Regulation of Bombyx mori–BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms
Abstract
1. Introduction
2. Techniques for Protein Interaction Research: From Classics to Frontiers
2.1. Traditional Protein Interaction Research Techniques
2.2. Emerging Protein Interaction Research Techniques
2.3. Technology Integration Strategies and Challenges
2.3.1. Current Challenges
2.3.2. Technology Integration Strategies
3. Identification and Functional Analysis of Bombyx mori Proteins Interacting with BmNPV and Their Roles in Viral Proliferation
3.1. Apoptosis and Cycle Regulation
3.1.1. Apoptosis Regulation
3.1.2. Cell Cycle Regulation
3.2. Regulation of Viral Protein Invasion and Transport
3.2.1. Viral Invasion and Membrane Fusion: Vesicle Protein-Mediated Adsorption and Membrane Fusion
3.2.2. Viral Transport and Nucleation: Microtubule Network-Dependent Intranuclear Transport
3.3. Non-Coding RNA Regulation
3.3.1. Mechanisms of Non-Coding RNA-Mediated Viral Proliferation Promotion
3.3.2. Non-Coding RNA-Mediated Host Antiviral Defense Mechanisms
3.3.3. Strategies for Identifying BmNPV-Derived ncRNAs and Their Host Targets
3.4. Metabolic Regulation
3.4.1. Mitochondrial Metabolic Antiviral Mechanisms
3.4.2. Mechanisms of Mitochondrial Virus Hijacking
3.4.3. Immunomodulation
4. Discussion
4.1. Comparative Insights with Closely Related Baculoviruses
4.2. Unresolved Challenges and Future Prospects
- (i)
- Identifying critical viral hijacking nodes by utilizing multi-omics technologies to pinpoint key host molecular targets manipulated by the virus.
- (ii)
- Developing targeted interventions by designing strategies against these targets, with safety and efficiency first validated in genetically modified silkworms or via genome editing.
- (iii)
- Assessing the economic impact by evaluating the effects of any interventions on economically vital traits such as silk yield and quality to balance host health and agricultural value.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, L. Insights Into the antiviral pathways of the Silkworm Bombyx mori. Front. Immunol. 2021, 12, 639092. [Google Scholar] [CrossRef] [PubMed]
- Biron, D.G.; Nedelkov, D.; Missé, D.; Holzmuller, P. Proteomics and Host Pathogen Interactions: A Bright Future? In Genetics and Evolution of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2017; pp. 263–303. [Google Scholar]
- Liu, P.G.; Li, W.G.; Wang, Y.Q. Progress of proteomics research in silkworm (Bombyx mori). Acta Agric. Zhejiangensis 2010, 22, 124–129. [Google Scholar]
- Chen, P.; Bao, X.Y.; Kang, T.T.; Dong, Z.Q.; Zhu, Y.; Pan, M.H.; Lu, C. Screening and identification of proteins interacting with Bombyx mori IAP and their effects on BmNPV proliferation. Sci. Agric. Sin. 2019, 52, 558–567. [Google Scholar]
- Nchongboh, G.C.; Wu, G.W.; Hong, N.; Wang, G.P. Protein-protein interactions between proteins of Citrus tristeza virus isolates. Virus Genes 2014, 49, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Fields, S.; Song, O.K. A novel genetic system to detect proteinprotein interactions. Nature 1989, 340, 245. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.Y.; Chen, L.B.; Wu, W.J.; Zhao, D.; Wang, Y.; Qin, X.; Zhang, C.X. Direct interactions between bidensovirus BmDNV-Z proteins and midgut proteins from the virus target Bombyx mori. FEBS J. 2013, 280, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Kong, X.S.; Wang, H.P.; Xu, W.F.; Wu, X.F. Screening of Proteins Interacting with P74 Oral Infection Factor of Bombyx mori Nucleopolyhedrovirus in Midgut Cells of Silkworm (Bombyx mori). Sci. Seric. 2018, 44, 8. [Google Scholar]
