, 10, e34393. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, F. Interpretable neural networks: Principles and applications. Front. Artif. Intell. 2023, 6, 974295. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Kumar, R.; Tiwari, S.K.; Ranjan, P. Machine learning approaches in the diagnosis of infectious diseases: A review. Bull. Electr. Eng. Inform. 2022, 11, 3509–3520. [Google Scholar] [CrossRef]
- Edeh, M.O.; Dalal, S.; Dhaou IBen Agubosim, C.C.; Umoke, C.C.; Richard-Nnabu, N.E.; Dahiya, N. Artificial Intelligence-Based Ensemble Learning Model for Prediction of Hepatitis C Disease. Front. Public Health 2022, 10, 892371. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.J.; Lee, I.K.; Liu, J.W. Analyses of clinical and laboratory characteristics of dengue adults at their hospital presentations based on the World Health Organization clinical-phase framework: Emphasizing risk of severe dengue in the elderly. J. Microbiol. Immunol. Infect. 2018, 51, 740–748. [Google Scholar] [CrossRef]
- Rajoub, B. Supervised and unsupervised learning. In Biomedical Signal Processing and Artificial Intelligence in Healthcare; Zgallai, W., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 51–89. [Google Scholar]
- Shortreed, S.M.; Laber, E.; Lizotte, D.J.; Stroup, T.S.; Pineau, J.; Murphy, S.A. Informing sequential clinical decision-making through reinforcement learning: An empirical study. Mach. Learn. 2011, 84, 109–136. [Google Scholar] [CrossRef]
- Phillips, P.J.; Hahn, C.A.; Fontana, P.C.; Yates, A.N.; Greene, K.; Broniatowski, D.A.; Przybocki, M.A. Four Principles of Explainable Artificial Intelligence; Report No.: 8312; National Institute of Standards and Technology (US): Gaithersburg, MD, USA, 2021; 43p. [CrossRef]
- Van Den Broeck, G.; Lykov, A.; Schleich, M.; Suciu, D. On the Tractability of SHAP Explanations. J. Artif. Intell. Res. 2022, 74, 851–886. [Google Scholar] [CrossRef]
- Liu, Y.; Fu, Y.; Peng, Y.; Ming, J. Clinical decision support tool for breast cancer recurrence prediction using SHAP value in cooperative game theory. Heliyon 2024, 10, e24876. [Google Scholar] [CrossRef]
- Hadash, S.; Willemsen, M.C.; Snijders, C.; Ijsselsteijn, W.A. Improving understandability of feature contributions in model-agnostic explainable AI tools. In Proceedings of the Conference on Human Factors in Computing Systems—Proceedings, New Orleans, LA, USA, 29 April–5 May 2022. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Molinaro, A.M. Diagnostic tests: How to estimate the positive predictive value. Neurooncol. Pract. 2015, 2, 161–165. [Google Scholar] [CrossRef][Green Version]
- Ghanem, M.; Ghaith, A.K.; El-Hajj, V.G.; Bhandarkar, A.; de Giorgio, A.; Elmi-Terander, A.; Bydon, M. Limitations in Evaluating Machine Learning Models for Imbalanced Binary Outcome Classification in Spine Surgery: A Systematic Review. Brain Sci. 2023, 13, 1723. [Google Scholar] [CrossRef]
- Chicco, D.; Tötsch, N.; Jurman, G. The matthews correlation coefficient (Mcc) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Min. 2021, 14, 1–22. [Google Scholar] [CrossRef]
- Thölke, P.; Mantilla-Ramos, Y.J.; Abdelhedi, H.; Maschke, C.; Dehgan, A.; Harel, Y.; Kemtur, A.; Mekki Berrada, L.; Sahraoui, M.; Young, T.; et al. Class imbalance should not throw you off balance: Choosing the right classifiers and performance metrics for brain decoding with imbalanced data. Neuroimage 2023, 277, 120253. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Overfitting, Model Tuning, and Evaluation of Prediction Performance. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer Nature: Dordrecht, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Ying, X. An Overview of Overfitting and its Solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Jurman, G.; Riccadonna, S.; Furlanello, C. A comparison of MCC and CEN error measures in multi-class prediction. PLoS ONE 2012, 7, e41882. [Google Scholar] [CrossRef]
- Wang, M.; Yang, B.; Liu, Y.; Yang, Y.; Ji, H.; Yang, C. Emerging infectious disease surveillance using a hierarchical diagnosis model and the Knox algorithm. Sci. Rep. 2023, 13, 19836. [Google Scholar] [CrossRef]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.F.; et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef]
- Shears, P. Poverty and infection in the developing world: Healthcare-related infections and infection control in the tropics. J. Hosp. Infect. 2007, 67, 217–224. [Google Scholar] [CrossRef]
- Faruk, M.O.; Jannat, S.N.; Rahman, M.S. Impact of environmental factors on the spread of dengue fever in Sri Lanka. Int. J. Environ. Sci. Technol. 2022, 19, 10637–10648. [Google Scholar] [CrossRef]
