Zooming in and out: Exploring RNA Viral Infections with Multiscale Microscopic Methods
Abstract
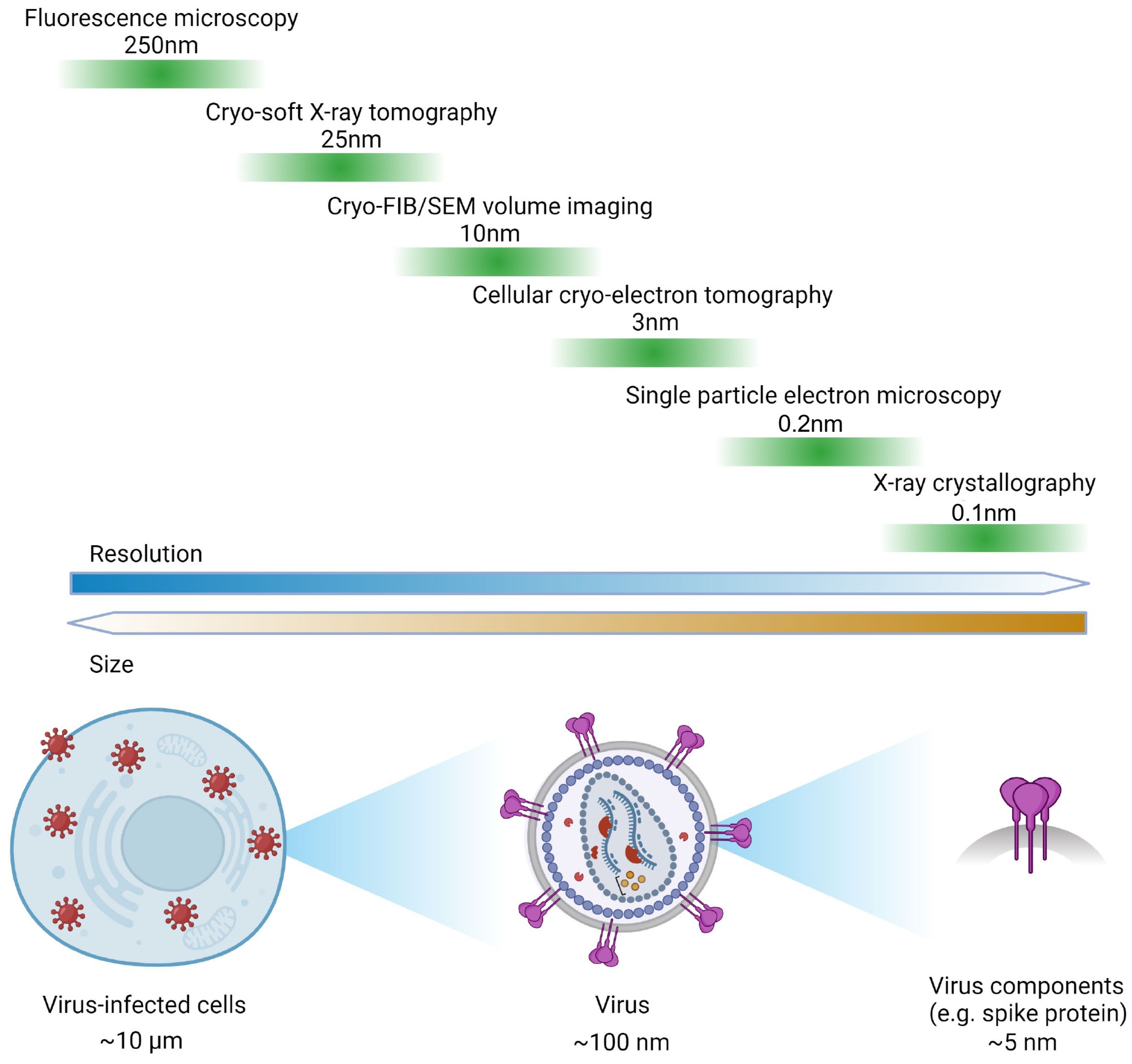
1. Introduction
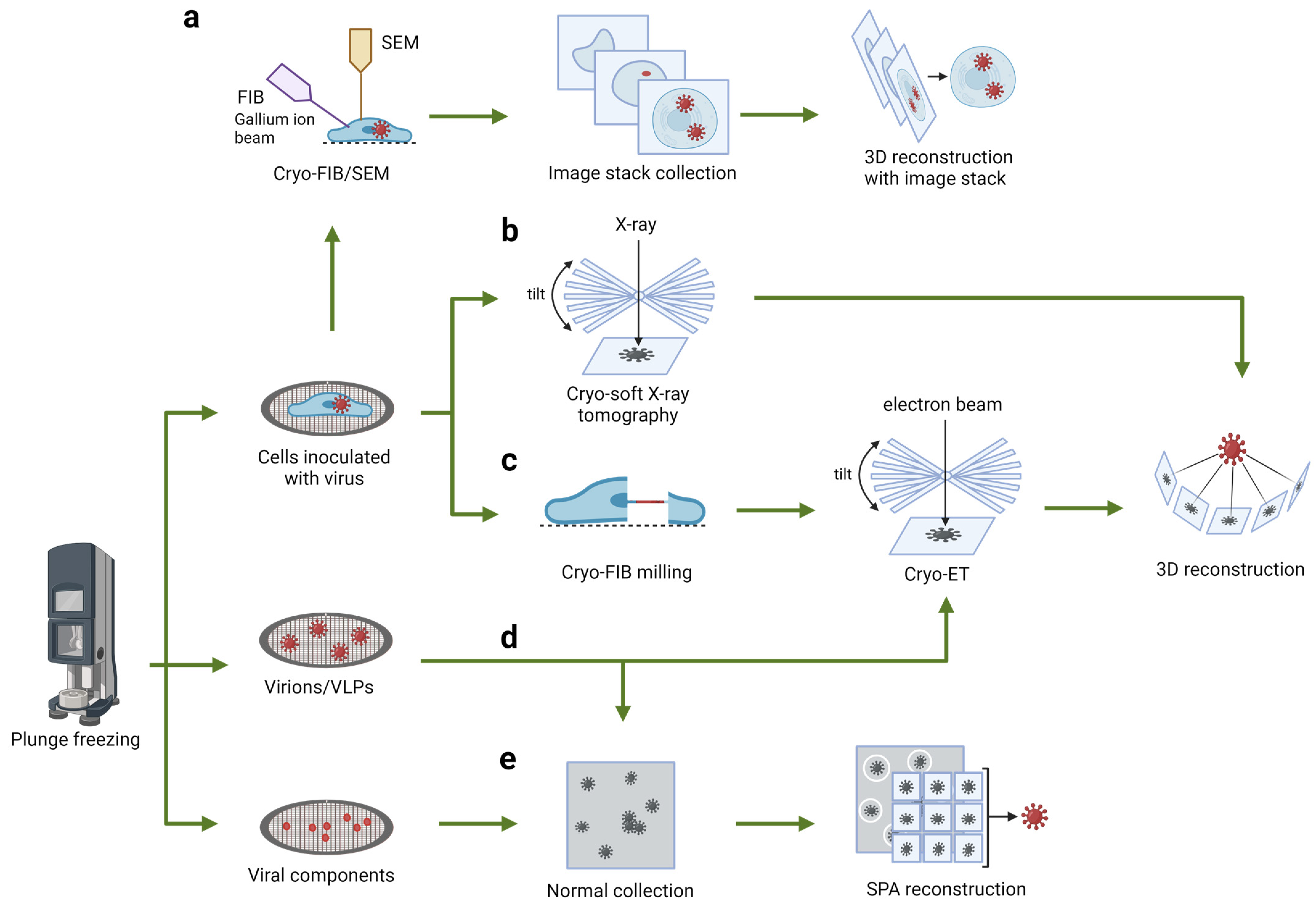
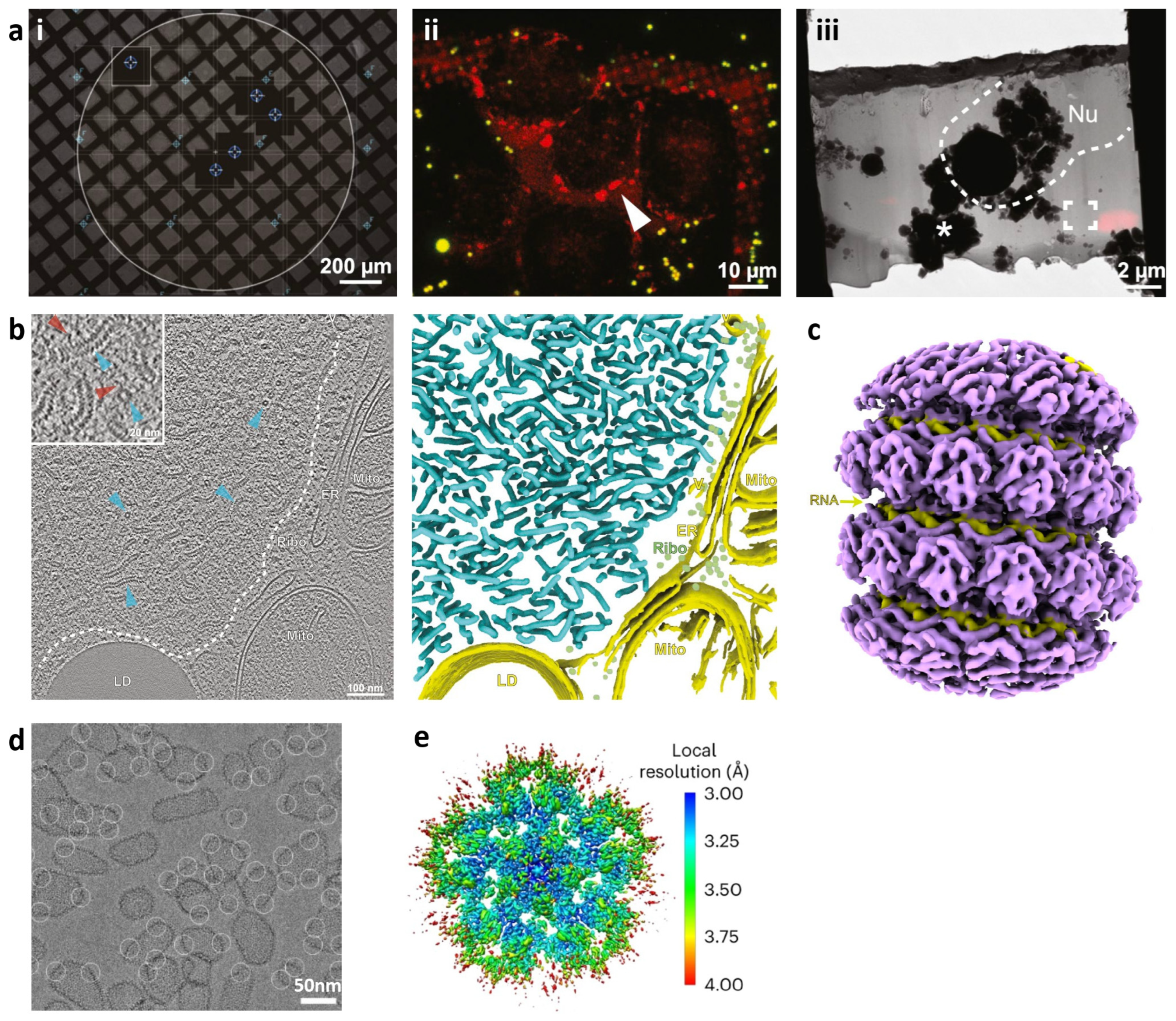
2. Seeing through the Whole Cell: Cryo-Soft X-ray Tomography
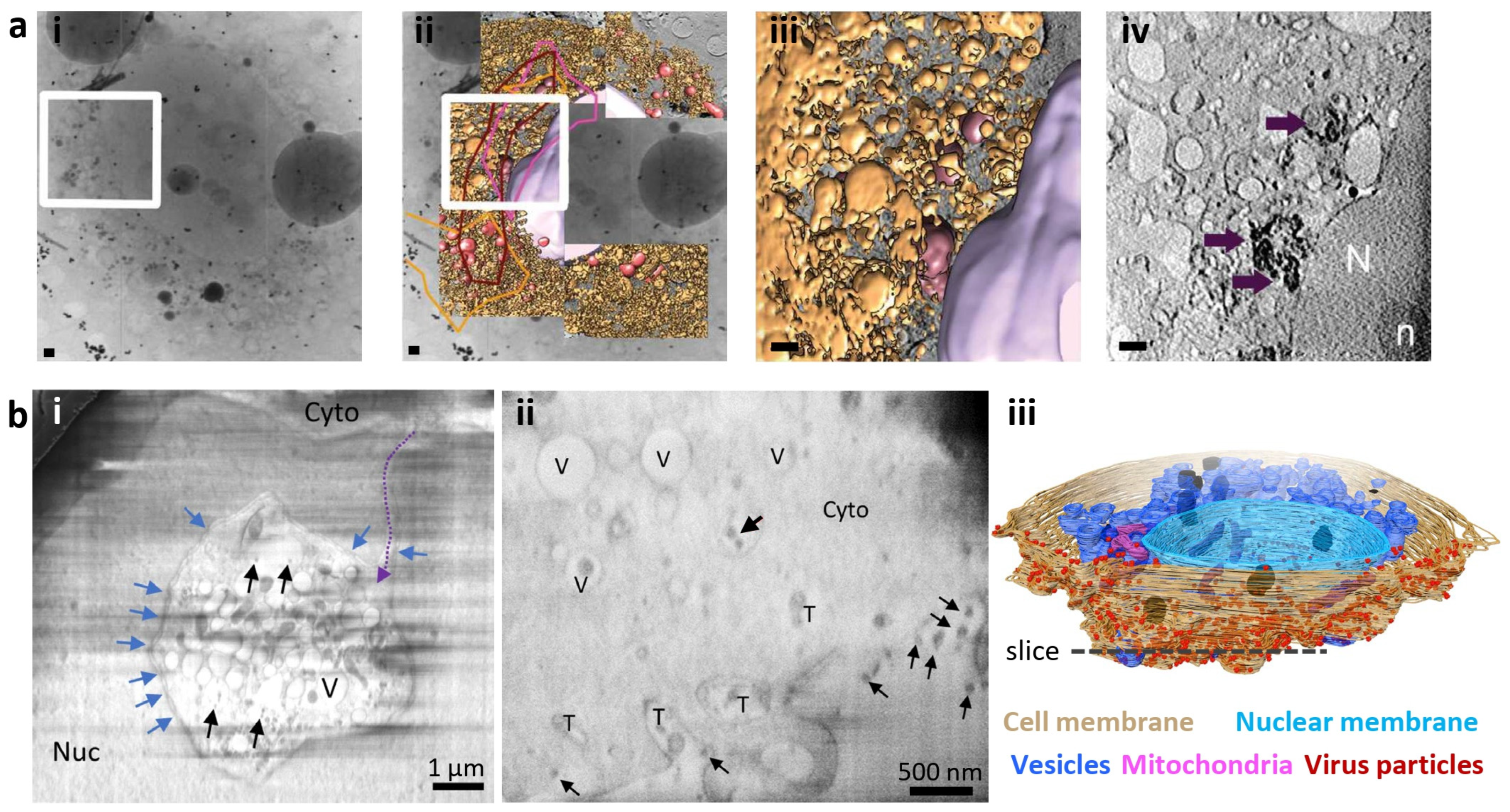
3. Peeling Back the Layers: Serial Cryo-FIB/SEM Volume Imaging
4. Zooming in on a Single Layer: Cellular Cryo-Electron Tomography
5. Spotlight on Key Components: Single-Particle Analysis
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Wasik, B.R.; Turner, P.E. On the biological success of viruses. Annu. Rev. Microbiol. 2013, 67, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef]
- Jacob, S.T.; Crozier, I.; Fischer, W.A., 2nd; Hewlett, A.; Kraft, C.S.; Vega, M.A.; Soka, M.J.; Wahl, V.; Griffiths, A.; Bollinger, L.; et al. Ebola virus disease. Nat. Rev. Dis. Primers 2020, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; McFadden, G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef]
- De Armas-Rillo, L.; Valera, M.S.; Marrero-Hernandez, S.; Valenzuela-Fernandez, A. Membrane dynamics associated with viral infection. Rev. Med. Virol. 2016, 26, 146–160. [Google Scholar] [CrossRef]
- Hernandez-Gonzalez, M.; Larocque, G.; Way, M. Viral use and subversion of membrane organization and trafficking. J. Cell Sci. 2021, 134, jcs252676. [Google Scholar] [CrossRef]
- Kondylis, P.; Schlicksup, C.J.; Zlotnick, A.; Jacobson, S.C. Analytical Techniques to Characterize the Structure, Properties, and Assembly of Virus Capsids. Anal. Chem. 2019, 91, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-electron microscopy of viruses. Nature 1984, 308, 32–36. [Google Scholar] [CrossRef]
- Perez-Berna, A.J.; Benseny-Cases, N.; Rodriguez, M.J.; Valcarcel, R.; Carrascosa, J.L.; Gastaminza, P.; Pereiro, E. Monitoring reversion of hepatitis C virus-induced cellular alterations by direct-acting antivirals using cryo soft X-ray tomography and infrared microscopy. Acta Crystallogr. D Struct. Biol. 2021, 77, 1365–1377. [Google Scholar] [CrossRef]
- Mendonca, L.; Howe, A.; Gilchrist, J.B.; Sheng, Y.; Sun, D.; Knight, M.L.; Zanetti-Domingues, L.C.; Bateman, B.; Krebs, A.S.; Chen, L.; et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021, 12, 4629. [Google Scholar] [CrossRef]