- Feng, M.; Kong, X.S.; Zhang, J.J.; Xu, W.F.; Wu, X.F. Identification of a novel host protein SINAL10 interacting with GP64 and its role in Bombyx mori nucleopolyhedrovirus infection. Virus Res. 2018, 247, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Nada, H.; Choi, Y.; Kim, S.; Jeong, K.S.; Meanwell, N.A.; Lee, K. New insights into protein–protein interaction modulators in drug discovery and therapeutic advance. Signal Transduct. Target. Ther. 2024, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Shen, Z.Y.; Hou, J.G.; Zhang, J.; Geng, T.; Tang, X.D.; Xu, L.; Guo, X.J. Identification of a protein interacting with the spore wall protein SWP26 of Nosema bombycis in a cultured BmN cell line of silkworm. Infect. Genet. Evol. 2013, 17, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, G.Y.; Hou, J.G.; Zhang, J.; Geng, T.; Tang, X.D.; Xu, L.; Guo, X.J. Potential Proteins Interactions with Bombyx mori Nucleopolyhedrovirus Revealed by Co-Immunoprecipitation. Insects 2022, 13, 575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Feng, M.; Fan, Y.; Xu, W.F.; Zheng, Q.; Wu, X.F. Interaction Network Among BV Structural Proteins of Bombyx mori Nucleopolyhedrovirus. In Proceedings of the 2018 Annual Academic Conference of the Chinese Society of Sericulture, Hangzhou, China, 23–26 May 2018. [Google Scholar]
- Wissmueller, S.; Font, J.; Liew, C.W.; Cram, E.C.; Schroeder, T.; Turner, J.; Crossley, M.; Mackay, J.P.; Matthews, J.M. Protein–protein interactions: Analysis of a false positive GST pulldown result. Proteins 2011, 79, 2365–2371. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Na, S.; Qin, L. Screening of Bombyx mori brain proteins interacting with protein tyrosine phosphatase of BmNPV. Arch. Insect Biochem. Physiol. 2020, 105, e21732. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, S.M.; Shyu, Y.J.; Duren, H.M.; Hu, C.D. Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in Caenorhabditis elegans. Methods 2008, 45, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, T.H.; Dong, X.L.; Wu, Y.F.; Yang, J.G.; Zhou, L.; Pan, C.X.; Pan, M.H. Identification of the Interactions of CDK11 with RNPS1 and 9G8 in the Silkworm (Bombyx mori). Sci. Agric. Sin. 2017, 50, 4398–4407. [Google Scholar]
- Lin, S.; Yin, H.T.; Zhao, Z.M.; Chen, Z.K.; Zhou, X.M.; Zhang, Z.D.; Guo, X.J.; Zhao, W.G.; Wu, P. LincRNAXR209691.3 could promote Bombyx mori nucleopolyhedrovirus replication by interacting with BmHSP70. Insect Mol. Biol. 2022, 32, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Seger, C.; Salzmann, L. After another decade: LC-MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.J.; Yu, M.H.; Dong, X.Y.; Wang, W.L.; Tian, T.; Yu, X.Y.; Tang, X.D. Protein composition analysis of polyhedra matrix of Bombyx mori nucleopolyhedrovirus (BmNPV) showed powerful capacity of polyhedra to encapsulate foreign proteins. Sci. Rep. 2017, 7, 8768. [Google Scholar] [CrossRef] [PubMed]
- Capelli, D.; Scognamiglio, V.; Montanari, R. Surface plasmon resonance technology: Recent advances, applications and experimental cases. Trends Anal. Chem. 2023, 163, 117079. [Google Scholar] [CrossRef]
- Wang, S.J.; Su, D.; Zhang, T. Research progress of surface plasmons mediated photothermal effects. Acta Phys. Sin. 2019, 68, 144401. [Google Scholar] [CrossRef]
- Jeremy, J.; Donald, D. Binding specificity of Bacillus thuringiensis Cry1Aa for purified, native Bombyx mori aminopeptidase N and cadherin-like receptors. BMC Biochem. 2001, 2, 12. [Google Scholar]
