Structural Insights into the Dynamic Assembly of a YFV sNS1 Tetramer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Construction, Expression, and Purification of YFV NS1
2.3. CryoEM Grid Preparation and Data Collection
2.4. CryoEM Data Processing of YFV NS1
2.5. Model Building and Refinement
3. Results
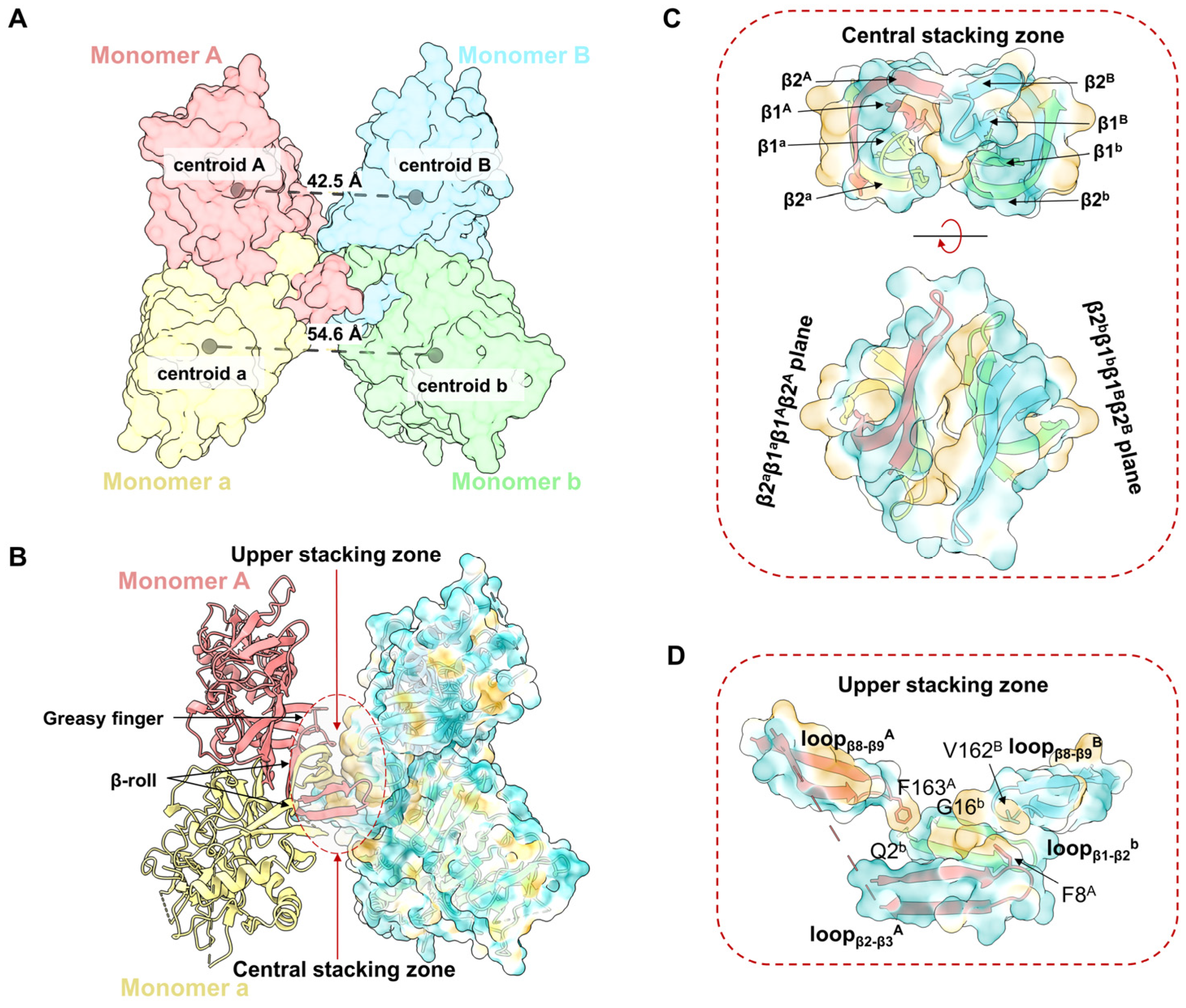
3.1. CryoEM Structures Analysis of YFV sNS1 Oligomers
3.2. The Assembly of YFV sNS1 Tetramer
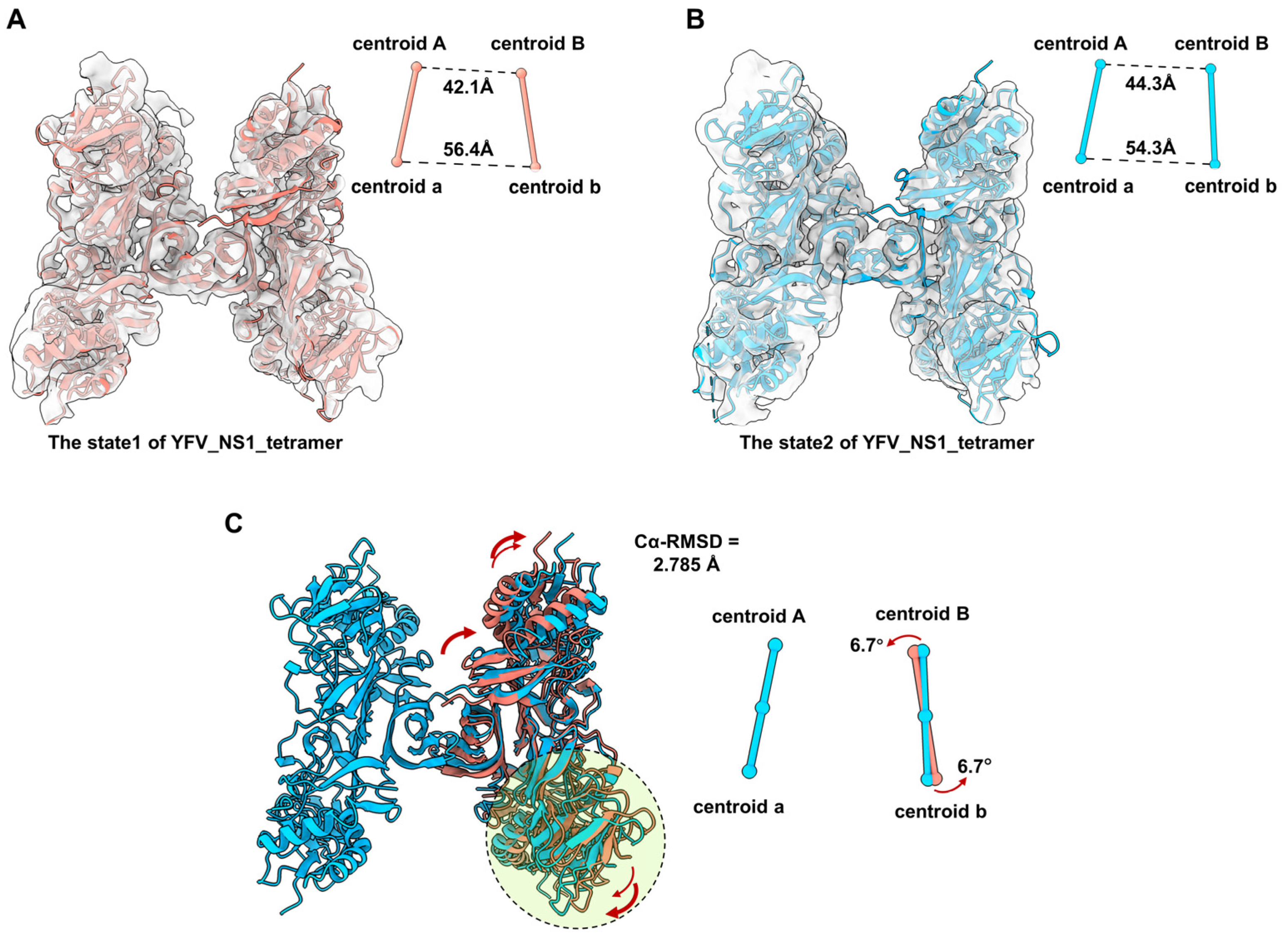
3.3. The YFV Tetramer Is a Semi-Stable Structure
3.4. The Dynamic Characteristics of YFV NS1 Tetramer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douam, F.; Ploss, A. Yellow Fever Virus: Knowledge Gaps Impeding the Fight Against an Old Foe. Trends Microbiol. 2018, 26, 913–928. [Google Scholar] [CrossRef]
- Barrett, A.D.T. The reemergence of yellow fever. Science 2018, 361, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Kleinert, R.D.V.; Montoya-Diaz, E.; Khera, T.; Welsch, K.; Tegtmeyer, B.; Hoehl, S.; Ciesek, S.; Brown, R.J.P. Yellow Fever: Integrating Current Knowledge with Technological Innovations to Identify Strategies for Controlling a Re-Emerging Virus. Viruses 2019, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Delatorre, E.; de Abreu, F.V.S.; Ribeiro, I.P.; Gómez, M.M.; Dos Santos, A.A.C.; Ferreira-de-Brito, A.; Neves, M.; Bonelly, I.; de Miranda, R.M.; Furtado, N.D.; et al. Distinct YFV Lineages Co-circulated in the Central-Western and Southeastern Brazilian Regions from 2015 to 2018. Front. Microbiol. 2019, 10, 1079. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, D.; Zhang, T.; Yu, N.; Ren, R.; Chen, Y.; Wang, C. Preparation and application of yellow fever virus NS1 protein-specific monoclonal antibodies. J. Med. Virol. 2021, 93, 3374–3382. [Google Scholar] [CrossRef]
- Stock, N.K.; Escadafal, C.; Achazi, K.; Cissé, M.; Niedrig, M. Development and characterization of polyclonal peptide antibodies for the detection of Yellow fever virus proteins. J. Virol. Methods 2015, 222, 110–116. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef]