- Sugeno, M.; Kawazu, E.C.; Kim, H.; Banouvong, V.; Pehlivan, N.; Gilfillan, D.; Kim, H.; Kim, Y. Association between environmental factors and dengue incidence in Lao People’s Democratic Republic: A nationwide time-series study. BMC Public Health 2023, 23, 2348. [Google Scholar] [CrossRef]
- Edelson, P.J.; Harold, R.; Ackelsberg, J.; Duchin, J.S.; Lawrence, S.J.; Manabe, Y.C.; Zahn, M.; LaRocque, R.C. Climate Change and the Epidemiology of Infectious Diseases in the United States. Clin. Infect. Dis. 2023, 76, 950–956. [Google Scholar] [CrossRef]
- Van de Vuurst, P.; Escobar, L.E. Climate change and infectious disease: A review of evidence and research trends. Infect. Dis. Poverty 2023, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Mora, C.; McKenzie, T.; Gaw, I.M.; Dean, J.M.; von Hammerstein, H.; Knudson, T.A.; Setter, R.O.; Smith, C.Z.; Webster, K.M.; Patz, J.A.; et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Change 2022, 12, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Buckee, C.; Noor, A.; Sattenspiel, L. Thinking clearly about social aspects of infectious disease transmission. Nature 2021, 595, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Changruenngam, S.; Bicout, D.J.; Modchang, C. How the individual human mobility spatio-temporally shapes the disease transmission dynamics. Sci. Rep. 2020, 10, 11325. [Google Scholar] [CrossRef]
- Zeng, D.; Cao, Z.; Neill, D.B. Artificial intelligence–enabled public health surveillance—From local detection to global epidemic monitoring and control. In Artificial Intelligence in Medicine: Technical Basis and Clinical Applications; Elsevier Applied Science: Amsterdam, The Netherlands, 2020; pp. 437–453. [Google Scholar]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, M.; Wu, Y.; Wang, J.; Hong, Y.; Huang, Y.; Yuan, L.; Ma, J.; Wang, K.; Wang, S.; et al. Cross-species tropism and antigenic landscapes of circulating SARS-CoV-2 variants. Cell Rep. 2022, 38, 110558. [Google Scholar] [CrossRef]
- Munjal, G.; Hanmandlu, M.; Srivastava, S. Phylogenetics Algorithms and Applications. In Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2019; pp. 187–194. [Google Scholar]
- Cahuantzi, R.; Lythgoe, K.A.; Hall, I.I.; Pellis, L.I.; House, T. Unsupervised identification of significant lineages of SARS-CoV-2 through scalable machine learning methods. Proc. Natl. Acad. Sci. USA 2024, 121, e2317284121. [Google Scholar] [CrossRef]
- Sipser, M. Introduction to The Theory of Computation, 3rd ed.; Cengage Learning: Boston, MA, USA, 2012. [Google Scholar]
- Sun, C.; Li, H.; Song, M.; Cai, D.; Zhang, B.; Hong, S. Continuous diagnosis and prognosis by controlling the update process of deep neural networks. Patterns 2023, 4, 100687. [Google Scholar] [CrossRef]
- Bilic, I.; Hess, M. Spatial Lifecourse Epidemiology and Infectious Disease Research. Trends Parasitol. 2020, 36, 232–235. [Google Scholar] [CrossRef]
- Haque, S.; Mengersen, K.; Barr, I.; Wang, L.; Yang, W.; Vardoulakis, S.; Bambrick, H.; Hu, W. Towards development of functional climate-driven early warning systems for climate-sensitive infectious diseases: Statistical models and recommendations. Environ. Res. 2024, 249, 118568. [Google Scholar] [CrossRef]
- Salim, N.A.M.; Wah, Y.B.; Reeves, C.; Smith, M.; Yaacob, W.F.W.; Mudin, R.N.; Dapari, R.; Sapri, N.N.F.F.; Haque, U. Prediction of dengue outbreak in Selangor Malaysia using machine learning techniques. Sci. Rep. 2021, 11, 939. [Google Scholar] [CrossRef] [PubMed]
- Iacus, S.M.; Santamaria, C.; Sermi, F.; Spyratos, S.; Tarchi, D.; Vespe, M. Human mobility and COVID-19 initial dynamics. Nonlinear Dyn. 2020, 101, 1901–1919. [Google Scholar] [CrossRef] [PubMed]
- Getz, W.M.; Salter, R.; Mgbara, W. Adequacy of SEIR models when epidemics have spatial structure: Ebola in Sierra Leone. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180282. [Google Scholar] [CrossRef] [PubMed]
- Katragadda, S.; Bhupatiraju, R.T.; Raghavan, V.; Ashkar, Z.; Gottumukkala, R. Examining the COVID-19 case growth rate due to visitor vs. local mobility in the United States using machine learning. Sci. Rep. 2022, 12, 12337. [Google Scholar] [CrossRef]
- Kiang, M.V.; Santillana, M.; Chen, J.T.; Onnela, J.P.; Krieger, N.; Engø-Monsen, K.; Ekapirat, N.; Areechokchai, D.; Prempree, P.; Maude, R.J.; et al. Incorporating human mobility data improves forecasts of Dengue fever in Thailand. Sci. Rep. 2021, 11, 923. [Google Scholar] [CrossRef]
