Fluoxetine and Sertraline Potently Neutralize the Replication of Distinct SARS-CoV-2 Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Plasmids
2.3. Preparation of VSV*∆G-Fluc Spike Pseudotype Virus-like Particles
2.4. Transduction In Vitro Experiments
2.5. Neutralization Assay
2.6. Quantification of SARS-CoV-2 RNA
2.7. Cytotoxicity Assay
2.8. Statistics
3. Results
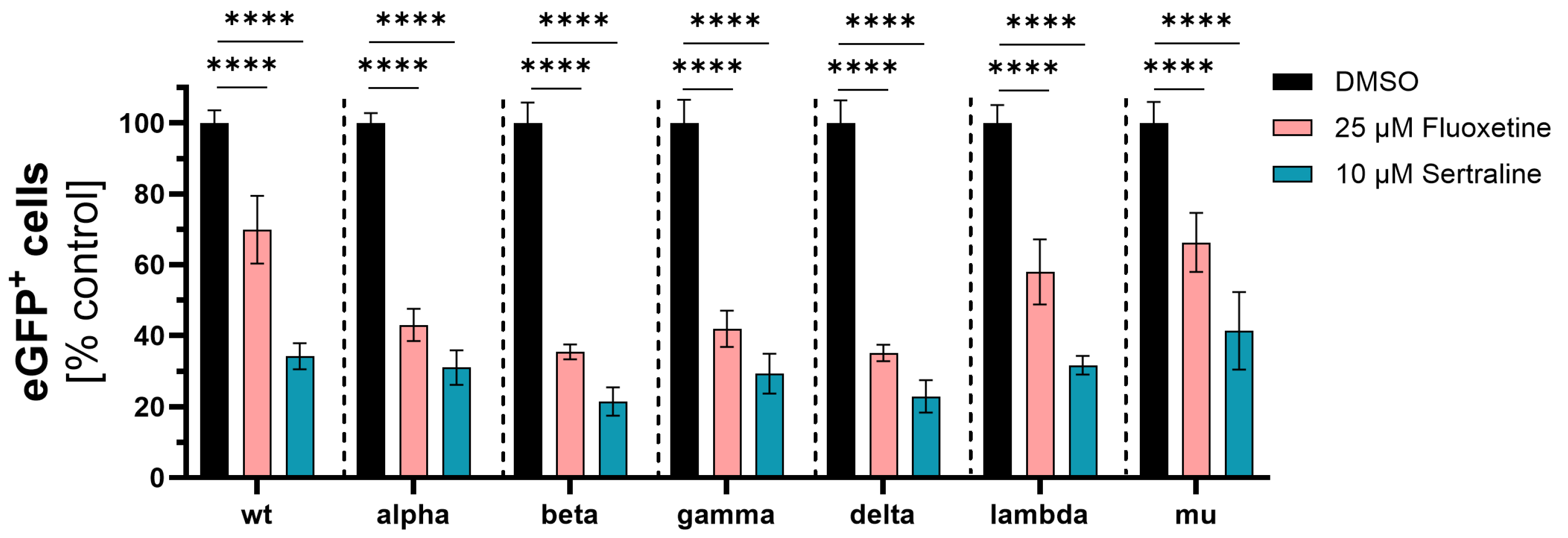
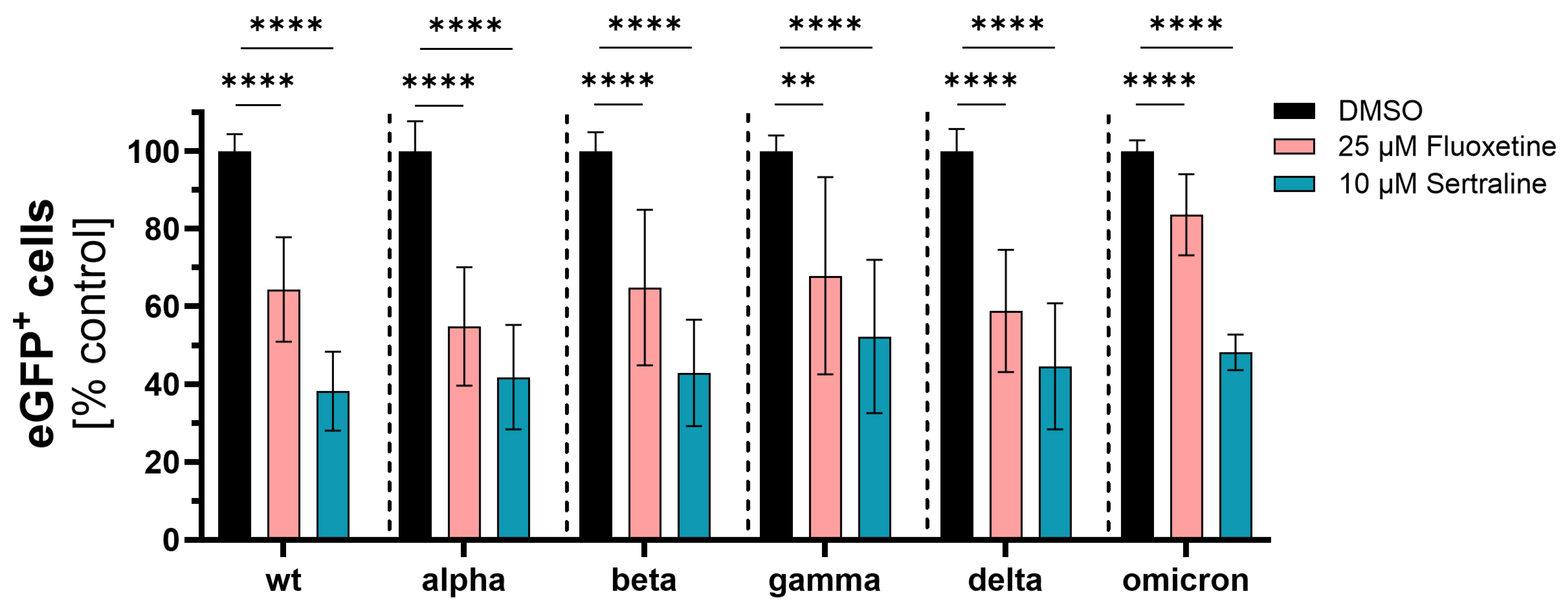
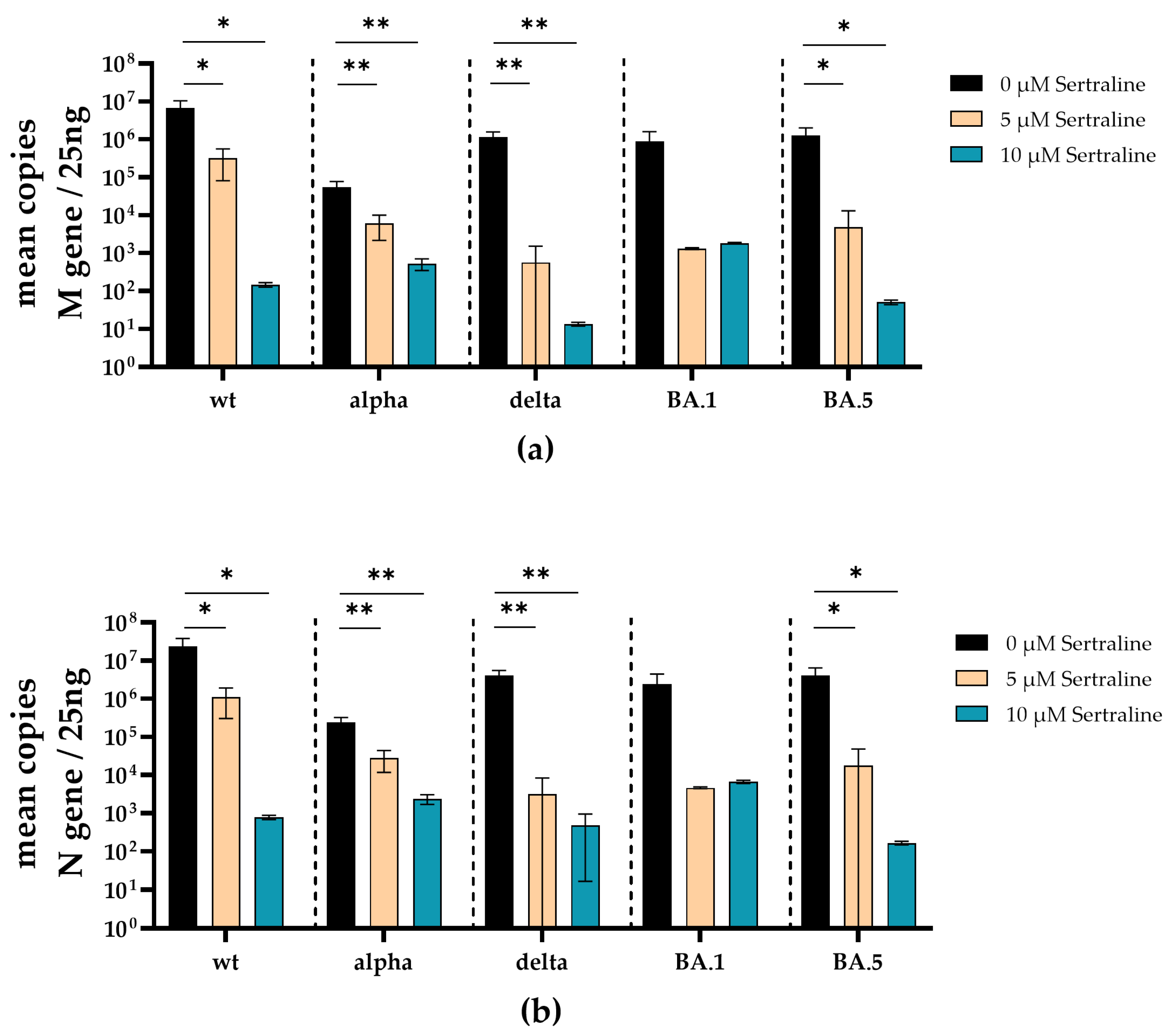