- Sumathy, R.; Rao, A.S.K.; Chandrakanth, N.; Gopalakrishnan, V.K. Insilico identification of protein-protein interactions in Silkworm, Bombyx mori. Bioinformation 2014, 10, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Koegl, M.; Uetz, P. Improving yeast two-hybrid screening systems. Brief. Funct. Genom. 2008, 6, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.-C.; Gstaiger, M.; Raught, B.; Aebersold, R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007, 8, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.Y.; Wang, C.; Shi, F.M. Research advances on protein interaction technology. Hubei Agric. Sci. 2019, 58, 5–10. [Google Scholar]
- Jason-Moller, L.; Murphy, M.; Bruno, J.A. Overview of Biacore Systems and Their Applications. Curr. Protoc. Protein Sci. 2006, 45, 19. [Google Scholar] [CrossRef] [PubMed]
- Olaru, A.; Bala, C.; Jaffrezic-Renault, N.; Aboul-Enein, H.Y. Surface Plasmon Resonance (SPR) Biosensors in Pharmaceutical Analysis. Crit. Rev. Anal. Chem. 2015, 45, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast Two-Hybrid, a Powerful Tool for Systems Biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.C.; Chen, W.X.; Li, X.-P. Research Progress of Protein-protein Interaction Technology A Research Review of the Technology of Protein-protein Interaction. Fujian J. Agric. Sci. 2016, 31, 1131–1138. [Google Scholar]
- Chen, Z.B.; Zhou, G.L.; Huang, J.X. Application of GST Pull-Down Technique in Study on Protein-Protein Interaction. Chin. J. Biol. 2014, 27, 1354–1358. [Google Scholar]
- Kodama, Y.; Hu, C.D. Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 2012, 53, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Magliery, T.J.; Wilson, C.G.M.; Pan, W.; Mishler, D.; Ghosh, I.; Hamilton, A.D.; Regan, L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: Scope and mechanism. J. Am. Chem. Soc. 2005, 127, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Li, Y. The application and development of liquid chromatography-mass spectrometry in clinical laboratory medicine. Chin. J. Lab. Med. 2016, 39, 650–653. [Google Scholar]
- Ashrafi, T.M.S.; Mohanty, G. Surface Plasmon Resonance Sensors: A Critical Review of Recent Advances, Market Analysis, and Future Directions. Plasmonics 2025, 163, 1–21. [Google Scholar] [CrossRef]
- Martins, Y.C.; Ziviani, A.; Nicolás, M.F.; de Vasconcelos, A.T.R. Large-Scale Protein Interactions Prediction by Multiple Evidence Analysis Associated With an In-Silico Curation Strategy. Front. Bioinform. 2021, 1, 731345. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.X.; Wang, J.H.; Yasen, A.; Fan, B.Y.; Hull, J.J.; Shen, X.J. Determination of Key Components in the Bombyx mori p53 Apoptosis Regulation Network Using Y2H-Seq. Insects 2023, 14, 362. [Google Scholar] [CrossRef] [PubMed]
- Shiseki, M.; Nagashima, M.; Pedeux, R.M.; Kitahama-Shiseki, M.; Miura, K.; Okamura, S.; Onogi, H.; Higashimoto, Y.; Appella, E.; Yokota, J.; et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003, 63, 2373–2378. [Google Scholar] [PubMed]
- Mo, Y.Q.; Gu, C.G.; Hu, M.; Wu, J.N.; Miao, M.; Yu, W. The Regulatory Role of ING5 Acetylation in Baculovirus Infection of Bombyx mori. Sci. Seric. 2024, 50, 232–241. [Google Scholar]
- Dong, Z.Q.; Zhang, X.L.; Xiao, M.; Li, K.J.; Wang, J.; Chen, P.; Hu, Z.G.; Lu, C.; Pan, M.H. Baculovirus LEF-11 interacts with BmIMPI to induce cell cycle arrest in the G2/M phase for viral replication. Pestic. Biochem. Physiol. 2022, 188, 105231. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Dong, Z.Q.; Xiao, Q.; Hu, Z.G.; Wang, J.; Chen, P.; Pan, M.H. Baculovirus apoptosis suppressor gene IAP1 promotes nuclear accumulation of CyclinB in Bombyx mori. Acta Microbiol. Sin. 2022, 62, 3478–3488. [Google Scholar]
- Qin, X.; Dong, Z.Q.; Zhu, Y.; Zhang, Q.; Yang, X.; Xiao, M.; Chen, P.; Lu, C.; Pan, M.H. Bombyx mori Nucleopolyhedrovirus (BmNPV) Induces G2/M Arrest to Promote Viral Multiplication by Depleting BmCDK1. Insects 2021, 12, 1098. [Google Scholar]