- Zhang, X.; Sridharan, S.; Zagoriy, I.; Eugster Oegema, C.; Ching, C.; Pflaesterer, T.; Fung, H.K.H.; Becher, I.; Poser, I.; Muller, C.W.; et al. Molecular mechanisms of stress-induced reactivation in mumps virus condensates. Cell 2023, 186, 1877–1894.e27. [Google Scholar] [CrossRef] [PubMed]
- Schirra, R.T.; Dos Santos, N.F.B.; Zadrozny, K.K.; Kucharska, I.; Ganser-Pornillos, B.K.; Pornillos, O. A molecular switch modulates assembly and host factor binding of the HIV-1 capsid. Nat. Struct. Mol. Biol. 2023, 30, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Groen, J.; Sorrentino, A.; Aballe, L.; Oliete, R.; Valcarcel, R.; Okolo, C.; Kounatidis, I.; Harkiolaki, M.; Perez-Berna, A.J.; Pereiro, E. A 3D Cartographic Description of the Cell by Cryo Soft X-ray Tomography. J. Vis. Exp. JoVE 2021, 169. [Google Scholar] [CrossRef]
- Garriga, D.; Chichon, F.J.; Calisto, B.M.; Ferrero, D.S.; Gastaminza, P.; Pereiro, E.; Perez-Berna, A.J. Imaging of Virus-Infected Cells with Soft X-ray Tomography. Viruses 2021, 13, 2109. [Google Scholar] [CrossRef]
- Harkiolaki, M.; Darrow, M.C.; Spink, M.C.; Kosior, E.; Dent, K.; Duke, E. Cryo-soft X-ray tomography: Using soft X-rays to explore the ultrastructure of whole cells. Emerg. Top. Life Sci. 2018, 2, 81–92. [Google Scholar] [CrossRef]
- Okolo, C.A. A guide into the world of high-resolution 3D imaging: The case of soft X-ray tomography for the life sciences. Biochem. Soc. Trans. 2022, 50, 649–663. [Google Scholar] [CrossRef]
- Carzaniga, R.; Domart, M.C.; Duke, E.; Collinson, L.M. Correlative cryo-fluorescence and cryo-soft X-ray tomography of adherent cells at European synchrotrons. Methods Cell Biol. 2014, 124, 151–178. [Google Scholar] [CrossRef]
- Loconte, V.; Chen, J.H.; Cortese, M.; Ekman, A.; Le Gros, M.A.; Larabell, C.; Bartenschlager, R.; Weinhardt, V. Using soft X-ray tomography for rapid whole-cell quantitative imaging of SARS-CoV-2-infected cells. Cell Rep. Methods 2021, 1, 100117. [Google Scholar] [CrossRef]
- Ekman, A.; Weinhardt, V.; Chen, J.H.; McDermott, G.; Le Gros, M.A.; Larabell, C. PSF correction in soft X-ray tomography. J. Struct. Biol. 2018, 204, 9–18. [Google Scholar] [CrossRef]