- Milinovich, G.J.; Avril, S.M.R.; Clements, A.C.A.; Brownstein, J.S.; Tong, S.; Hu, W. Using internet search queries for infectious disease surveillance: Screening diseases for suitability. BMC Infect. Dis. 2014, 14, 690. [Google Scholar] [CrossRef]
- Uda, K.; Hagiya, H.; Yorifuji, T.; Koyama, T.; Tsuge, M.; Yashiro, M.; Tsukahara, H. Correlation between national surveillance and search engine query data on respiratory syncytial virus infections in Japan. BMC Public Health 2022, 22, 1517. [Google Scholar] [CrossRef]
- Jang, B.; Kim, Y.; Il Kim, G.; Wook Kim, J. Deep similarity analysis and forecasting of actual outbreak of major infectious diseases using Internet-Sourced data. J. Biomed. Inform. 2022, 133, 104148. [Google Scholar] [CrossRef]
- Kim, Y.; Park, C.R.; Ahn, J.P.; Jang, B. COVID-19 outbreak prediction using Seq2Seq + Attention and Word2Vec keyword time series data. PLoS ONE 2023, 18, e0284298. [Google Scholar] [CrossRef]
- Kim, J.; Ahn, I. Infectious disease outbreak prediction using media articles with machine learning models. Sci. Rep. 2021, 11, 4413. [Google Scholar] [CrossRef]
- Gupta, A.; Katarya, R. Social media based surveillance systems for healthcare using machine learning: A systematic review. J. Biomed. Inform. 2020, 108, 103500. [Google Scholar] [CrossRef] [PubMed]
- Ayorinde, A.; Ghosh, I.; Ali, I.; Zahair, I.; Olarewaju, O.; Singh, M.; Meehan, E.; Anjorin, S.S.; Rotheram, S.; Barr, B.; et al. Health inequalities in infectious diseases: A systematic overview of reviews. BMJ Open 2023, 13, e067429. [Google Scholar] [CrossRef] [PubMed]
- Tizzoni, M.; Nsoesie, E.O.; Gauvin, L.; Karsai, M.; Perra, N.; Bansal, S. Addressing the socioeconomic divide in computational modeling for infectious diseases. Nat. Commun. 2022, 13, 2897. [Google Scholar] [CrossRef]
- Kananura, R.M. Machine learning predictive modelling for identification of predictors of acute respiratory infection and diarrhoea in Uganda’s rural and urban settings. PLoS Global. Public Health 2022, 2, e0000430. [Google Scholar] [CrossRef]
- Kalayou, M.H.; Kassaw, A.A.K.; Shiferaw, K.B. Empowering child health: Harnessing machine learning to predict acute respiratory infections in Ethiopian under-fives using demographic and health survey insights. BMC Infect. Dis. 2024, 24, 338. [Google Scholar] [CrossRef]
- Sebastianelli, A.; Spiller, D.; Carmo, R.; Wheeler, J.; Nowakowski, A.; Jacobson, L.V.; Kim, D.; Barlevi, H.; Cordero, Z.E.R.; Colón-González, F.J.; et al. A reproducible ensemble machine learning approach to forecast dengue outbreaks. Sci. Rep. 2024, 14, 3807. [Google Scholar] [CrossRef]
- Kim, M.; Chae, K.; Lee, S.; Jang, H.J.; Kim, S. Automated classification of online sources for infectious disease occurrences using machine-learning-based natural language processing approaches. Int. J. Environ. Res. Public Health 2020, 17, 9467. [Google Scholar] [CrossRef]
- Budd, J.; Miller, B.S.; Manning, E.M.; Lampos, V.; Zhuang, M.; Edelstein, M.; Rees, G.; Emery, V.C.; Stevens, M.M.; Keegan, N.; et al. Digital technologies in the public-health response to COVID-19. Nat. Med. 2020, 26, 1183–1192. [Google Scholar] [CrossRef]
- Eze, P.U.; Geard, N.; Mueller, I.; Chades, I. Anomaly Detection in Endemic Disease Surveillance Data Using Machine Learning Techniques. Healthcare 2023, 11, 1896. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Tuyet-Hanh, T.T.; Mulhall, J.; Minh, H.V.; Duong, T.Q.; Chien, N.V.; Nhung, N.T.T.; Lan, V.H.; Minh, H.B.; Cuong, D.; et al. Deep learning models for forecasting dengue fever based on climate data in Vietnam. PLoS Negl. Trop. Dis. 2022, 16, e0010509. [Google Scholar] [CrossRef]
- Zhang, D.; Ge, Y.; Wu, X.; Liu, H.; Zhang, W.; Lai, S. Data-Driven Models Informed by Spatiotemporal Mobility Patterns for Understanding Infectious Disease Dynamics. ISPRS Int. J. Geoinf. 2023, 12, 266. [Google Scholar] [CrossRef]
- Finazzi, F. Replacing discontinued Big Tech mobility reports: A penetration-based analysis. Sci. Rep. 2023, 13, 935. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, Y.; Zou, Y.; Tu, J.; Tang, W.; Yu, R.; Yang, S.; Huang, P. Associations of socioeconomic status with infectious diseases mediated by lifestyle, environmental pollution and chronic comorbidities: A comprehensive evaluation based on UK Biobank. Infect. Dis. Poverty 2023, 12, 5. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Chies, J.A.B. Pathogen Pollution: Viral Diseases Associated with Poor Sanitation in Brazil. Hygiene 2023, 3, 441–449. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: Infectious Disease Surveillance Using Artificial Intelligence (AI) and its Role in Epidemic and Pandemic Preparedness. Med. Sci. Monit. 2023, 29, e941209. [Google Scholar] [CrossRef]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, Y.; Dong, X.; Jin, Y.; Cheng, S.; Zhang, N.; Xu, S.; Gu, S.; Wu, Y.; Yang, J.; et al. Artificial intelligence-based preoperative prediction system for diagnosis and prognosis in epithelial ovarian cancer: A multicenter study. Front. Oncol. 2022, 12, 975703. [Google Scholar] [CrossRef]