- Hao, B.; Nan, W.; Xu, Y.; Liu, L.; Liu, N.; Huang, J. Two Cholesterol Recognition Amino Acid Consensus Motifs of GP64 with Uncleaved Signal Peptide Are Required for Bombyx mori Nucleopolyhedrovirus Infection. Microbiol. Spectr. 2021, 9, e0172521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Fei, S.; Awais, M.M.; Zheng, H.; Feng, M.; Sun, J. Heat Shock Protein 75 (TRAP1) facilitate the proliferation of the Bombyx mori nucleopolyhedrovirus. Int. J. Biol. Macromol. 2021, 175, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.L.; Liu, T.H.; Wang, W.; Pan, C.X.; Wu, Y.F.; Du, G.Y.; Chen, P.; Lu, C.L.; Pan, M.H. BmREEPa Is a Novel Gene that Facilitates BmNPV Entry into Silkworm Cells. PLoS ONE 2015, 10, e0144575. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.L.; Wu, Y.F.; Liu, T.H.; Wang, W.; Pan, C.X.; Adur, M.; Zhang, M.J.; Pan, M.H.; Lu, C. Bombyx mori protein BmREEPa and BmPtchd could form a complex with BmNPV envelope protein GP64. Biochem. Biophys. Res. Commun. 2017, 490, 1254–1259. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Fan, Y.P.; Wei, J.H.; Mei, X.G.; He, Q.; Zhang, Y.H.; Li, T.; Long, M.X.; Chen, J.; Bao, J.L.; et al. Baculovirus Utilizes Cholesterol Transporter NIEMANN–Pick C1 for Host Cell Entry. Front. Microbiol. 2019, 10, 2825. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.P.; Bao, J.L.; Fu, X.Y.; Wu, P.F.; Chen, J.Y.; Huang, Y.; Wei, J.H.; Pan, G.Q.; Li, C.F.; Zhou, Z.Y. The NPC Families Mediate BmNPV Entry. Microbiol. Spectr. 2022, 10, e0091722. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.N.; Fan, Y.P.; Yang, Z.H.; Wei, J.H.; Pan, G.Q.; Li, C.F. Identification of Key Sites for BmNPC2 Interaction withBmNPV GP64. Acta Sericologica Sin. 2025, 1–14. [Google Scholar] [CrossRef]
- Hu, N.; Dong, Z.Q.; Chen, T.T.; Pan, M.H. Overexpression of Bombyx mori nucleopolyhedrovirus nucleocapsidprotein VP39 inhibits the proliferation of BmNPV in BmN-SWU1. Acta Entomol. Sin. 2015, 58, 1222–1228. [Google Scholar]
- Wu, P.; Shang, Q.; Huang, H.L.; Zhang, S.L.; Zhong, J.B.; Hou, Q.R.; Guo, X.J. Quantitative proteomics analysis provides insight into the biological role of Hsp90 in BmNPV infection in Bombyx mori. J Proteom. 2019, 203, 103379. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Q.; Jiang, Y.M.; Pan, M.H. Screening and identification of candidate proteins interacting with bmhsp60 in the silkworm (Bombyx mori). J. Integr. Agric. 2019, 52, 376–384. [Google Scholar]
- Pan, J.S.; Li, X.D.; Li, R.S.; Chen, Y.; Memon, F.U.; Wu, K.; Hu, J.H.; Xie, X.L.; Deng, J.H.; Xu, R.T.; et al. Baculovirus protein kinase 1 activates AMPK-protein phosphatase 5 axis to hijack transcription factor EB for self-proliferation. Int. J. Biol. Macromol. 2025, 298, 139884. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lv, J.L.; Cheng, S.; Su, Z.H.; Qin, S.; Sun, X.; Tang, X.D.; Liu, Q.N.; Li, M.-W.; Wang, X.Y. Bombyx mori transcription factor, E74A, beneficially affects BmNPV infection through direct interaction. Pest. Manag. Sci. 2022, 78, 5302–5312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, Y.F.; Chen, P.; Liu, T.H.; Dong, Z.Q.; Lu, C.; Pan, M.H. Bombyx mori cell division cycle protein 37 promotes the proliferation of BmNPV. Pestic. Biochem. Physiol. 2021, 178, 104923. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.Y.; Cao, Y.Q.; Ge, S.C.; Xu, A.Y.; Qian, H.Y.; Li, G. Identification of key genes involved in resistance to early stage of BmNPV infection in silkworms. Viruses 2022, 14, 2405. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, H.L.; Zhang, Z.L.; Xu, L.; Shen, Z.Y.; Tang, X.D. Verification of Interaction Between BmNPV Immediate-early ProteinPE38 and BmSRPK of Silkworm, Bom byxmori. Acta Sericologica Sin. 2017, 43, 0428–0435. [Google Scholar]