- Parkinson, D.Y.; Knoechel, C.; Yang, C.; Larabell, C.A.; Le Gros, M.A. Automatic alignment and reconstruction of images for soft X-ray tomography. J. Struct. Biol. 2012, 177, 259–266. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Chen, L.; Guan, Y.; Tian, L.; Xiong, Y.; Liu, G.; Tian, Y. Quantitative imaging of Candida utilis and its organelles by soft X-ray Nano-CT. J. Microsc. 2018, 270, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Kunne, S.; Fish, T.M.; Dobbie, I.M.; Harkiolaki, M.; Paul-Gilloteaux, P. Protocol for image registration of correlative soft X-ray tomography and super-resolution structured illumination microscopy images. STAR Protoc. 2021, 2, 100529. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.; Goellner, S.; Acosta, E.G.; Neufeldt, C.J.; Oleksiuk, O.; Lampe, M.; Haselmann, U.; Funaya, C.; Schieber, N.; Ronchi, P.; et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017, 18, 2113–2123. [Google Scholar] [CrossRef]
- Kounatidis, I.; Stanifer, M.L.; Phillips, M.A.; Paul-Gilloteaux, P.; Heiligenstein, X.; Wang, H.; Okolo, C.A.; Fish, T.M.; Spink, M.C.; Stuart, D.I.; et al. 3D Correlative Cryo-Structured Illumination Fluorescence and Soft X-ray Microscopy Elucidates Reovirus Intracellular Release Pathway. Cell 2020, 182, 515–530.e517. [Google Scholar] [CrossRef]
- Nahas, K.L.; Connor, V.; Scherer, K.M.; Kaminski, C.F.; Harkiolaki, M.; Crump, C.M.; Graham, S.C. Near-native state imaging by cryo-soft-X-ray tomography reveals remodelling of multiple cellular organelles during HSV-1 infection. PLoS Pathog. 2022, 18, e1010629. [Google Scholar] [CrossRef]
- Okolo, C.; Jadhav, A.; Fish, T.; Nahas, K.; Watts, A.; Harkiolaki, M. Applications of Soft X-ray Tomography for the Direct Observation of Native Cellular Events. Microsc. Microanal. 2023, 29, 1147–1148. [Google Scholar] [CrossRef] [PubMed]
- Perez-Berna, A.J.; Rodriguez, M.J.; Chichon, F.J.; Friesland, M.F.; Sorrentino, A.; Carrascosa, J.L.; Pereiro, E.; Gastaminza, P. Structural Changes In Cells Imaged by Soft X-ray Cryo-Tomography During Hepatitis C Virus Infection. ACS Nano 2016, 10, 6597–6611. [Google Scholar] [CrossRef]
- Castro, V.; Perez-Berna, A.J.; Calvo, G.; Pereiro, E.; Gastaminza, P. Three-Dimensional Remodeling of SARS-CoV2-Infected Cells Revealed by Cryogenic Soft X-ray Tomography. ACS Nano 2023, 17, 22708–22721. [Google Scholar] [CrossRef]