- Boehm, K.M.; Aherne, E.A.; Ellenson, L.; Nikolovski, I.; Alghamdi, M.; Vázquez-García, I.; Zamarin, D.; Long Roche, K.; Liu, Y.; Patel, D.; et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat. Cancer 2022, 3, 723–733. [Google Scholar] [CrossRef]
- Hira, M.T.; Razzaque, M.A.; Angione, C.; Scrivens, J.; Sawan, S.; Sarkar, M. Integrated multi-omics analysis of ovarian cancer using variational autoencoders. Sci. Rep. 2021, 11, 6265. [Google Scholar] [CrossRef]
- Kebede, S.R.; Waldamichael, F.G.; Debelee, T.G.; Aleme, M.; Bedane, W.; Mezgebu, B.; Merga, Z.C. Dual view deep learning for enhanced breast cancer screening using mammography. Sci. Rep. 2024, 14, 3839. [Google Scholar] [CrossRef]
- Hendrix, W.; Hendrix, N.; Scholten, E.T.; Mourits, M.; Trap-de Jong, J.; Schalekamp, S.; Korst, M.; van Leuken, M.; van Ginneken, B.; Prokop, M.; et al. Deep learning for the detection of benign and malignant pulmonary nodules in non-screening chest CT scans. Commun. Med. 2023, 3, 156. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadek, T.; Yusuf, N. Ultraviolet Radiation Biological and Medical Implications. Curr. Issues Mol. Biol. 2024, 46, 1924–1942. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.Y.; Tuboi, S.; de Jesus Lopes de Abreu, A.; Abud, D.A.; Lobao Neto, A.A.; Pereira, R.; Siqueira, J.B. A machine learning model to assess potential misdiagnosed dengue hospitalization. Heliyon 2023, 9, e16634. [Google Scholar] [CrossRef] [PubMed]
- AlMohimeed, A.; Saleh, H.; El-Rashidy, N.; Saad, R.M.A.; El-Sappagh, S.; Mostafa, S. Diagnosis of COVID-19 Using Chest X-ray Images and Disease Symptoms Based on Stacking Ensemble Deep Learning. Diagnostics 2023, 13, 1968. [Google Scholar] [CrossRef]
- Medeiros, E.P.; Machado, M.R.; de Freitas, E.D.G.; da Silva, D.S.; de Souza, R.W.R. Applications of machine learning algorithms to support COVID-19 diagnosis using X-rays data information. Expert Syst. Appl. 2024, 238, 122029. [Google Scholar] [CrossRef]
- Park, M.; Lee, Y.; Kim, S.; Kim, Y.J.; Kim, S.Y.; Kim, Y.; Kim, H.M. Distinguishing nontuberculous mycobacterial lung disease and Mycobacterium tuberculosis lung disease on X-ray images using deep transfer learning. BMC Infect. Dis. 2023, 23, 32. [Google Scholar] [CrossRef]
- Hussein, A.M.A.; Sharifai, A.G.; Alia, O.M.; Abualigah, L.; Almotairi, K.H.; Abujayyab, S.K.M.; Gandomi, A.H. Auto-detection of the coronavirus disease by using deep convolutional neural networks and X-ray photographs. Sci. Rep. 2024, 14, 534. [Google Scholar] [CrossRef]
- Sanida, M.V.; Sanida, T.; Sideris, A.; Dasygenis, M. An Advanced Deep Learning Framework for Multi-Class Diagnosis from Chest X-ray Images. J 2024, 7, 48–71. [Google Scholar] [CrossRef]
- Kazemzadeh, S.; Yu, J.; Jamshy, S.; Pilgrim, R.; Nabulsi, Z.; Chen, C.; Beladia, N.; Lau, C.; McKinney, S.M.; Hughes, T.; et al. Deep Learning Detection of Active Pulmonary Tuberculosis at Chest Radiography Matched the Clinical Performance of Radiologists. Radiology 2023, 306, 124–137. [Google Scholar] [CrossRef]
- Sanghvi, H.A.; Patel, R.H.; Agarwal, A.; Gupta, S.; Sawhney, V.; Pandya, A.S. A deep learning approach for classification of COVID and pneumonia using DenseNet-201. Int. J. Imaging Syst. Technol. 2023, 33, 18–38. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Rahman, A.; AlGhamdi, F.; AlDakheel, S.; Hakami, H.; AlJumah, A.; AlIbrahim, Z.; Youldash, M.; Alam Khan, M.A.; Basheer Ahmed, M.I. Joint Diagnosis of Pneumonia, COVID-19, and Tuberculosis from Chest X-ray Images: A Deep Learning Approach. Diagnostics 2023, 13, 2562. [Google Scholar] [CrossRef]
- Pan, C.T.; Kumar, R.; Wen, Z.H.; Wang, C.H.; Chang, C.Y.; Shiue, Y.L. Improving Respiratory Infection Diagnosis with Deep Learning and Combinatorial Fusion: A Two-Stage Approach Using Chest X-ray Imaging. Diagnostics 2024, 14, 500. [Google Scholar] [CrossRef]
- Abdulahi, A.R.T.; Ogundokun, R.O.; Adenike, A.R.; Shah, M.A.; Ahmed, Y.K. PulmoNet: A novel deep learning based pulmonary diseases detection model. BMC Med. Imaging 2024, 24, 51. [Google Scholar] [CrossRef]