- van den Elsen, K.; Chew, B.L.A.; Ho, J.S.; Luo, D. Flavivirus nonstructural proteins and replication complexes as antiviral drug targets. Curr. Opin. Virol. 2023, 59, 101305. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; d’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Mégret, F.; et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef]
- Avirutnan, P.; Fuchs, A.; Hauhart, R.E.; Somnuke, P.; Youn, S.; Diamond, M.S.; Atkinson, J.P. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J. Exp. Med. 2010, 207, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Guardo, H.; Glasner, D.R.; Espinosa, D.A.; Biering, S.B.; Patana, M.; Ratnasiri, K.; Wang, C.; Beatty, P.R.; Harris, E. Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell Rep. 2019, 26, 1598–1613.e8. [Google Scholar] [CrossRef]
- Wessel, A.W.; Dowd, K.A.; Biering, S.B.; Zhang, P.; Edeling, M.A.; Nelson, C.A.; Funk, K.E.; DeMaso, C.R.; Klein, R.S.; Smith, J.L.; et al. Levels of Circulating NS1 Impact West Nile Virus Spread to the Brain. J. Virol. 2021, 95, e0084421. [Google Scholar] [CrossRef] [PubMed]
- Scaturro, P.; Cortese, M.; Chatel-Chaix, L.; Fischl, W.; Bartenschlager, R. Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins. PLoS Pathog. 2015, 11, e1005277. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, F.T.G.; Warnes, C.M.; Manuli, E.R.; Ng, A.; D’Elia Zanella, L.; Ho, Y.L.; Bhat, S.; Romano, C.M.; Beatty, P.R.; Biering, S.B.; et al. Yellow fever disease severity and endothelial dysfunction are associated with elevated serum levels of viral NS1 protein and syndecan-1. medRxiv 2023. [Google Scholar] [CrossRef]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef]
- Edeling, M.A.; Diamond, M.S.; Fremont, D.H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 4285–4290. [Google Scholar] [CrossRef]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Han, M.; Qi, J.; Hilgenfeld, R.; Luo, T.; Shi, Y.; Gao, G.F.; Song, H. Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus. Sci. China Life Sci. 2017, 60, 1403–1406. [Google Scholar] [CrossRef]