- Biggins, S.W.; Bambha, K.M.; Terrault, N.A.; Inadomi, J.; Shiboski, S.; Dodge, J.L.; Gralla, J.; Rosen, H.R.; Roberts, J.P. Projected future increase in aging hepatitis C virus-infected liver transplant candidates: A potential effect of hepatocellular carcinoma. Liver Transplant. 2012, 18, 1471–1478. [Google Scholar] [CrossRef]
- Thirukkumaran, C.; Morris, D.G. Oncolytic viral therapy using reovirus. Methods Mol. Biol. 2009, 542, 607–634. [Google Scholar] [CrossRef]
- Scher, N.; Rechav, K.; Paul-Gilloteaux, P.; Avinoam, O. In situ fiducial markers for 3D correlative cryo-fluorescence and FIB-SEM imaging. iScience 2021, 24, 102714. [Google Scholar] [CrossRef] [PubMed]
- Schertel, A.; Snaidero, N.; Han, H.M.; Ruhwedel, T.; Laue, M.; Grabenbauer, M.; Mobius, W. Cryo FIB-SEM: Volume imaging of cellular ultrastructure in native frozen specimens. J. Struct. Biol. 2013, 184, 355–360. [Google Scholar] [CrossRef]
- Zhu, Y.; Sun, D.; Schertel, A.; Ning, J.; Fu, X.; Gwo, P.P.; Watson, A.M.; Zanetti-Domingues, L.C.; Martin-Fernandez, M.L.; Freyberg, Z.; et al. Serial cryoFIB/SEM Reveals Cytoarchitectural Disruptions in Leigh Syndrome Patient Cells. Structure 2021, 29, 82–87.e3. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.H.; Mitchell, P.G.; Galaz-Montoya, J.G.; Hecksel, C.W.; Sontag, E.M.; Gangadharan, V.; Marshman, J.; Mankus, D.; Bisher, M.E.; Lytton-Jean, A.K.R.; et al. Multi-scale 3D Cryo-Correlative Microscopy for Vitrified Cells. Structure 2020, 28, 1231–1237.e3. [Google Scholar] [CrossRef]
- Sviben, S.; Gal, A.; Hood, M.A.; Bertinetti, L.; Politi, Y.; Bennet, M.; Krishnamoorthy, P.; Schertel, A.; Wirth, R.; Sorrentino, A.; et al. A vacuole-like compartment concentrates a disordered calcium phase in a key coccolithophorid alga. Nat. Commun. 2016, 7, 11228. [Google Scholar] [CrossRef] [PubMed]
- Akiva, A.; Nelkenbaum, O.; Schertel, A.; Yaniv, K.; Weiner, S.; Addadi, L. Intercellular pathways from the vasculature to the forming bone in the zebrafish larval caudal fin: Possible role in bone formation. J. Struct. Biol. 2019, 206, 139–148. [Google Scholar] [CrossRef]
- Vidavsky, N.; Masic, A.; Schertel, A.; Weiner, S.; Addadi, L. Mineral-bearing vesicle transport in sea urchin embryos. J. Struct. Biol. 2015, 192, 358–365. [Google Scholar] [CrossRef]