- Topff, L.; Sánchez-García, J.; López-González, R.; Pastor, A.J.; Visser, J.J.; Huisman, M.; Guiot, J.; Beets-Tan, R.G.H.; Alberich-Bayarri, A.; Fuster-Matanzo, A.; et al. A deep learning-based application for COVID-19 diagnosis on CT: The Imaging COVID-19 AI initiative. PLoS ONE 2023, 18, e0285121. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, X.; Jia, Y.; Xu, J. The communication of artificial intelligence and deep learning in computer tomography image recognition of epidemic pulmonary infectious diseases. PLoS ONE 2024, 19, e0297578. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.C.; Diao, K.Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef]
- Islam, M.R.; Nahiduzzaman, M. Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach. Expert Syst. Appl. 2022, 195, 116554. [Google Scholar] [CrossRef]
- Kathamuthu, N.D.; Subramaniam, S.; Le, Q.H.; Muthusamy, S.; Panchal, H.; Sundararajan, S.C.M.; Alrubaie, A.J.; Maher Abdul Zahra, M. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Adv. Eng. Softw. 2023, 175, 103317. [Google Scholar] [CrossRef]
- Choudhary, T.; Gujar, S.; Goswami, A.; Mishra, V.; Badal, T. Deep learning-based important weights-only transfer learning approach for COVID-19 CT-scan classification. Appl. Intell. 2023, 53, 7201–7215. [Google Scholar] [CrossRef]
- Bala, D.; Hossain, M.S.; Hossain, M.A.; Abdullah, M.I.; Rahman, M.M.; Manavalan, B.; Gu, N.; Islam, M.S.; Huang, Z. MonkeyNet: A robust deep convolutional neural network for monkeypox disease detection and classification. Neural Netw. 2023, 161, 757–775. [Google Scholar] [CrossRef]
- Yotsu, R.R.; Ding, Z.; Hamm, J.; Blanton, R.E. Deep learning for AI-based diagnosis of skin-related neglected tropical diseases: A pilot study. PLoS Negl. Trop. Dis. 2023, 17, e0011230. [Google Scholar] [CrossRef]
- Almufareh, M.F.; Tehsin, S.; Humayun, M.; Kausar, S. A Transfer Learning Approach for Clinical Detection Support of Monkeypox Skin Lesions. Diagnostics 2023, 13, 1503. [Google Scholar] [CrossRef]
- Hassan, M.; Ali, S.; Saleem, M.; Sanaullah, M.; Fahad, L.G.; Kim, J.Y.; Alquhayz, H.; Tahir, S.F. Diagnosis of dengue virus infection using spectroscopic images and deep learning. PeerJ Comput. Sci. 2022, 8, e985. [Google Scholar] [CrossRef]
- Liou, N.; De, T.; Urbanski, A.; Chieng, C.; Kong, Q.; David, A.L.; Khasriya, R.; Yakimovich, A.; Horsley, H. A clinical microscopy dataset to develop a deep learning diagnostic test for urinary tract infection. Sci. Data 2024, 11, 155. [Google Scholar] [CrossRef]
- Mayrose, H.; Sampathila, N.; Muralidhar Bairy, G.; Nayak, T.; Belurkar, S.; Saravu, K. An Explainable Artificial Intelligence Integrated System for Automatic Detection of Dengue From Images of Blood Smears Using Transfer Learning. IEEE Access 2024, 12, 41750–41762. [Google Scholar] [CrossRef]
- Lundin, J.; Suutala, A.; Holmström, O.; Henriksson, S.; Valkamo, S.; Kaingu, H.; Kinyua, F.; Muinde, M.; Lundin, M.; Diwan, V.; et al. Diagnosis of soil-transmitted helminth infections with digital mobile microscopy and artificial intelligence in a resource-limited setting. PLoS Negl. Trop. Dis. 2024, 18, e0012041. [Google Scholar] [CrossRef]
- Giraud, C. Introduction to High-Dimensional Statistics; Taylor & Francis: London, UK, 2021. [Google Scholar] [CrossRef]
- Kaagaard, M.D.; Matos, L.O.; Evangelista, M.V.P.; Wegener, A.; Holm, A.E.; Vestergaard, L.S.; Do Valle, S.C.N.; Silvestre, O.M.; Lacerda, M.V.G.; de Souza, R.M.; et al. Frequency of pleural effusion in dengue patients by severity, age and imaging modality: A systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 327. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Bhat, V.; Sampathila, N.; Umakanth, S.; Chadaga, R. A Decision Support System for Diagnosis of COVID-19 from Non-COVID-19 Influenza-like Illness Using Explainable Artificial Intelligence. Bioengineering 2023, 10, 439. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Gunčar, G.; Kukar, M.; Smole, T.; Moškon, S.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Notar, M.; Köster, M.; et al. Differentiating viral and bacterial infections: A machine learning model based on routine blood test values. Heliyon 2024, 10, e29372. [Google Scholar] [CrossRef]
- Goodman-Meza, D.; Rudas, A.; Chiang, J.N.; Adamson, P.C.; Ebinger, J.; Sun, N.; Botting, P.; Fulcher, J.A.; Saab, F.G.; Brook, R.; et al. A machine learning algorithm to increase COVID-19 inpatient diagnostic capacity. PLoS ONE 2020, 15, e0239474. [Google Scholar] [CrossRef]
