Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens Used in Assay Comparison
2.2. RNA Extractions and cDNA Synthesis
2.3. Pan-Coronavirus PCR Protocols Selected for Comparison
2.4. Statistical Analysis
3. Results
3.1. Clinical and Environmental Specimens Used in Evaluating the Assay
3.2. Comparative Analysis of Different Pan-CoV PCR Assays in Human Specimens
3.3. Comparative Analysis of Different Pan-CoV PCR Assays in Wastewater Specimens
3.4. Comparative Analysis of Different Pan-CoV PCR Assays in Bat Specimens
3.5. Comparative Analysis of Different Pan-CoV PCR Assays in Avian Specimens
3.6. Comparative Analysis of Different Pan-CoV PCR Assays in CoV Detection from Each Genus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Lau, S.K.; To, K.K.; Cheng, V.C.; Woo, P.C.; Yuen, K.Y. Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015, 28, 465–522. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral. Res. 2014, 101, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Tsoi, H.W.; Wong, B.H.; Wong, S.S.; Leung, S.Y.; Chan, K.H.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Li, K.S.; Poon, R.W.; Wong, B.H.; Tsoi, H.W.; Yip, B.C.; Huang, Y.; Chan, K.H.; Yuen, K.Y. Molecular diversity of coronaviruses in bats. Virology 2006, 351, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Sintunawa, C.; Kaewpom, T.; Khongnomnan, K.; Olival, K.J.; Epstein, J.H.; Rodpan, A.; Sangsri, P.; Intarut, N.; Chindamporn, A.; et al. Group C betacoronavirus in bat guano fertilizer, Thailand. Emerg. Infect. Dis. 2013, 19, 1349–1351. [Google Scholar] [CrossRef]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; Alhakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef]
- WHO Headquarters (HQ). WHO-Convened Global Study of Origins of SARS-CoV-2: China Part; World Health Organization: Geneva, Switzerland, 2021; 14 January–10 February 2021. [Google Scholar]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.-L.; Shi, W.-F.; Zhang, W.; Zhu, Y.; Zhang, Y.-W.; Xie, Q.-M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Scientific Advisory Group on the Origins of Novel Pathogens (SAGO). Preliminary Report for the Scientific Advisory Group for the Origins of Novel Pathogens (SAGO); World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Holbrook, M.G.; Anthony, S.J.; Navarrete-Macias, I.; Bestebroer, T.; Munster, V.J.; van Doremalen, N. Updated and Validated Pan-Coronavirus PCR Assay to Detect All Coronavirus Genera. Viruses 2021, 13, 599. [Google Scholar] [CrossRef]
- Drzewnioková, P.; Festa, F.; Panzarin, V.; Lelli, D.; Moreno, A.; Zecchin, B.; De Benedictis, P.; Leopardi, S. Best Molecular Tools to Investigate Coronavirus Diversity in Mammals: A Comparison. Viruses 2021, 13, 1975. [Google Scholar] [CrossRef]
- Xiu, L.; Binder, R.A.; Alarja, N.A.; Kochek, K.; Coleman, K.K.; Than, S.T.; Bailey, E.S.; Bui, V.N.; Toh, T.-H.; Erdman, D.D.; et al. A RT-PCR assay for the detection of coronaviruses from four genera. J. Clin. Virol. 2020, 128, 104391. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Jung, K.; Wang, Q.; Saif, L.J.; Vlasova, A.N. Development of a one-step RT-PCR assay for detection of pancoronaviruses (α-, β-, γ-, and δ-coronaviruses) using newly designed degenerate primers for porcine and avian `fecal samples. J. Virol. Methods 2018, 256, 116–122. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Moës, E.; Keyaerts, E.; Li, S.; Van Ranst, M. A pancoronavirus RT-PCR assay for detection of all known coronaviruses. Methods Mol. Biol. 2008, 454, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Buathong, R.; Iamsirithawon, S.; Chaifoo, W.; Ponpinit, T.; Ruchisrisarod, C.; Sonpee, C.; Katasrila, P.; Yomrat, S.; Ghai, S.; et al. Identification of a Novel Pathogen Using Family-Wide PCR: Initial Confirmation of COVID-19 in Thailand. Front. Public Health 2020, 8, 555013. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Firth, C.; Street, C.; Henriquez, J.A.; Petrosov, A.; Tashmukhamedova, A.; Hutchison, S.K.; Egholm, M.; Osinubi, M.O.; Niezgoda, M.; et al. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. mBio 2010, 1, e00208-10. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Müller, M.A.; Costabel, U.; Timm, J.; Binger, T.; Meyer, B.; Kreher, P.; Lattwein, E.; Eschbach-Bludau, M.; Nitsche, A.; et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012, 17, 20334. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Johnson, C.K.; Greig, D.J.; Kramer, S.; Che, X.; Wells, H.; Hicks, A.L.; Joly, D.O.; Wolfe, N.D.; Daszak, P.; et al. Global patterns in coronavirus diversity. Virus Evol. 2017, 3, vex012. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Masangkay, J.S.; Nagata, N.; Morikawa, S.; Mizutani, T.; Fukushi, S.; Alviola, P.; Omatsu, T.; Ueda, N.; Iha, K.; et al. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg. Infect. Dis. 2010, 16, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Tangwangvivat, R.; Wacharapluesadee, S.; Pinyopornpanish, P.; Petcharat, S.; Hearn, S.M.; Thippamom, N.; Phiancharoen, C.; Hirunpatrawong, P.; Duangkaewkart, P.; Supataragul, A.; et al. SARS-CoV-2 Variants Detection Strategies in Wastewater Samples Collected in the Bangkok Metropolitan Region. Viruses 2023, 15, 876. [Google Scholar] [CrossRef]
- Hess, A.S.; Shardell, M.; Johnson, J.K.; Thom, K.A.; Strassle, P.; Netzer, G.; Harris, A.D. Methods and recommendations for evaluating and reporting a new diagnostic test. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2111–2116. [Google Scholar] [CrossRef]
- Arsham, H. Test for Equality of Several Population Proportions. Available online: https://home.ubalt.edu/ntsbarsh/business-stat/otherapplets/ProporTest.htm (accessed on 19 March 2024).