- Liao, J.X.; Zhang, C.; Ru, W.J.; Wang, D.; Zhang, W.P. Effects of overexpression and inhibited expression of thymosin, an actin-interacting protein from Bombyx mori, on BmNPV proliferation and replication. Arch. Insect Biochem. Physiol. 2018, 98, e21449. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Shen, Z.Y.; Wang, M.D.; Zhou, X.M.; Xu, T.; Jiao, X.H.; Wang, L.L.; Guo, X.J.; Wu, P. Lnc557 promotes Bombyx mori nucleopolyhedrovirus replication by interacting with BmELAVL1 to enhance its stability and expression. Pestic. Biochem. Physiol. 2024, 204, 106046. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Zhang, S.L.; Yin, H.T.; Zhao, Z.M.; Chen, Z.K.; Shen, M.M.; Zhang, Z.D.; Guo, X.J.; Wu, P. Cellular Lnc_209997 suppresses Bombyx mori nucleopolyhedrovirus replication by targeting miR-275-5p in Bombyx mori. Insect Mol. Biol. 2022, 31, 308–316. [Google Scholar]
- Cao, H.H.; Kong, W.W.; Ling, B.; Wang, Z.Y.; Zhang, Y.; Guo, Z.X.; Liu, S.H.; Xu, J.P. Bmo-miR-3351 modulates glutathione content and inhibits BmNPV proliferation by targeting BmGSTe6 in Bombyx mori. Insect Sci. 2024, 31, 1378–1396. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.H.; Kong, W.W.; Chen, X.Y.; Ayaz, S.; Hou, C.P.; Wang, Y.S.; Liu, S.-H.; Xu, J.-P. Bmo-miR-6498-5p suppresses Bombyx mori nucleopolyhedrovirus infection by down-regulating BmPLPP2 to modulate pyridoxal phosphate content in Bombyx mori. Insect Mol. Biol. 2024, 33, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.Z.; Yassour, M.; Adiconis, X.; Nusbaum, C.; Thompson, D.A.; Friedman, N.; Gnirke, A.; Regev, A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 2010, 7, 709–715. [Google Scholar] [CrossRef] [PubMed]
- He, P.A.; Nie, Z.; Chen, J.Q.; Chen, J.; Lv, Z.B.; Sheng, Q.; Zhou, S.P.; Gao, X.L.; Kong, L.Y.; Wu, X.F.; et al. Identification and characteristics of microRNAs from Bombyx mori. BMC Genom. 2008, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Q.; Jiang, Y.M.; Chen, P.; Pan, M.H. Inhibition of BmNPV proliferation by Bombyx mori ADP/ATP transportase (BmANT). Acta Microbiol. Sin. 2019, 59, 1474–1483. [Google Scholar]
- Wang, X.Y.; Wu, K.H.; Pang, H.L.; Xu, P.Z.; Li, M.W.; Zhang, G.Z. Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera). Int. J. Mol. Sci. 2019, 20, 4325. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.B.; Zhu, H.D.; Huang, Z.H.; Cao, H.H.; Ayaz, S.; Yang, J.Y.; Chen, X.Y.; Zhang, Y.; Liu, S.H.; Xu, J.P. BmNPV p35 regulates apoptosis in Bombyx mori via a novel target of interaction with the BmVDAC2-BmRACK1 complex. Insect Biochem. Mol. Biol. 2024, 169, 104125. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Q.; Hu, N.; Dong, F.F.; Chen, T.T.; Jiang, Y.M.; Chen, P.; Lu, C.; Pan, M.H. Baculovirus LEF-11 Hijack Host ATPase ATAD3A to Promote Virus Multiplication in Bombyx mori cells. Sci. Rep. 2017, 7, 46187. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.C.; Li, Z.Q.; Zhang, W.C.; Guo, H.; Lan, W.Q.; Shen, G.W.; Xia, Q.Y.; Zhao, P. SUMOylation of Translationally Regulated Tumor Protein Modulates Its Immune Function. Front. Immunol. 2022, 13, 807097. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.H.; Mao, Y.X.; Xu, Y.X. Screening for Proteins Interacting with Bombyx mori Serine Protease Inhibitor Serpin-3. Jiangsu Agric. Sci. 2015, 43, 37–40. [Google Scholar]
- Cao, H.H.; Wang, Y.L.; Toufeeq, S.; Kong, W.W.; Ayaz, S.; Liu, S.H.; Wang, J.; Xu, J.P. Bombyx mori serpin 3 is involved in innate immunity by interacting with serine protease 7 to regulate prophenoloxidase activation. J. Invertebr. Pathol. 2024, 207, 108188. [Google Scholar] [CrossRef] [PubMed]