- Poonsiri, T.; Wright, G.S.A.; Diamond, M.S.; Turtle, L.; Solomon, T.; Antonyuk, S.V. Structural Study of the C-Terminal Domain of Nonstructural Protein 1 from Japanese Encephalitis Virus. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Flamand, M.; Megret, F.; Mathieu, M.; Lepault, J.; Rey, F.A.; Deubel, V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 1999, 73, 6104–6110. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Landsberg, M.J.; Bletchly, C.; Rothnagel, R.; Waddington, L.; Hankamer, B.; Young, P.R. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J. Gen. Virol. 2012, 93, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Ooi, J.S.G.; Tan, A.W.K.; Ng, T.S.; Dejnirattisai, W.; Mongkolsapaya, J.; Fibriansah, G.; Shi, J.; Kostyuchenko, V.A.; Screaton, G.R.; et al. CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever. Nat. Commun. 2022, 13, 6756. [Google Scholar] [CrossRef] [PubMed]
- Benfrid, S.; Park, K.H.; Dellarole, M.; Voss, J.E.; Tamietti, C.; Pehau-Arnaudet, G.; Raynal, B.; Brûlé, S.; England, P.; Zhang, X.; et al. Dengue virus NS1 protein conveys pro-inflammatory signals by docking onto high-density lipoproteins. EMBO Rep. 2022, 23, e53600. [Google Scholar] [CrossRef]
- Chew, B.L.A.; Ngoh, A.Q.; Phoo, W.W.; Chan, K.W.K.; Ser, Z.; Tulsian, N.K.; Lim, S.S.; Weng, M.J.G.; Watanabe, S.; Choy, M.M.; et al. Secreted Dengue Virus NS1 from Infection Is Predominantly Dimeric and in Complex with High-Density Lipoprotein; Cold Spring Harbor Laboratory: New York, NY, USA, 2022. [Google Scholar] [CrossRef]
- Chew, B.L.A.; Ngoh, A.Q.; Phoo, W.W.; Weng, M.J.G.; Sheng, H.J.; Chan, K.W.K.; Tan, E.Y.J.; Gelbart, T.; Xu, C.; Tan, G.S.; et al. Structural basis of Zika virus NS1 multimerization and human antibody recognition. NPJ Viruses 2024, 2, 14. [Google Scholar] [CrossRef]
- Syzdykova, L.R.; Binke, S.; Keyer, V.V.; Shevtsov, A.B.; Zaripov, M.M.; Zhylkibayev, A.A.; Ramanculov, E.M.; Shustov, A.V. Fluorescent tagging the NS1 protein in yellow fever virus: Replication-capable viruses which produce the secretory GFP-NS1 fusion protein. Virus Res. 2021, 294, 198291. [Google Scholar] [CrossRef] [PubMed]
- Ceconi, M.; Ariën, K.K.; Delputte, P. Diagnosing arthropod-borne flaviviruses: Non-structural protein 1 (NS1) as a biomarker. Trends Microbiol. 2023, 32, 678–696. [Google Scholar] [CrossRef] [PubMed]