- Dumoux, M.; Glen, T.; Smith, J.L.R.; Ho, E.M.L.; Perdigao, L.M.A.; Pennington, A.; Klumpe, S.; Yee, N.B.Y.; Farmer, D.A.; Lai, P.Y.A.; et al. Cryo-plasma FIB/SEM volume imaging of biological specimens. eLife 2023, 12, e83623. [Google Scholar] [CrossRef]
- Baena, V.; Conrad, R.; Friday, P.; Fitzgerald, E.; Kim, T.; Bernbaum, J.; Berensmann, H.; Harned, A.; Nagashima, K.; Narayan, K. FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist. Viruses 2021, 13, 611. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grunewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef] [PubMed]
- Chmielewski, D.; Schmid, M.F.; Simmons, G.; Jin, J.; Chiu, W. Chikungunya virus assembly and budding visualized in situ using cryogenic electron tomography. Nat. Microbiol. 2022, 7, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Briggs, J.A. Cryo-Electron Tomography and Subtomogram Averaging. Methods Enzymol. 2016, 579, 329–367. [Google Scholar] [CrossRef] [PubMed]
- Lucic, V.; Rigort, A.; Baumeister, W. Cryo-electron tomography: The challenge of doing structural biology in situ. J. Cell Biol. 2013, 202, 407–419. [Google Scholar] [CrossRef]
- Schaffer, M.; Mahamid, J.; Engel, B.D.; Laugks, T.; Baumeister, W.; Plitzko, J.M. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 2017, 197, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Liu, Y.; Zhang, R.; Xin, H.L. A joint deep learning model to recover information and reduce artifacts in missing-wedge sinograms for electron tomography and beyond. Sci. Rep. 2019, 9, 12803. [Google Scholar] [CrossRef]
- Hagen, W.J.H.; Wan, W.; Briggs, J.A.G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017, 197, 191–198. [Google Scholar] [CrossRef]
- Himes, B.A.; Zhang, P. emClarity: Software for high-resolution cryo-electron tomography and subtomogram averaging. Nat. Methods 2018, 15, 955–961. [Google Scholar] [CrossRef]
- Ni, T.; Frosio, T.; Mendonca, L.; Sheng, Y.; Clare, D.; Himes, B.A.; Zhang, P. High-resolution in situ structure determination by cryo-electron tomography and subtomogram averaging using emClarity. Nat. Protoc. 2022, 17, 421–444. [Google Scholar] [CrossRef]
- Bharat, T.A.; Scheres, S.H. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat. Protoc. 2016, 11, 2054–2065. [Google Scholar] [CrossRef]