3.1. Initial Investigation of the Antiviral Activity of Fluoxetine and Sertraline against SARS-CoV-2
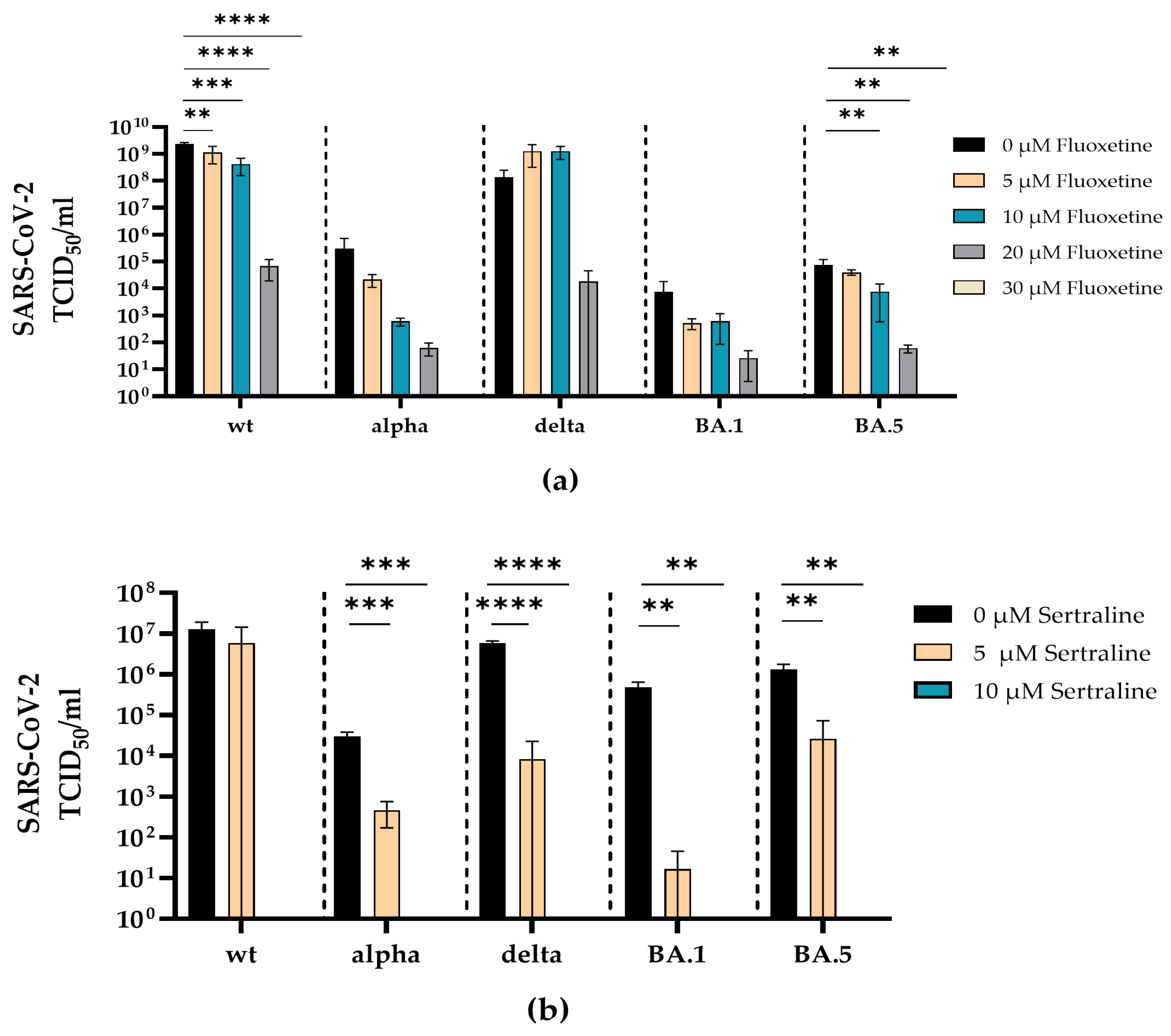
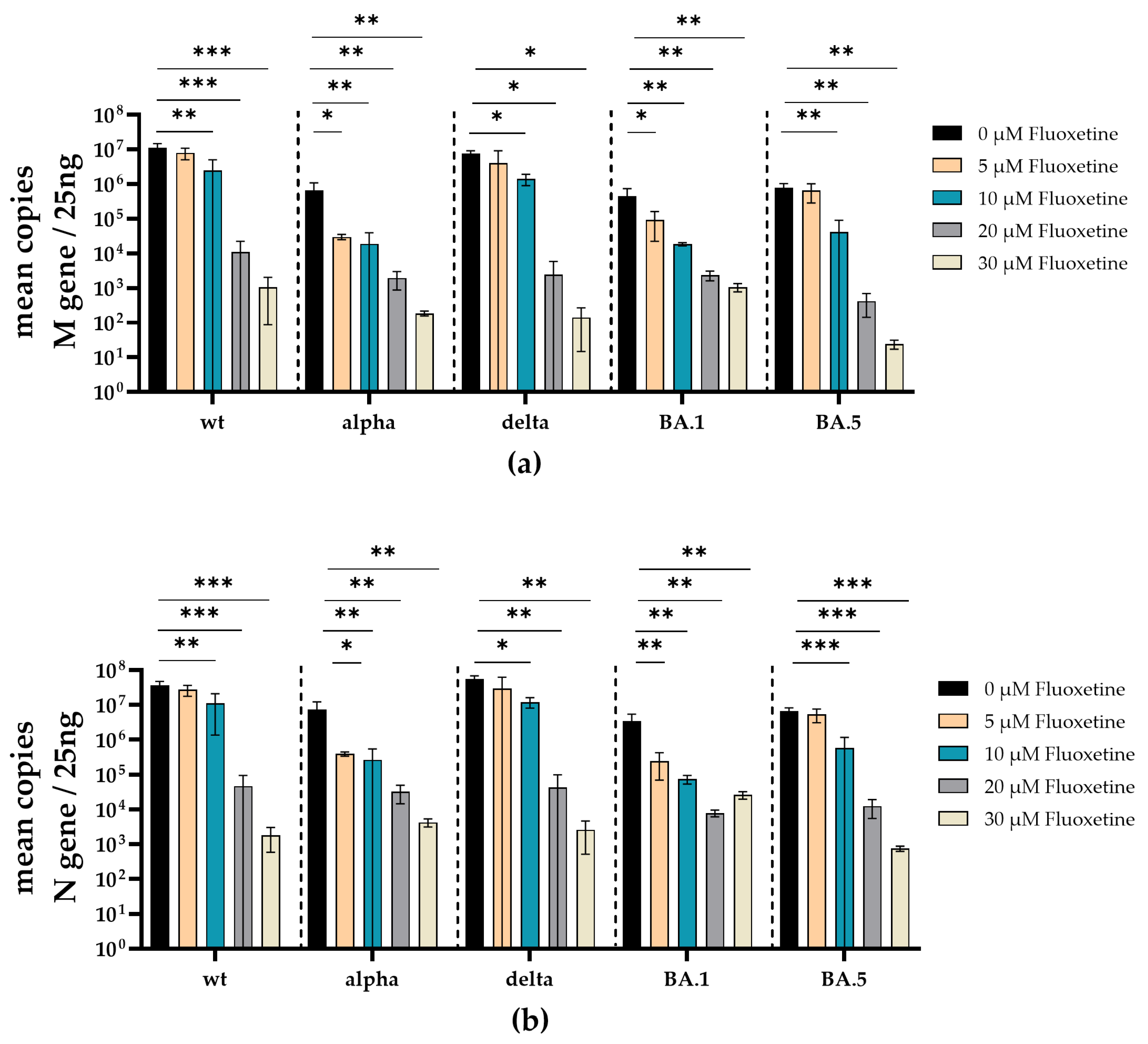
3.2. Fluoxetine and Sertraline Effectively Blocks SARS-CoV-2 Infection
3.3. Fluoxetine and Sertraline Showed No Toxic Effects at SARS-CoV-2 Neutralizing Concentrations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Worldometers. Available online: https://www.worldometers.info/coronavirus/ (accessed on 8 January 2024).