- Akey, D.L.; Brown, W.C.; Jose, J.; Kuhn, R.J.; Smith, J.L. Structure-guided insights on the role of NS1 in flavivirus infection. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015, 37, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]


| YFV_NS1 Focused Dimer (EMDB: 39898) (PDB: 8ZB9) | YFV_NS1_tetramer (EMDB: 39899) (PDB: 8ZBA) | |
|---|---|---|
| Data and processing | v | |
| Magnification | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å2) | 50 | 50 |
| Defocus range (μm) | −1.2~−1.8 | −1.2~−1.8 |
| Pixel size (Å) | 0.85 | 0.85 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 4,793,330 | 4,793,330 |
| Final particle images (no.) | 215,543 | 215,543 |
| Map resolution (Å) | 3.31 | 3.57 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range | 3.0–4.5 | 3.0–5.0 |
| Refinement | ||
| Initial model used (PDB code) | - | - |
| Map sharpening B factor (Å2) | −172.8 | −183.0 |
| Model composition | ||
| Non-hydrogen atoms | 4912 | 9804 |
| Protein residues | 623 | 1244 |
| Ligands | - | - |
| B factor (Å2) | ||
| Protein | 54.32 | 54.32 |
| Ligand | - | - |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.007 |
| Bond angles (°) | 0.830 | 0.893 |
| Validation | ||
| MolProbity score | 2.33 | 2.40 |
| Clash score | 14.87 | 7.10 |
| Rotamers outliers (%) | 0.00 | 0.92 |
| Ramachandra plot | ||
| Favored | 85.50 | 89.28 |
| Allowed | 14.33 | 14.98 |
| Outliers | 0.16 | 0.00 |
| Model-map FSC (0.143) | 3.43 | 3.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Q.; Chen, Q.; Zhang, W.; Jiao, H.; Yu, L.; Hu, H. Structural Insights into the Dynamic Assembly of a YFV sNS1 Tetramer. Viruses 2024, 16, 1212. https://doi.org/10.3390/v16081212
Pan Q, Chen Q, Zhang W, Jiao H, Yu L, Hu H. Structural Insights into the Dynamic Assembly of a YFV sNS1 Tetramer. Viruses. 2024; 16(8):1212. https://doi.org/10.3390/v16081212
Chicago/Turabian StylePan, Qi, Qiang Chen, Wanqin Zhang, Haizhan Jiao, Lei Yu, and Hongli Hu. 2024. "Structural Insights into the Dynamic Assembly of a YFV sNS1 Tetramer" Viruses 16, no. 8: 1212. https://doi.org/10.3390/v16081212
APA StylePan, Q., Chen, Q., Zhang, W., Jiao, H., Yu, L., & Hu, H. (2024). Structural Insights into the Dynamic Assembly of a YFV sNS1 Tetramer. Viruses, 16(8), 1212. https://doi.org/10.3390/v16081212