- Henderson, L.D.; Beeby, M. High-Throughput Electron Cryo-tomography of Protein Complexes and Their Assembly. Methods Mol. Biol. 2018, 1764, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Zheng, S.; Wolff, G.; Greenan, G.; Chen, Z.; Faas, F.G.A.; Barcena, M.; Koster, A.J.; Cheng, Y.; Agard, D.A. AreTomo: An integrated software package for automated marker-free, motion-corrected cryo-electron tomographic alignment and reconstruction. J. Struct. Biol. X 2022, 6, 100068. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Stagg, S.M. Automated batch fiducial-less tilt-series alignment in Appion using Protomo. J. Struct. Biol. 2015, 192, 270–278. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, T.; Zhong, L.; Zhang, W.; Zhang, Y.; Yu, X.; Yuan, S.; Ni, T. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature 2024, 633, 224–231. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef] [PubMed]
- Bharat, T.A.M.; Russo, C.J.; Lowe, J.; Passmore, L.A.; Scheres, S.H.W. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure 2015, 23, 1743–1753. [Google Scholar] [CrossRef]
- Wan, W.; Kolesnikova, L.; Clarke, M.; Koehler, A.; Noda, T.; Becker, S.; Briggs, J.A.G. Structure and assembly of the Ebola virus nucleocapsid. Nature 2017, 551, 394–397. [Google Scholar] [CrossRef]
- Wan, W.; Clarke, M.; Norris, M.J.; Kolesnikova, L.; Koehler, A.; Bornholdt, Z.A.; Becker, S.; Saphire, E.O.; Briggs, J.A. Ebola and Marburg virus matrix layers are locally ordered assemblies of VP40 dimers. eLife 2020, 9, e59225. [Google Scholar] [CrossRef]
- Schur, F.K.; Obr, M.; Hagen, W.J.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Krausslich, H.G.; Briggs, J.A. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef]
- Liu, C.; Mendonca, L.; Yang, Y.; Gao, Y.; Shen, C.; Liu, J.; Ni, T.; Ju, B.; Liu, C.; Tang, X.; et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28, 1218–1224.e4. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Sung, P.Y.; Martynowycz, M.W.; Gonen, T.; Roy, P.; Zhou, Z.H. RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells. Cell 2024, 187, 2236–2249.e17. [Google Scholar] [CrossRef] [PubMed]
- Ni, T.; Mendonca, L.; Zhu, Y.; Howe, A.; Radecke, J.; Shah, P.M.; Sheng, Y.; Krebs, A.S.; Duyvesteyn, H.M.E.; Allen, E.; et al. ChAdOx1 COVID vaccines express RBD open prefusion SARS-CoV-2 spikes on the cell surface. iScience 2023, 26, 107882. [Google Scholar] [CrossRef]