- Toufeeq, S.; Wang, J.; Zhang, S.Z.; Li, B.; Hu, P.; Zhu, L.B.; You, L.L.; Xu, J.P. Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori. Insects 2019, 10, 399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Z.; Wang, J.; Zhu, L.B.; Toufeeq, S.; Xu, X.; You, L.L.; Li, B.; Hu, P.; Xu, J.P. Quantitative label-free proteomic analysis reveals differentially expressed proteins in the digestive juice of resistant versus susceptible silkworm strains and their predicted impacts on BmNPV infection. J. Proteom. 2020, 210, 103527. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Sang, Q.; Pan, G.Q.; Li, C.F.; Zhou, Z.Y. A Toll-Sptzle Pathway in the Immune Response of Bombyx mori. Insects 2020, 11, 586. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Xu, J.H.; Wang, L.L.; Guo, P.C.; Tang, Z.C.; Sun, X.T.; Tang, X.; Wang, W.; Wang, L.Y.; Cao, Y.; et al. Serpin-1a and serpin-6 regulate the Toll pathway immune homeostasis by synergistically inhibiting the Sptzle-processing enzyme CLIP2 in silkworm, Bombyx mori. PLoS Pathog. 2023, 19, e1011740. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, J.J.; Buchon, N.; Blissard, G.W. Identification of Cellular Genes Involved in Baculovirus GP64 Trafficking to the Plasma Membrane. J. Virol. 2022, 96, e0021522. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Wu, X.F. A Review on Interactions Between Insect Baculovirus Envelope Protein GP64 and Host Cell Surface Factors. Acta Sericologica Sin. 2014, 40, 911–916. [Google Scholar]
- Lu, S.; Ge, G.; Qi, Y. Ha-VP39 binding to actin and the influence of F-actin on assembly of progeny virions. Arch. Virol. 2004, 149, 2187–2198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Li, Y.; Zhao, S.D.; Wu, X.F. Identification of A functional region in Bombyx mori nucleopolyhedrovirus VP39 that is essential for nuclear actin polymerization. Virology 2020, 550, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Benning, F.M.C.; Jenni, S.; Garcia, C.Y.; Nguyen, T.H.; Zhang, X.W.; Chao, L.H. Helical reconstruction of VP39 reveals principles for baculovirus nucleocapsid assembly. Nat. Commun. 2024, 15, 250. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.K.; Lin, S.; Wu, Y.X.; Zhao, Z.M.; Zhou, X.M.; Sadiq, S.; Zhang, Z.D.; Guo, X.J.; Wu, P. Hsp90 could promote BmNPV proliferation by interacting with Actin-4 and enhance its expression. Dev. Comp. Immunol. 2023, 142, 104667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Ma, S.Y.; Gu, C.G.; Hu, M.; Miao, M.; Quan, Y.P.; Yu, W. K64 acetylation of heat shock protein 90 suppresses nucleopolyhedrovirus replication in Bombyx mori. Arch. Insect Biochem. Physiol. 2024, 115, e22079. [Google Scholar] [CrossRef] [PubMed]

| Methodologies | Advantages | Limitations | Applicable Scenarios | Refs |
|---|---|---|---|---|
| Y2H | High throughput; can screen for unknown interacting proteins | High false positive rate; inability to detect membrane proteins or modified proteins | Large-scale screening of interactions; suitable for initial screening of intranuclear interactions | [31] |
| Co-IP | Close to physiological conditions; detectable natural complexes | Relies on specific antibodies; unsuitable for large-scale screening of interacting proteins | Validation of known interactions; applicable to physiological condition validation | [32] |
| GST pulldown | Controllable; suitable for in vitro validation | Non-physiological conditions; may bind non-specifically | In vitro validation of interactions; recombinant protein interaction studies | [33] |