- Ming, D.K.; Tuan, N.M.; Hernandez, B.; Sangkaew, S.; Vuong, N.L.; Chanh, H.Q.; Chau, N.V.V.; Simmons, C.P.; Wills, B.; Georgiou, P.; et al. The Diagnosis of Dengue in Patients Presenting With Acute Febrile Illness Using Supervised Machine Learning and Impact of Seasonality. Front. Digit. Health 2022, 4, 849641. [Google Scholar] [CrossRef]
- Aguirre, U.; Urrechaga, E. Diagnostic performance of machine learning models using cell population data for the detection of sepsis: A comparative study. Clin. Chem. Lab. Med. 2023, 61, 356–365. [Google Scholar] [CrossRef]
- Tay, J.; Yen, Y.H.; Rivera, K.; Chou, E.H.; Wang, C.H.; Chou, F.Y.; Sun, J.T.; Han, S.T.; Tsai, T.P.; Chen, Y.C.; et al. Development and External Validation of Clinical Features-based Machine Learning Models for Predicting COVID-19 in the Emergency Department. West. J. Emerg. Med. 2024, 25, 67–78. [Google Scholar] [CrossRef]
- Park, D.J.; Park, M.W.; Lee, H.; Kim, Y.J.; Kim, Y.; Park, Y.H. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci. Rep. 2021, 11, 7567. [Google Scholar] [CrossRef]
- Dantas, L.F.; Peres, I.T.; Bastos, L.S.L.; Marchesi, J.F.; De Souza, G.F.G.; Gelli, J.G.M.; Baião, F.A.; MacAira, P.; Hamacher, S.; Bozza, F.A. App-based symptom tracking to optimize SARS-CoV-2 testing strategy using machine learning. PLoS ONE 2021, 16, e0248920. [Google Scholar] [CrossRef]
- Vu, D.M.; Krystosik, A.R.; Ndenga, B.A.; Mutuku, F.M.; Ripp, K.; Liu, E.; Bosire, C.M.; Heath, C.; Chebii, P.; Maina, P.W.; et al. Detection of acute dengue virus infection, with and without concurrent malaria infection, in a cohort of febrile children in Kenya, 2014–2019, by clinicians or machine learning algorithms. PLoS Global. Public Health 2023, 3, e0001950. [Google Scholar] [CrossRef]
- Hong, S.; Lynn, H.S. Accuracy of random-forest-based imputation of missing data in the presence of non-normality, non-linearity, and interaction. BMC Med. Res. Methodol. 2020, 20, 199. [Google Scholar] [CrossRef]
- Moallemi, S.; Lloyd, A.R.; Rodrigo, C. Early biomarkers for prediction of severe manifestations of dengue fever: A systematic review and a meta-analysis. Sci. Rep. 2023, 13, 17485. [Google Scholar] [CrossRef]
- Savage, T.; Nayak, A.; Gallo, R.; Rangan, E.; Chen, J.H. Diagnostic reasoning prompts reveal the potential for large language model interpretability in medicine. NPJ Digit. Med. 2024, 7, 20. [Google Scholar] [CrossRef]
- Schwartz, I.S.; Link, K.E.; Daneshjou, R.; Cortés-Penfield, N. Black Box Warning: Large Language Models and the Future of Infectious Diseases Consultation. Clin. Infect. Dis. 2024, 78, 860–866. [Google Scholar] [CrossRef]
- Cheng, K.; Li, Z.; He, Y.; Guo, Q.; Lu, Y.; Gu, S.; Wu, H. Potential Use of Artificial Intelligence in Infectious Disease: Take ChatGPT as an Example. Ann. Biomed. Eng. 2023, 51, 1130–1135. [Google Scholar] [CrossRef]
- Chiu, W.H.K.; Ko, W.S.K.; Cho, W.C.S.; Hui, S.Y.J.; Chan, W.C.L.; Kuo, M.D. Evaluating the Diagnostic Performance of Large Language Models on Complex Multimodal Medical Cases. J. Med. Internet Res. 2024, 26, e53724. [Google Scholar] [CrossRef]
- Hansebout, R.R.; Cornacchi, S.D.; Haines, T.; Goldsmith, C.H. How to use an article about prognosis. Can. J. Surg. 2009, 52, 328–336. [Google Scholar]
- D’Abramo, A.; Rinaldi, F.; Vita, S.; Mazzieri, R.; Corpolongo, A.; Palazzolo, C.; Ascoli Bartoli, T.; Faraglia, F.; Giancola, M.L.; Girardi, E.; et al. A machine learning approach for early identification of patients with severe imported malaria. Malar. J. 2024, 23, 46. [Google Scholar] [CrossRef]
- Chaw, J.K.; Chaw, S.H.; Quah, C.H.; Sahrani, S.; Ang, M.C.; Zhao, Y.; Ting, T.T. A predictive analytics model using machine learning algorithms to estimate the risk of shock development among dengue patients. Healthc. Anal. 2024, 5, 100290. [Google Scholar] [CrossRef]
- Lourenço, A.A.; Amaral, P.H.R.; Paim, A.A.O.; Marques, G.F.; Gomes-de-Pontes, L.; da Mata, C.P.S.M.; da Fonseca, F.G.; Pérez, J.C.G.; Coelho-dos-Reis, J.G.A. Algorithms for predicting COVID outcome using ready-to-use laboratorial and clinical data. Front. Public Health 2024, 12, 1347334. [Google Scholar] [CrossRef]
- Ming, D.K.; Hernandez, B.; Sangkaew, S.; Vuong, N.L.; Lam, P.K.; Nguyet, N.M.; Tam, D.T.H.; Trung, D.T.; Tien, N.T.H.; Tuan, N.M.; et al. Applied machine learning for the risk-stratification and clinical decision support of hospitalised patients with dengue in Vietnam. PLoS Digital. Health 2022, 18, e0000005. [Google Scholar] [CrossRef]
- Queipo, M.; Barbado, J.; Torres, A.M.; Mateo, J. Approaching Personalized Medicine: The Use of Machine Learning to Determine Predictors of Mortality in a Population with SARS-CoV-2 Infection. Biomedicines 2024, 12, 409. [Google Scholar] [CrossRef]