- Panneer, S.; Kantamaneni, K.; Akkayasamy, V.S.; Susairaj, A.X.; Panda, P.K.; Acharya, S.S.; Rice, L.; Liyanage, C.; Pushparaj, R.R.B. The Great Lockdown in the Wake of COVID-19 and Its Implications: Lessons for Low and Middle-Income Countries. Int. J. Environ. Res. Public Health 2022, 19, 610. [Google Scholar] [CrossRef]
- Stamm, T.A.; Partheymüller, J.; Mosor, E.; Ritschl, V.; Kritzinger, S.; Alunno, A.; Eberl, J.-M. Determinants of COVID-19 vaccine fatigue. Nat. Med. 2023, 29, 1164–1171. [Google Scholar] [CrossRef]
- Bormann, M.; Brochhagen, L.; Alt, M.; Otte, M.; Thümmler, L.; van de Sand, L.; Kraiselburd, I.; Thomas, A.; Gosch, J.; Braß, P.; et al. Immune responses in COVID-19 patients during breakthrough infection with SARS-CoV-2 variants Delta, Omicron-BA.1 and Omicron-BA.5. Front. Immunol. 2023, 14, 1150667. [Google Scholar] [CrossRef]
- Bergwerk, M.; Gonen, T.; Lustig, Y.; Amit, S.; Lipsitch, M.; Cohen, C.; Mandelboim, M.; Levin, E.G.; Rubin, C.; Indenbaum, V.; et al. Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 2021, 385, 1474–1484. [Google Scholar] [CrossRef]
- Dash, N.R.; Barqawi, H.J.; Obaideen, A.A.; Al Chame, H.Q.; Samara, K.A.; Qadri, R.; Eldesouki, S. COVID-19 Breakthrough Infection Among Vaccinated Population in the United Arab Emirates. J. Epidemiol. Glob. Health 2023, 13, 67–90. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Chen, S.; Zhan, Q.; Wu, D.; Yang, C.; He, X.; Qiu, M.; Zhang, N.; Li, Z.; et al. Sertraline Is an Effective SARS-CoV-2 Entry Inhibitor Targeting the Spike Protein. J. Virol. 2022, 96, e01245-22. [Google Scholar] [CrossRef]
- Manganaro, R.; Zonsics, B.; Bauer, L.; Lopez, M.L.; Donselaar, T.; Zwaagstra, M.; Saporito, F.; Ferla, S.; Strating, J.R.P.M.; Coutard, B.; et al. Synthesis and antiviral effect of novel fluoxetine analogues as enterovirus 2C inhibitors. Antivir. Res. 2020, 178, 104781. [Google Scholar] [CrossRef]
- Messacar, K.; Sillau, S.; Hopkins, S.E.; Otten, C.; Wilson-Murphy, M.; Wong, B.; Santoro, J.D.; Treister, A.; Bains, H.K.; Torres, A.; et al. Safety, tolerability, and efficacy of fluoxetine as an antiviral for acute flaccid myelitis. Neurology 2019, 92, E2118–E2126. [Google Scholar] [CrossRef]
- Tseng, K.C.; Hsu, B.Y.; Ling, P.; Lu, W.W.; Lin, C.W.; Kung, S.H. Antidepressant Sertraline Is a Broad-Spectrum Inhibitor of Enteroviruses Targeting Viral Entry through Neutralization of Endolysosomal Acidification. Viruses 2022, 14, 109. [Google Scholar] [CrossRef]
- Zuo, J.; Quinn, K.K.; Kye, S.; Cooper, P.; Damoiseaux, R.; Krogstad, P. Fluoxetine Is a Potent Inhibitor of Coxsackievirus Replication. Antimicrob. Agents Chemother. 2012, 56, 4838–4844. [Google Scholar] [CrossRef]
- Patel, K.; Lim, S.G.; Cheng, C.W.; Lawitz, E.; Tillmann, H.L.; Chopra, N.; Altmeyer, R.; Randle, J.C.R.; McHutchison, J.G. Open-label Phase 1b pilot study to assess the antiviral efficacy of simvastatin combined with sertraline in chronic hepatitis C patients. Antivir. Ther. 2011, 16, 1341–1346. [Google Scholar] [CrossRef]
- Herring, S.; Oda, J.M.; Wagoner, J.; Kirchmeier, D.; O’Connor, A.; Nelson, E.A.; Huang, Q.F.; Liang, Y.Y.; DeWald, L.E.; Johansen, L.M.; et al. Inhibition of Arenaviruses by Combinations of Orally Available Approved Drugs. Antimicrob. Agents Chemother. 2021, 65, e01146-20. [Google Scholar] [CrossRef]
- Albouz, S.; Boutry, J.M.; Dubois, G.; Bourdon, R.; Hauw, J.J.; Baumann, N. Lipid and lysosomal enzymes in human fibroblasts cultured with perhexiline maleate. Naunyn-Schmiedebergs Arch. Pharmacol. 1981, 317, 173–177. [Google Scholar] [CrossRef]
- Hurwitz, R.; Ferlinz, K.; Sandhoff, K. The Tricyclic Antidepressant Desipramine Causes Proteolytic Degradation of Lysosomal Sphingomyelinase in Human Fibroblasts. Biol. Chem. Hoppe-Seyler 1994, 375, 447–450. [Google Scholar] [CrossRef]
- Kornhuber, J.; Tripal, P.; Reichel, M.; Mühle, C.; Rhein, C.; Muehlbacher, M.; Groemer, T.W.; Gulbins, E. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): A novel pharmacological group of drugs with broad clinical applications. Cell Physiol. Biochem. 2010, 26, 9–20. [Google Scholar] [CrossRef]