| BiFC | Visualizing subcellular localization; can be used to study weak or transient interactions between proteins | Irreversible; fluorescence maturation may be delayed | Localization and validation of intracellular interactions | [34,35] |
| MS | High throughput, high specificity, and high flexibility | Equipment is expensive; complexity of data analysis | Structural analysis of protein complexes; localization of interaction interfaces | [36] |
| SPR | No marking is required; affinity and kinetic parameters can be determined | Equipment is expensive; protein needs to be purified | Quantitative analysis of the strength of interactions | [37] |
| AI-driven protein interaction prediction | Highly accurate prediction of protein structure | Limited support for dynamic interactions | Structure prediction and functional studies of protein complexes | [38] |
| Proliferative Effect on BmNPV | Interacting Proteins | Mechanisms | Refs. |
|---|---|---|---|
| Inhibits | ING5/P53 | Promotes clearance of infected cells by accelerating apoptosis by reducing mitochondrial membrane potential | [39] |
| Promotes | Acetylated ING5/P53 | Reduces P53 protein stability and inhibits its pro-apoptotic function | [41] |
| Promotes | BmIAP/BmPP5 | Protein phosphorylation modification regulates apoptosis | [4] |
| Promotes | LEF-11/BmIMPI | Inhibition of CDK1/cyclin B activity leads to cell cycle arrest in G2/M phase | [42] |
| Promotes | IAP1/cyclin B | Inducing aberrant accumulation of cyclin B in the nucleus and specific blockade of the G2/M phase prolong the time window for viral assembly | [43] |
| Promotes | IAP1/BmCDK1 | Reduces BmCDK1 levels, inhibits cell cycle progression, and supports viral replication and proliferation | [44] |
| Proliferative Effect on BmNPV | Interacting Proteins | Mechanisms | Refs |
|---|---|---|---|
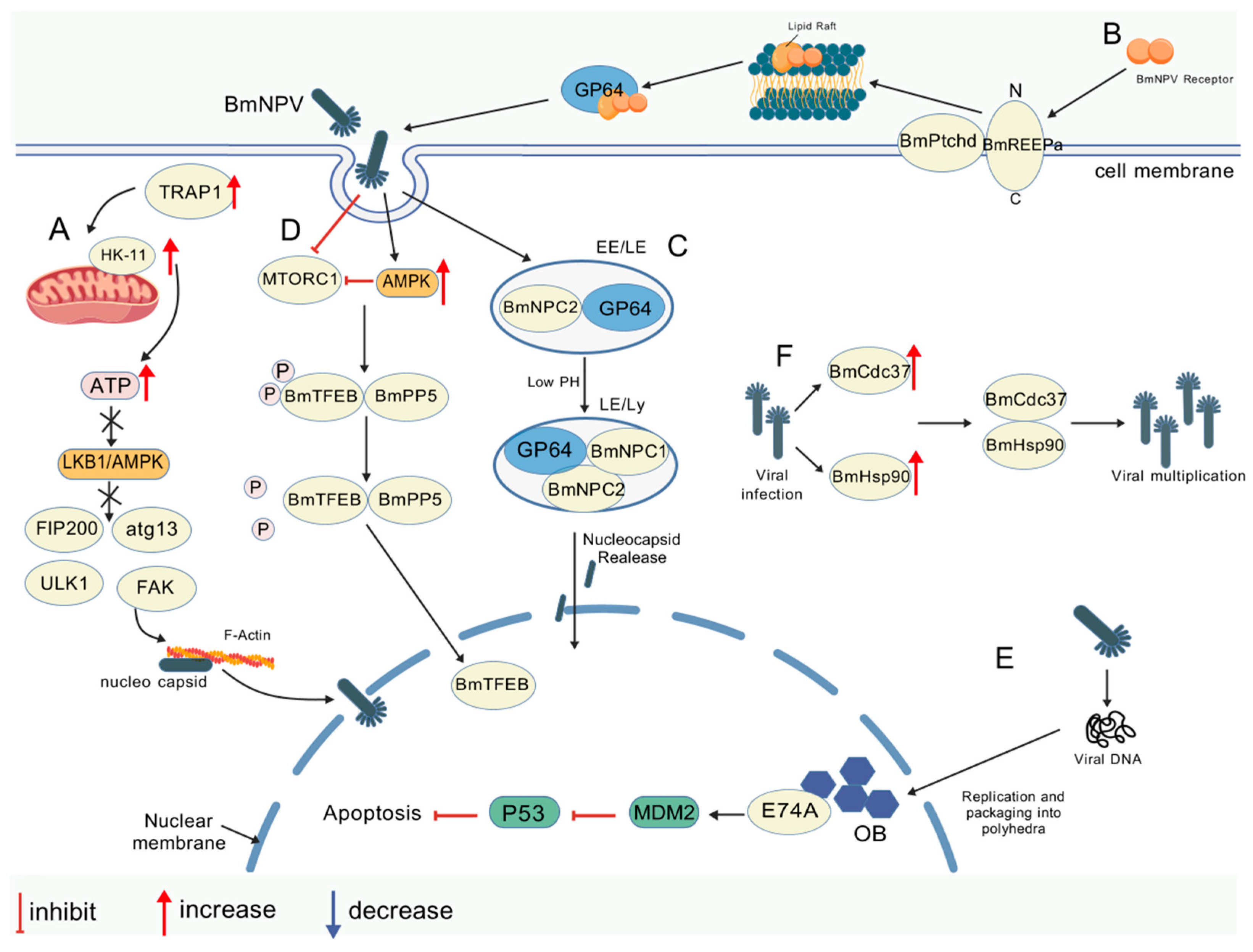
| Promotes | GP64/SINAL10 | K63-linked ubiquitination modification to stabilize GP64 conformation and enhance membrane fusion efficiency | [9] |
| Promotes | GP64/TRAP1 | Enhanced membrane fusion efficiency | [46] |
| Promotes | GP64/BmREEPa-BmPtchd complex | Activates AMPK signaling via dephosphorylation, drives nuclear translocation, and upregulates viral proliferation-related genes | [47,48] |
| Mutant site Inhibits | GP64/NPC1-NPC2 receptor complex | Mediated viral endocytosis synergizes with membrane fusion and mutated reciprocal sites to inhibit proliferation | [50,51] |
| Promotes | SEC61/viral protein | Mediates translocation of viral proteins to the endoplasmic reticulum, supports viral replication, and promotes translation and assembly of viral proteins | [12] |
| Inhibits | GP64/FABP1 | Antagonizes E3 ubiquitinase activity, inhibits GP64 membrane fusion, and inhibits viral membrane fusion and invasion | [12] |
| Inhibits | VP39/F-actin | Interaction with F-actin may interfere with viral transport (mechanism not defined) | [52] |
| Promotes | BmHsp90/BmTbce | Regulation of nucleocapsid–microtubule transport drives nuclear import for viral genome replication/transcription | [53] |
| Promotes | BmHsp90/BmGolga5 | Interactions disrupt Golgi apparatus function, impacting viral protein processing/transport and viral particle assembly/release | [54] |
| Promotes | PK1/BmPP5/BmTFEB | Activates AMPK signaling via dephosphorylation, drives nuclear translocation, and upregulates viral proliferation-related genes | [55] |
| Promotes | BmE74A/viral protein | Directly binds to viral proteins, enhances viral gene expression, and promotes early viral gene transcription | [56] |
| Promotes | BmCdc37/BmHsp90 | Enhances Hsp90 activity as a molecular chaperone, supports viral protein folding, and maintains viral protein function and stability | [57] |