- Lu, X.; Chen, Y.; Zhang, G.; Zeng, X.; Lai, L.; Qu, C. Comparative Analysis of Machine Learning Models for Prediction of Acute Liver Injury in Sepsis Patients. J. Emerg. Trauma Shock 2024, 17, 91–101. [Google Scholar] [CrossRef]
- Yilmaz, G.; Sezer, S.; Bastug, A.; Singh, V.; Gopalan, R.; Aydos, O.; Ozturk, B.Y.; Gokcinar, D.; Kamen, A.; Gramz, J.; et al. Concordance and generalization of an AI algorithm with real-world clinical data in the pre-omicron and omicron era. Heliyon 2024, 10, e25410. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Jeong, Y.J.; Kim, Y.H.; Kim, J.Y.; Kim, J.H.; Kim, E.Y.; Lim, J.K.; Kim, J.; Kim, Z.; Kim, K.; et al. Development and Validation of a Robust and Interpretable Early Triaging Support System for Patients Hospitalized With COVID-19: Predictive Algorithm Modeling and Interpretation Study. J. Med. Internet Res. 2024, 26, e52134. [Google Scholar] [CrossRef] [PubMed]
- Rui, F.; Yeo, Y.H.; Xu, L.; Zheng, Q.; Xu, X.; Ni, W.; Tan, Y.; Zeng, Q.L.; He, Z.; Tian, X.; et al. Development of a machine learning-based model to predict hepatic inflammation in chronic hepatitis B patients with concurrent hepatic steatosis: A cohort study. EClinicalMedicine 2024, 68, 102419. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Cui, X.; Zhao, Z.; Wu, R.; Zhang, Q.; Xue, L.; Zhang, H.; Ge, Q.; Leng, Y. A generalizable and easy-to-use COVID-19 stratification model for the next pandemic via immune-phenotyping and machine learning. Front. Immunol. 2024, 15, 1372539. [Google Scholar] [CrossRef]
- Park, H.; Choi, C.M.; Kim, S.H.; Kim, S.H.; Kim, D.K.; Jeong, J.B. In-hospital real-time prediction of COVID-19 severity regardless of disease phase using electronic health records. PLoS ONE 2024, 19, e0294362. [Google Scholar] [CrossRef]
- Zargari Marandi, R.; Leung, P.; Sigera, C.; Murray, D.D.; Weeratunga, P.; Fernando, D.; Rodrigo, C.; Rajapakse, S.; Macpherson, C.R. Development of a machine learning model for early prediction of plasma leakage in suspected dengue patients. PLoS Negl. Trop. Dis. 2023, 17, e0010758. [Google Scholar] [CrossRef]
- Zhang, G.; Shao, F.; Yuan, W.; Wu, J.; Qi, X.; Gao, J.; Shao, R.; Tang, Z.; Wang, T. Predicting sepsis in-hospital mortality with machine learning: A multi-center study using clinical and inflammatory biomarkers. Eur. J Med. Res. 2024, 29, 156. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, Y.; Zhang, W.; Lu, H.; Lin, S.; Zhang, W.; Xia, L.; Hu, H.; Song, Y.; Xu, F. Proteome analysis develops novel plasma proteins classifier in predicting the mortality of COVID-19. Cell Prolif. 2024, 57, e13617. [Google Scholar] [CrossRef]
- Xu, W.; Sun, N.N.; Gao, H.N.; Chen, Z.Y.; Yang, Y.; Ju, B.; Tang, L.L. Risk factors analysis of COVID-19 patients with ARDS and prediction based on machine learning. Sci. Rep. 2021, 11, 2933. [Google Scholar] [CrossRef]
- Huang, S.W.; Tsai, H.P.; Hung, S.J.; Ko, W.C.; Wang, J.R. Assessing the risk of dengue severity using demographic information and laboratory test results with machine learning. PLoS Negl. Trop. Dis. 2020, 14, e0008960. [Google Scholar] [CrossRef]
- Hien, N.T.K.; Tsai, F.J.; Chang, Y.H.; Burton, W.; Phuc, P.T.; Nguyen, P.A.; Harnod, D.; Lam, C.S.; Lu, T.C.; Chen, C.I.; et al. Unveiling the future of COVID-19 patient care: Groundbreaking prediction models for severe outcomes or mortality in hospitalized cases. Front. Med. 2023, 10, 1289968. [Google Scholar] [CrossRef] [PubMed]
- Yong, L.; Zhenzhou, L. Deep learning-based prediction of in-hospital mortality for sepsis. Sci. Rep. 2024, 14, 372. [Google Scholar] [CrossRef] [PubMed]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Chadaga, R.; Umakanth, S.; Bhat, D.; G S, S.K. Explainable artificial intelligence approaches for COVID-19 prognosis prediction using clinical markers. Sci. Rep. 2024, 14, 1783. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.E.; Saul, S.; Rao, A.M.; Robinson, M.L.; Agudelo Rojas, O.L.; Sanz, A.M.; Verghese, M.; Solis, D.; Sibai, M.; Huang, C.H.; et al. An 8-gene machine learning model improves clinical prediction of severe dengue progression. Genome Med. 2022, 14, 33. [Google Scholar] [CrossRef]
- Chen, L.; Hua, J.; He, X. Bioinformatics analysis identifies a key gene HLA_DPA1 in severe influenza-associated immune infiltration. BMC Genom. 2024, 25, 257. [Google Scholar] [CrossRef]
- Natali, E.N.; Horst, A.; Meier, P.; Greiff, V.; Nuvolone, M.; Babrak, L.M.; Fink, K.; Miho, E. The dengue-specific immune response and antibody identification with machine learning. NPJ Vaccines 2024, 9, 16. [Google Scholar] [CrossRef]