- Zimniak, M.; Kirschner, L.; Hilpert, H.; Geiger, N.; Danov, O.; Oberwinkler, H.; Steinke, M.; Sewald, K.; Seibel, J.; Bodem, J. The serotonin reuptake inhibitor Fluoxetine inhibits SARS-CoV-2 in human lung tissue. Sci. Rep. 2021, 11, 5890. [Google Scholar] [CrossRef]
- Kutkat, O.; Moatasim, Y.; Al-Karmalawy, A.A.; Abulkhair, H.S.; Gomaa, M.R.; El-Taweel, A.N.; Abo Shama, N.M.; GabAllah, M.; Mahmoud, D.B.; Kayali, G.; et al. Robust antiviral activity of commonly prescribed antidepressants against emerging coronaviruses: In vitro and in silico drug repurposing studies. Sci. Rep. 2022, 12, 12920. [Google Scholar] [CrossRef]
- Péricat, D.; Leon-Icaza, S.A.; Sanchez Rico, M.; Mühle, C.; Zoicas, I.; Schumacher, F.; Planès, R.; Mazars, R.; Gros, G.; Carpinteiro, A.; et al. Antiviral and Anti-Inflammatory Activities of Fluoxetine in a SARS-CoV-2 Infection Mouse Model. Int. J. Mol. Sci. 2022, 23, 13623. [Google Scholar] [CrossRef]
- Lenze, E.J.; Mattar, C.; Zorumski, C.F.; Stevens, A.; Schweiger, J.; Nicol, G.E.; Miller, J.P.; Yang, L.; Yingling, M.; Avidan, M.S.; et al. Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients with Symptomatic COVID-19: A Randomized Clinical Trial. JAMA 2020, 324, 2292–2300. [Google Scholar] [CrossRef]
- Calusic, M.; Marcec, R.; Luksa, L.; Jurkovic, I.; Kovac, N.; Mihaljevic, S.; Likic, R. Safety and efficacy of fluvoxamine in COVID-19 ICU patients: An open label, prospective cohort trial with matched controls. Br. J. Clin. Pharmacol. 2022, 88, 2065–2073. [Google Scholar] [CrossRef]
- Seftel, D.; Boulware, D.R. Prospective Cohort of Fluvoxamine for Early Treatment of Coronavirus Disease 19. Open Forum. Infect. Dis. 2021, 8, ofab050. [Google Scholar] [CrossRef]
- Reis, G.; Silva, E.; Silva, D.C.M.; Thabane, L.; Milagres, A.C.; Ferreira, T.S.; Dos Santos, C.V.Q.; Campos, V.H.S.; Nogueira, A.M.R.; de Almeida, A.; et al. Effect of Early Treatment with Ivermectin among Patients with COVID-19. N. Engl. J. Med. 2022, 386, 1721–1731. [Google Scholar] [CrossRef]
- Carpinteiro, A.; Edwards, M.J.; Hoffmann, M.; Kochs, G.; Gripp, B.; Weigang, S.; Adams, C.; Carpinteiro, E.; Gulbins, A.; Keitsch, S.; et al. Pharmacological Inhibition of Acid Sphingomyelinase Prevents Uptake of SARS-CoV-2 by Epithelial Cells. Cell Rep. Med. 2020, 1, 100142. [Google Scholar] [CrossRef]
- Carpinteiro, A.; Gripp, B.; Hoffmann, M.; Pöhlmann, S.; Hoertel, N.; Edwards, M.J.; Kamler, M.; Kornhuber, J.; Becker, K.A.; Gulbins, E. Inhibition of acid sphingomyelinase by ambroxol prevents SARS-CoV-2 entry into epithelial cells. J. Biol. Chem. 2021, 296, 100701. [Google Scholar] [CrossRef]
- Hoertel, N. Do the Selective Serotonin Reuptake Inhibitor Antidepressants Fluoxetine and Fluvoxamine Reduce Mortality among Patients with COVID-19? JAMA Netw. Open 2021, 4, e2136510. [Google Scholar] [CrossRef]
- Hoertel, N.; Sánchez-Rico, M.; Cougoule, C.; Gulbins, E.; Kornhuber, J.; Carpinteiro, A.; Becker, K.A.; Reiersen, A.M.; Lenze, E.J.; Seftel, D.; et al. Repurposing antidepressants inhibiting the sphingomyelinase acid/ceramide system against COVID-19: Current evidence and potential mechanisms. Mol. Psychiatry 2021, 26, 7098–7099. [Google Scholar] [CrossRef]
- Widera, M.; Wilhelm, A.; Toptan, T.; Raffel, J.M.; Kowarz, E.; Roesmann, F.; Grozinger, F.; Siemund, A.L.; Luciano, V.; Kulp, M.; et al. Generation of a Sleeping Beauty Transposon-Based Cellular System for Rapid and Sensitive Screening for Compounds and Cellular Factors Limiting SARS-CoV-2 Replication. Front. Microbiol. 2021, 12, 701198. [Google Scholar] [CrossRef]
- Thümmler, L.; Gäckler, A.; Bormann, M.; Ciesek, S.; Widera, M.; Rohn, H.; Fisenkci, N.; Otte, M.; Alt, M.; Dittmer, U.; et al. Cellular and Humoral Immunity against Different SARS-CoV-2 Variants Is Detectable but Reduced in Vaccinated Kidney Transplant Patients. Vaccines 2022, 10, 1348. [Google Scholar] [CrossRef]
- WHO. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 9 January 2024).