- Watanabe, Y.; Mendonca, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 nCoV-19/AZD1222 Vaccine. ACS Cent. Sci. 2021, 7, 594–602. [Google Scholar] [CrossRef]
- Baldwin, C.L.; Yirsaw, A.; Gillespie, A.; Le Page, L.; Zhang, F.; Damani-Yokota, P.; Telfer, J.C. gammadelta T cells in livestock: Responses to pathogens and vaccine potential. Transbound. Emerg. Dis. 2020, 67 (Suppl. S2), 119–128. [Google Scholar] [CrossRef] [PubMed]
- Sutton, G.; Sun, D.; Fu, X.; Kotecha, A.; Hecksel, C.W.; Clare, D.K.; Zhang, P.; Stuart, D.I.; Boyce, M. Assembly intermediates of orthoreovirus captured in the cell. Nat. Commun. 2020, 11, 4445. [Google Scholar] [CrossRef]
- Shah, P.N.M.; Gilchrist, J.B.; Forsberg, B.O.; Burt, A.; Howe, A.; Mosalaganti, S.; Wan, W.; Radecke, J.; Chaban, Y.; Sutton, G.; et al. Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography. Cell Host Microbe 2023, 31, 604–615.e4. [Google Scholar] [CrossRef]
- Elmlund, D.; Elmlund, H. Cryogenic electron microscopy and single-particle analysis. Annu. Rev. Biochem. 2015, 84, 499–517. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, T.; You, X.; Zhang, F.; Sui, S.F.; Wan, X.; Zhang, X. Determining protein structures in cellular lamella at pseudo-atomic resolution by GisSPA. Nat. Commun. 2023, 14, 1282. [Google Scholar] [CrossRef] [PubMed]
- Highland, C.M.; Tan, A.; Ricana, C.L.; Briggs, J.A.G.; Dick, R.A. Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM. Proc. Natl. Acad. Sci. USA 2023, 120, e2220545120. [Google Scholar] [CrossRef] [PubMed]
- Bester, S.M.; Wei, G.; Zhao, H.; Adu-Ampratwum, D.; Iqbal, N.; Courouble, V.V.; Francis, A.C.; Annamalai, A.S.; Singh, P.K.; Shkriabai, N.; et al. Structural and mechanistic bases for a potent HIV-1 capsid inhibitor. Science 2020, 370, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Ge, J.; Zheng, L.; Zhang, Y.; Gao, Y.; Wang, T.; Huang, Y.; Yang, Y.; Gao, S.; Li, M.; et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell 2021, 184, 184–193.e110. [Google Scholar] [CrossRef] [PubMed]
- Rapp, M.; Shapiro, L.; Frank, J. Contributions of single-particle cryoelectron microscopy toward fighting COVID-19. Trends Biochem. Sci. 2022, 47, 117–123. [Google Scholar] [CrossRef]
- Stacey, J.C.V.; Tan, A.; Lu, J.M.; James, L.C.; Dick, R.A.; Briggs, J.A.G. Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding. Proc. Natl. Acad. Sci. USA 2023, 120, e2220557120. [Google Scholar] [CrossRef]