| Promotes | PE38/BmeIF4E/BmSRPK | Interacts with translation factors and splicing kinases to promote early viral gene expression | [58] |
| Promotes | P74/JAB-MPN structural domain protein | Binding of midgut cell JAB-MPN protein mediates ODV invasion to promote virus infection and spread in the midgut | [59] |
| Inhibits | BmTHY/actin | Interferes with viral transport by binding actin; inhibits capsid migration and replication | [60] |
| Proliferative Effect on BmNPV | Interacting Proteins | Mechanisms | Refs |
|---|---|---|---|
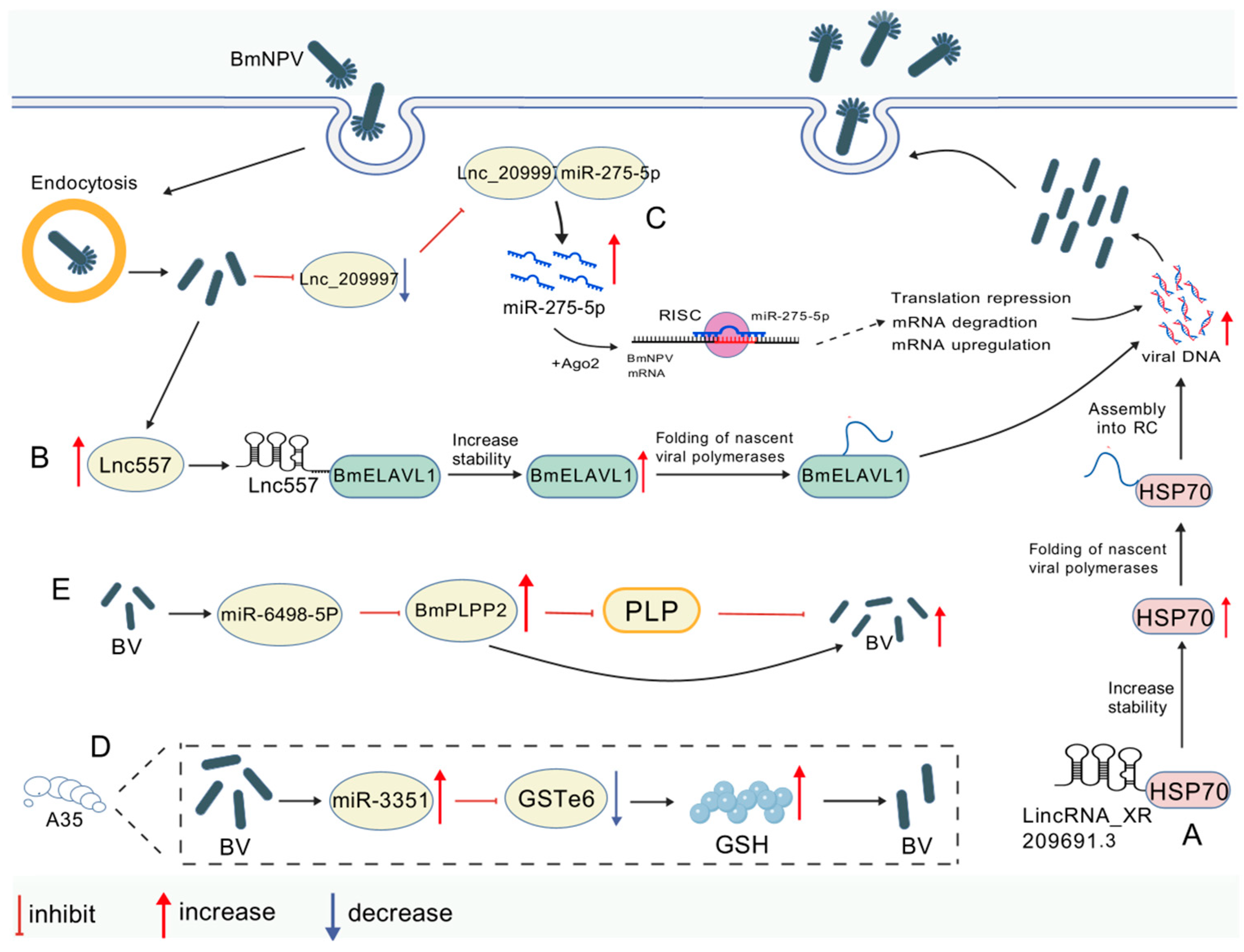
| Promotes | lincRNA_XR209691.3/BmHSP70 | Binding to the actin structural domain of BmHSP70 enhances its stability, optimizes viral protein folding efficiency, and promotes viral replication | [18] |
| Promotes | lnc557/BmELAVL1 | Promotes viral mRNA (e.g., ie-1, gp64) stability and accelerates viral structural protein synthesis | [61] |
| Promotes | Lnc_209997/miR-275-5p | BmNPV infection downregulates Lnc_209997, releasing miR-275-5p to promote viral proliferation via signaling pathways | [62] |
| Inhibits | miR-3351/BmGSTe6 (A35) | Targets BmGSTe6 to regulate glutathione metabolism and inhibit viral proliferation | [63] |
| Inhibits | miR-6498-5p/BmPLPP2 | miR-6498-5p inhibits viral replication by maintaining higher PLP levels through the inhibition of BmPLPP2 | [64] |
| Proliferative Effect on BmNPV | Interacting Proteins | Mechanisms | Refs |
|---|---|---|---|
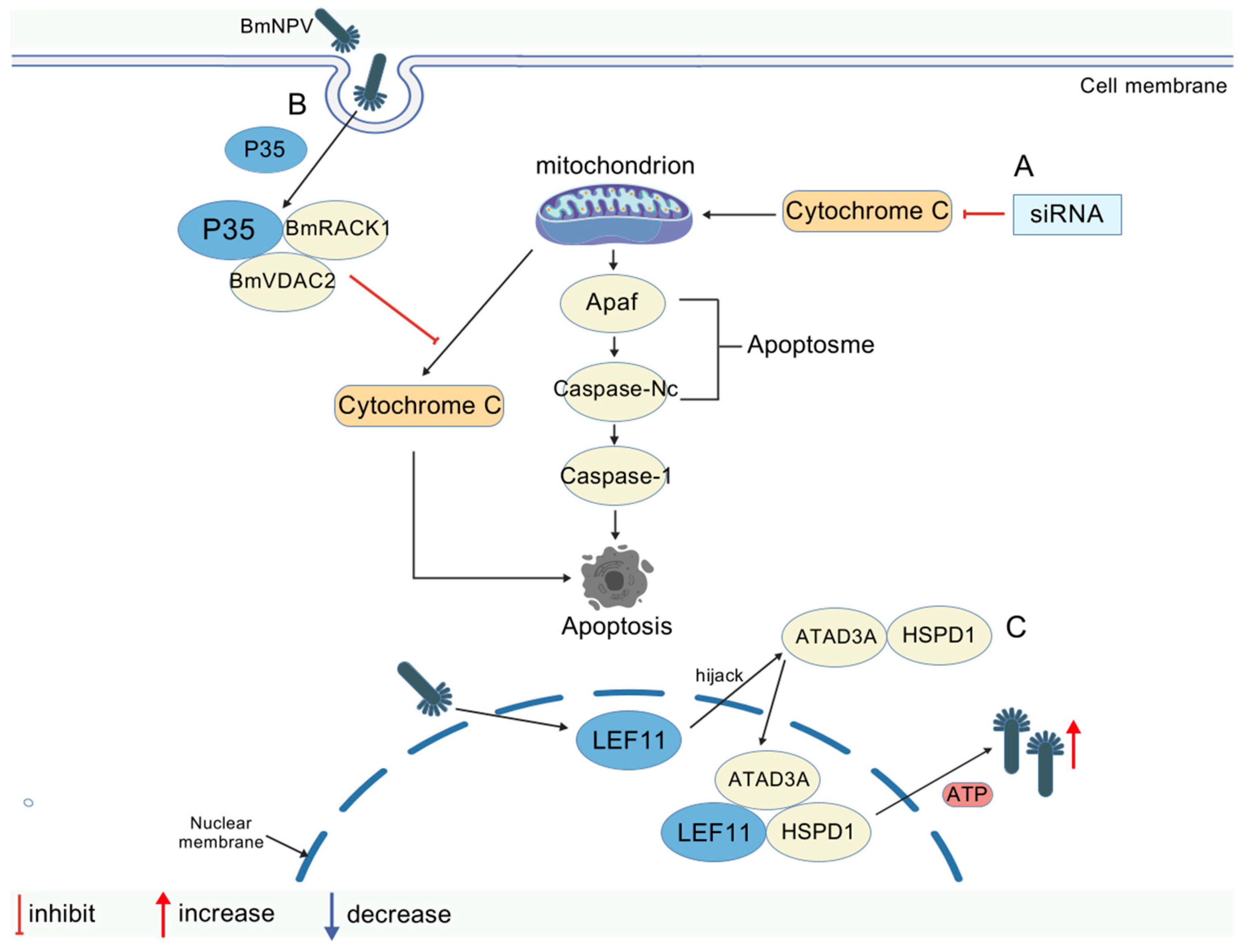
| Inhibits | BmANT/BmHSP60 | Interacts with BmHSP60 to downregulate its expression and inhibit viral replication; BmANT leads to abnormal accumulation of ANT and triggers ATP/ADP transport disorders | [54,67] |
| Inhibits | Bmcytc/Bmapaf/Bmcaspase-Nc | Release triggers the mitochondrial apoptotic pathway (Bmapaf/Bmcaspase-Nc) to clear infected cells | [68] |
| Promotes | p35/BmVDAC2-BmRACK1 | Blocks cytochrome c release, inhibits mitochondria-dependent apoptosis, and promotes viral proliferation | [69] |
| Promotes | LEF-11/ATAD3A/hspd1 (hsp60) | Activation of ATPase activity and enhancement of mitochondrial OXPHOS support viral DNA replication | [70] |
| Proliferative Effect on BmNPV | Interacting Components | Mechanisms | Refs |
|---|---|---|---|
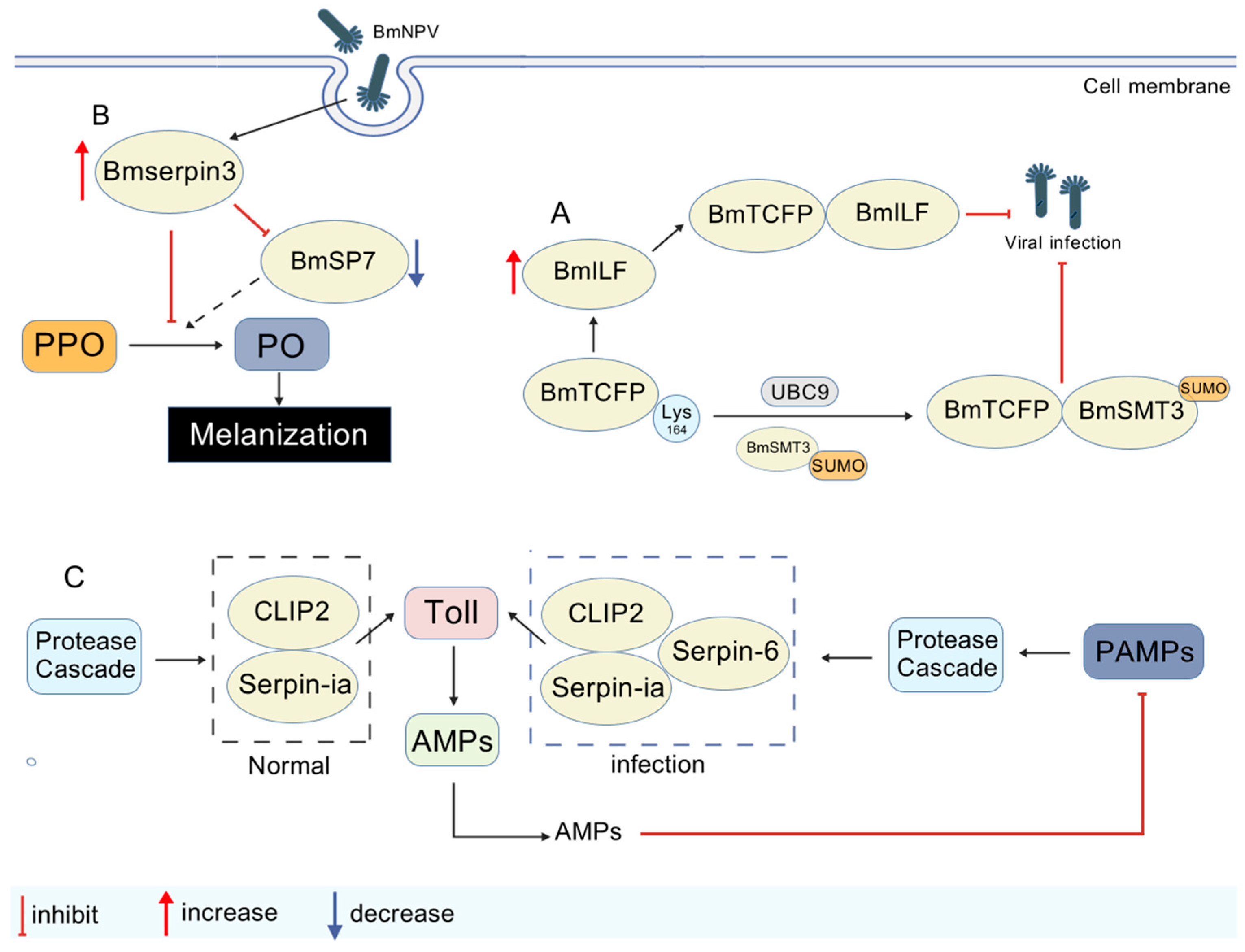
| Inhibit | BmTCTP/BmILF | Activates downstream immune signaling and significantly inhibits viral replication | [71] |
| Inhibit | BmSerpin3/storage protein | Involved in immunomodulation, regulating immune homeostasis, and indirectly suppressing viruses | [72] |
| Inhibit | BmSerpin3/SP7 | Inhibition of SP7 modulates the PPO activation cascade and balances the intensity of the blackening reaction | [73] |
| - | serpin-1a/serpin-6/CLIP2 | Serpin-1a and serpin-6 synergize to precisely regulate CLIP2 activity and balance immune activation | [77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, D.; Liu, B.; Cui, M.; Qian, H.; Li, G. Regulation of Bombyx mori–BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms. Viruses 2025, 17, 1017. https://doi.org/10.3390/v17071017
Guo D, Liu B, Cui M, Qian H, Li G. Regulation of Bombyx mori–BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms. Viruses. 2025; 17(7):1017. https://doi.org/10.3390/v17071017
Chicago/Turabian StyleGuo, Dan, Bowen Liu, Mingxing Cui, Heying Qian, and Gang Li. 2025. "Regulation of Bombyx mori–BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms" Viruses 17, no. 7: 1017. https://doi.org/10.3390/v17071017
APA StyleGuo, D., Liu, B., Cui, M., Qian, H., & Li, G. (2025). Regulation of Bombyx mori–BmNPV Protein Interactions: Study Strategies and Molecular Mechanisms. Viruses, 17(7), 1017. https://doi.org/10.3390/v17071017