- Heilingloh, C.S.; Aufderhorst, U.W.; Schipper, L.; Dittmer, U.; Witzke, O.; Yang, D.; Zheng, X.; Sutter, K.; Trilling, M.; Alt, M.; et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control 2020, 48, 1273–1275. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Berger Rentsch, M.; Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 2011, 6, e25858. [Google Scholar] [CrossRef]
- Becker, K.A.; Carpinteiro, A.; Hoffmann, M.; Pöhlmann, S.; Kornhuber, J.; Gulbins, E. Ex vivo assay to evaluate the efficacy of drugs targeting sphingolipids in preventing SARS-CoV-2 infection of nasal epithelial cells. STAR Protoc. 2021, 2, 100356. [Google Scholar] [CrossRef]
- Zettl, F.; Meister, T.L.; Vollmer, T.; Fischer, B.; Steinmann, J.; Krawczyk, A.; V’kovski, P.; Todt, D.; Steinmann, E.; Pfaender, S.; et al. Rapid Quantification of SARS-CoV-2-Neutralizing Antibodies Using Propagation-Defective Vesicular Stomatitis Virus Pseudotypes. Vaccines 2020, 8, 386. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Arora, P.; Sidarovich, A.; Moldenhauer, A.S.; Winkler, M.S.; et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 36, 109415. [Google Scholar] [CrossRef]
- Fred, S.M.; Kuivanen, S.; Ugurlu, H.; Casarotto, P.C.; Levanov, L.; Saksela, K.; Vapalahti, O.; Castrén, E. Antidepressant and Antipsychotic Drugs Reduce Viral Infection by SARS-CoV-2 and Fluoxetine Shows Antiviral Activity Against the Novel Variants in vitro. Front. Pharmacol. 2021, 12, 755600. [Google Scholar] [CrossRef]
- Geiger, N.; Kersting, L.; Schlegel, J.; Stelz, L.; Fähr, S.; Diesendorf, V.; Roll, V.; Sostmann, M.; König, E.-M.; Reinhard, S.; et al. The Acid Ceramidase Is a SARS-CoV-2 Host Factor. Cells 2022, 11, 2532. [Google Scholar] [CrossRef]
- Schloer, S.; Brunotte, L.; Goretzko, J.; Mecate-Zambrano, A.; Korthals, N.; Gerke, V.; Ludwig, S.; Rescher, U. Targeting the endolysosomal host-SARS-CoV-2 interface by clinically licensed functional inhibitors of acid sphingomyelinase (FIASMA) including the antidepressant fluoxetine. Emerg. Microbes Infect. 2020, 9, 2245–2255. [Google Scholar] [CrossRef]
- Kornhuber, J.; Hoertel, N.; Gulbins, E. The acid sphingomyelinase/ceramide system in COVID-19. Mol. Psychiatry 2022, 27, 307–314. [Google Scholar] [CrossRef]
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854. [Google Scholar] [CrossRef]
- Beckmann, N.; Becker, K.A. Ceramide and Related Molecules in Viral Infections. Int. J. Mol. Sci. 2021, 22, 5676. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Suzuki, T.; Hashimoto, K. Mechanisms of action of fluvoxamine for COVID-19: A historical review. Mol. Psychiatry 2022, 27, 1898–1907. [Google Scholar] [CrossRef] [PubMed]
- Grassme, H.; Riehle, A.; Wilker, B.; Gulbins, E. Rhinoviruses infect human epithelial cells via ceramide-enriched membrane platforms. J. Biol. Chem. 2005, 280, 26256–26262. [Google Scholar] [CrossRef]
- Miller, M.E.; Adhikary, S.; Kolokoltsov, A.A.; Davey, R.A. Ebolavirus requires acid sphingomyelinase activity and plasma membrane sphingomyelin for infection. J. Virol. 2012, 86, 7473–7483. [Google Scholar] [CrossRef]
- Hoertel, N.; Rezaei, K.; Sánchez-Rico, M.; Delgado-Álvarez, A.; Kornhuber, J.; Gulbins, E.; Olfson, M.; Ouazana-Vedrines, C.; Carpinteiro, A.; Cougoule, C.; et al. Medications Modulating the Acid Sphingomyelinase/Ceramide System and 28-Day Mortality among Patients with SARS-CoV-2: An Observational Study. Pharmaceuticals 2023, 16, 1107. [Google Scholar] [CrossRef] [PubMed]