- Sugita, Y.; Matsunami, H.; Kawaoka, Y.; Noda, T.; Wolf, M. Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 A resolution. Nature 2018, 563, 137–140. [Google Scholar] [CrossRef]
- Lazic, I.; Wirix, M.; Leidl, M.L.; de Haas, F.; Mann, D.; Beckers, M.; Pechnikova, E.V.; Muller-Caspary, K.; Egoavil, R.; Bosch, E.G.T.; et al. Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution. Nat. Methods 2022, 19, 1126–1136. [Google Scholar] [CrossRef]
- Pei, X.; Zhou, L.; Huang, C.; Boyce, M.; Kim, J.S.; Liberti, E.; Hu, Y.; Sasaki, T.; Nellist, P.D.; Zhang, P.; et al. Cryogenic electron ptychographic single particle analysis with wide bandwidth information transfer. Nat. Commun. 2023, 14, 3027. [Google Scholar] [CrossRef]
- Ni, T.; Zhu, Y.; Yang, Z.; Xu, C.; Chaban, Y.; Nesterova, T.; Ning, J.; Bocking, T.; Parker, M.W.; Monnie, C.; et al. Structure of native HIV-1 cores and their interactions with IP6 and CypA. Sci. Adv. 2021, 7, eabj5715. [Google Scholar] [CrossRef]
- Mendonca, L.; Sun, D.; Ning, J.; Liu, J.; Kotecha, A.; Olek, M.; Frosio, T.; Fu, X.; Himes, B.A.; Kleinpeter, A.B.; et al. CryoET structures of immature HIV Gag reveal six-helix bundle. Commun. Biol. 2021, 4, 481. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.; Hagen, W.J.; Rumlova, M.; Ruml, T.; Muller, B.; Krausslich, H.G.; Briggs, J.A. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 2015, 517, 505–508. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, C.-A.; Shen, Y.; Zhang, P. Zooming in and out: Exploring RNA Viral Infections with Multiscale Microscopic Methods. Viruses 2024, 16, 1504. https://doi.org/10.3390/v16091504
Lyu C-A, Shen Y, Zhang P. Zooming in and out: Exploring RNA Viral Infections with Multiscale Microscopic Methods. Viruses. 2024; 16(9):1504. https://doi.org/10.3390/v16091504
Chicago/Turabian StyleLyu, Cheng-An, Yao Shen, and Peijun Zhang. 2024. "Zooming in and out: Exploring RNA Viral Infections with Multiscale Microscopic Methods" Viruses 16, no. 9: 1504. https://doi.org/10.3390/v16091504
APA StyleLyu, C.-A., Shen, Y., & Zhang, P. (2024). Zooming in and out: Exploring RNA Viral Infections with Multiscale Microscopic Methods. Viruses, 16(9), 1504. https://doi.org/10.3390/v16091504