- Carney, M.; Pelaia, T.M.; Chew, T.; Teoh, S.; Phu, A.; Kim, K.; Wang, Y.; Iredell, J.; Zerbib, Y.; McLean, A.; et al. Host transcriptomics and machine learning for secondary bacterial infections in patients with COVID-19: A prospective, observational cohort study. Lancet Microbe 2024, 5, e272–e281. [Google Scholar] [CrossRef] [PubMed]
- Miyakis, S.; Karamanof, G.; Liontos, M.; Mountokalakis, T.D. Factors contributing to inappropriate ordering of tests in an academic medical department and the effect of an educational feedback strategy. Postgrad. Med. J. 2006, 82, 823–829. [Google Scholar] [CrossRef]
- Falcone, M.; Bauer, M.; Ferrer, R.; Gavazzi, G.; Gonzalez del Castillo, J.; Pilotto, A.; Schuetz, P. Biomarkers for risk stratification and antibiotic stewardship in elderly patients. Aging Clin. Exp. Res. 2023, 35, 925–935. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Chadaga, R. A machine learning and explainable artificial intelligence approach for predicting the efficacy of hematopoietic stem cell transplant in pediatric patients. Healthc. Anal. 2023, 3, 100170. [Google Scholar] [CrossRef]
- Collins, G.S.; Moons, K.G.M.; Dhiman, P.; Riley, R.D.; Beam, A.L.; Van Calster, B.; Ghassemi, M.; Liu, X.; Reitsma, J.B.; van Smeden, M.; et al. TRIPOD+AI statement: Updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 2024, 385, e078378. [Google Scholar] [CrossRef] [PubMed]
- Cruz Rivera, S.; Liu, X.; Chan, A.W.; Denniston, A.K.; Calvert, M.J.; SPIRIT-AI and CONSORT-AI Working Group; SPIRIT-AI and CONSORT-AI Steering Group; SPIRIT-AI and CONSORT-AI Consensus Group. Guidelines for clinical trial protocols for interventions involving artificial intelligence: The SPIRIT-AI extension. Nat. Med. 2020, 26, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Sander, R.M.; Limon, A. Twelve key challenges in medical machine learning and solutions. Intell. Based Med. 2022, 6, 100068. [Google Scholar] [CrossRef]
- Rockenschaub, P.; Akay, E.M.; Carlisle, B.G.; Hilbert, A.; Wendland, J.; Meyer-Eschenbach, F.; Näher, A.F.; Frey, D.; Madai, V.I. External validation of AI-based scoring systems in the ICU: A systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2025, 25, 5. [Google Scholar] [CrossRef] [PubMed]
- Amini, A.; Soleimany, A.P.; Schwarting, W.; Bhatia, S.N.; Rus, D. Uncovering and mitigating algorithmic bias through learned latent structure. In AIES 2019—Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society; Association for Computing Machinery, Inc.: New York, NY, USA, 2019; pp. 289–295. [Google Scholar]
- Li, J.; Guo, S.; Ma, R.; He, J.; Zhang, X.; Rui, D.; Ding, Y.; Li, Y.; Jian, L.; Cheng, J.; et al. Comparison of the effects of imputation methods for missing data in predictive modelling of cohort study datasets. BMC Med. Res. Methodol. 2024, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Moehring, A.; Banerjee, O.; Salz, T.; Agarwal, N.; Rajpurkar, P. Heterogeneity and predictors of the effects of AI assistance on radiologists. Nat. Med. 2024, 30, 837–849. [Google Scholar] [CrossRef]
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Chadaga, R.; Prabhu, S.; K S, S.; Jagdale, A.S.; Bhat, D. A decision support system for osteoporosis risk prediction using machine learning and explainable artificial intelligence. Heliyon 2023, 9, e22456. [Google Scholar] [CrossRef]
- Boussina, A.; Shashikumar, S.P.; Malhotra, A.; Owens, R.L.; El-Kareh, R.; Longhurst, C.A.; Quintero, K.; Donahue, A.; Chan, T.C.; Nemati, S.; et al. Impact of a deep learning sepsis prediction model on quality of care and survival. NPJ Digit. Med. 2024, 7, 14. [Google Scholar] [CrossRef]
- Poddar, M.; Marwaha, J.S.; Yuan, W.; Romero-Brufau, S.; Brat, G.A. An operational guide to translational clinical machine learning in academic medical centers. NPJ Digit. Med. 2024, 7, 129. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheah, B.C.J.; Vicente, C.R.; Chan, K.R. Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis. Viruses 2025, 17, 882. https://doi.org/10.3390/v17070882
Cheah BCJ, Vicente CR, Chan KR. Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis. Viruses. 2025; 17(7):882. https://doi.org/10.3390/v17070882
Chicago/Turabian StyleCheah, Brandon C. J., Creuza Rachel Vicente, and Kuan Rong Chan. 2025. "Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis" Viruses 17, no. 7: 882. https://doi.org/10.3390/v17070882
APA StyleCheah, B. C. J., Vicente, C. R., & Chan, K. R. (2025). Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis. Viruses, 17(7), 882. https://doi.org/10.3390/v17070882