- Reis, G.; Dos Santos Moreira Silva, E.A.; Medeiros Silva, D.C.; Thabane, L.; de Souza Campos, V.H.; Ferreira, T.S.; Quirino Dos Santos, C.V.; Ribeiro Nogueira, A.M.; Figueiredo Guimaraes Almeida, A.P.; Cançado Monteiro Savassi, L.; et al. Oral Fluvoxamine with Inhaled Budesonide for Treatment of Early-Onset COVID-19: A Randomized Platform Trial. Ann. Intern. Med. 2023, 176, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wei, Y.; Hung, C.T.; Jiang, X.; Li, C.; Jia, K.M.; Leung, E.Y.M.; Yam, C.H.K.; Chow, T.Y.; Zhao, S.; et al. Relationship between antidepressants and severity of SARS-CoV-2 Omicron infection: A retrospective cohort study using real-world data. Lancet Reg. Health West. Pac. 2023, 34, 100716. [Google Scholar] [CrossRef] [PubMed]
- Jittamala, P.; Boyd, S.; Schilling, W.H.; Watson, J.A.; Ngamprasertchai, T.; Siripoon, T.; Luvira, V.; Batty, E.M.; Wongnak, P.; Esper, L.M.; et al. Antiviral efficacy of fluoxetine in early symptomatic COVID-19: An open-label, randomised, controlled, adaptive platform trial (PLATCOV). medRxiv 2024. [Google Scholar] [CrossRef]
- Reis, G.; Dos Santos Moreira-Silva, E.A.; Silva, D.C.M.; Thabane, L.; Milagres, A.C.; Ferreira, T.S.; Dos Santos, C.V.Q.; de Souza Campos, V.H.; Nogueira, A.M.R.; de Almeida, A.; et al. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: The TOGETHER randomised, platform clinical trial. Lancet Glob. Health 2022, 10, e42–e51. [Google Scholar] [CrossRef]
- Oskotsky, T.; Maric, I.; Tang, A.; Oskotsky, B.; Wong, R.J.; Aghaeepour, N.; Sirota, M.; Stevenson, D.K. Mortality Risk Among Patients with COVID-19 Prescribed Selective Serotonin Reuptake Inhibitor Antidepressants. JAMA Netw Open 2021, 4, e2133090. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thümmler, L.; Beckmann, N.; Sehl, C.; Soddemann, M.; Braß, P.; Bormann, M.; Brochhagen, L.; Elsner, C.; Hoertel, N.; Cougoule, C.; et al. Fluoxetine and Sertraline Potently Neutralize the Replication of Distinct SARS-CoV-2 Variants. Viruses 2024, 16, 545. https://doi.org/10.3390/v16040545
Thümmler L, Beckmann N, Sehl C, Soddemann M, Braß P, Bormann M, Brochhagen L, Elsner C, Hoertel N, Cougoule C, et al. Fluoxetine and Sertraline Potently Neutralize the Replication of Distinct SARS-CoV-2 Variants. Viruses. 2024; 16(4):545. https://doi.org/10.3390/v16040545
Chicago/Turabian StyleThümmler, Laura, Nadine Beckmann, Carolin Sehl, Matthias Soddemann, Peer Braß, Maren Bormann, Leonie Brochhagen, Carina Elsner, Nicolas Hoertel, Céline Cougoule, and et al. 2024. "Fluoxetine and Sertraline Potently Neutralize the Replication of Distinct SARS-CoV-2 Variants" Viruses 16, no. 4: 545. https://doi.org/10.3390/v16040545
APA StyleThümmler, L., Beckmann, N., Sehl, C., Soddemann, M., Braß, P., Bormann, M., Brochhagen, L., Elsner, C., Hoertel, N., Cougoule, C., Ciesek, S., Widera, M., Dittmer, U., Lindemann, M., Horn, P. A., Witzke, O., Kadow, S., Kamler, M., Gulbins, E., ... Krawczyk, A. (2024). Fluoxetine and Sertraline Potently Neutralize the Replication of Distinct SARS-CoV-2 Variants. Viruses, 16(4), 545. https://doi.org/10.3390/v16040545