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef] [PubMed]
- Stadhouders, R.; Pas, S.D.; Anber, J.; Voermans, J.; Mes, T.H.; Schutten, M. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J. Mol. Diagn. 2010, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Duengkae, P.; Rodpan, A.; Kaewpom, T.; Maneeorn, P.; Kanchanasaka, B.; Yingsakmongkon, S.; Sittidetboripat, N.; Chareesaen, C.; Khlangsap, N.; et al. Diversity of coronavirus in bats from Eastern Thailand. Virol. J. 2015, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Duengkae, P.; Chaiyes, A.; Kaewpom, T.; Rodpan, A.; Yingsakmongkon, S.; Petcharat, S.; Phengsakul, P.; Maneeorn, P.; Hemachudha, T. Longitudinal study of age-specific pattern of coronavirus infection in Lyle’s flying fox (Pteropus lylei) in Thailand. Virol. J. 2018, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiu, L.; Binder, R.A.; Toh, T.-H.; Lee, J.S.-Y.; Ting, J.; Than, S.T.; Qi, W.; Coleman, K.K.; Perera, D.; et al. A pan-coronavirus RT-PCR assay for rapid viral screening of animal, human, and environmental specimens. One Health 2021, 13, 100274. [Google Scholar] [CrossRef] [PubMed]
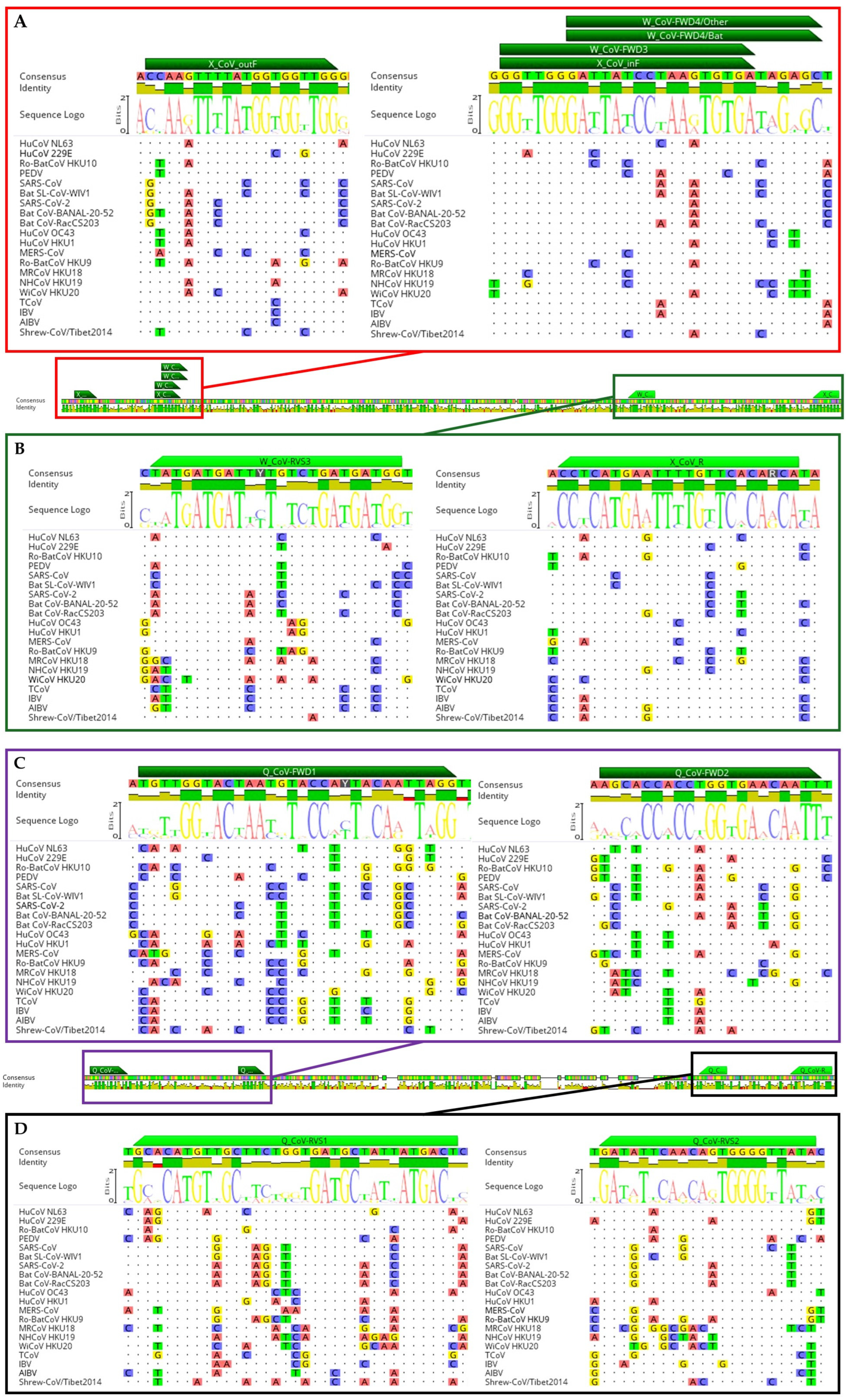
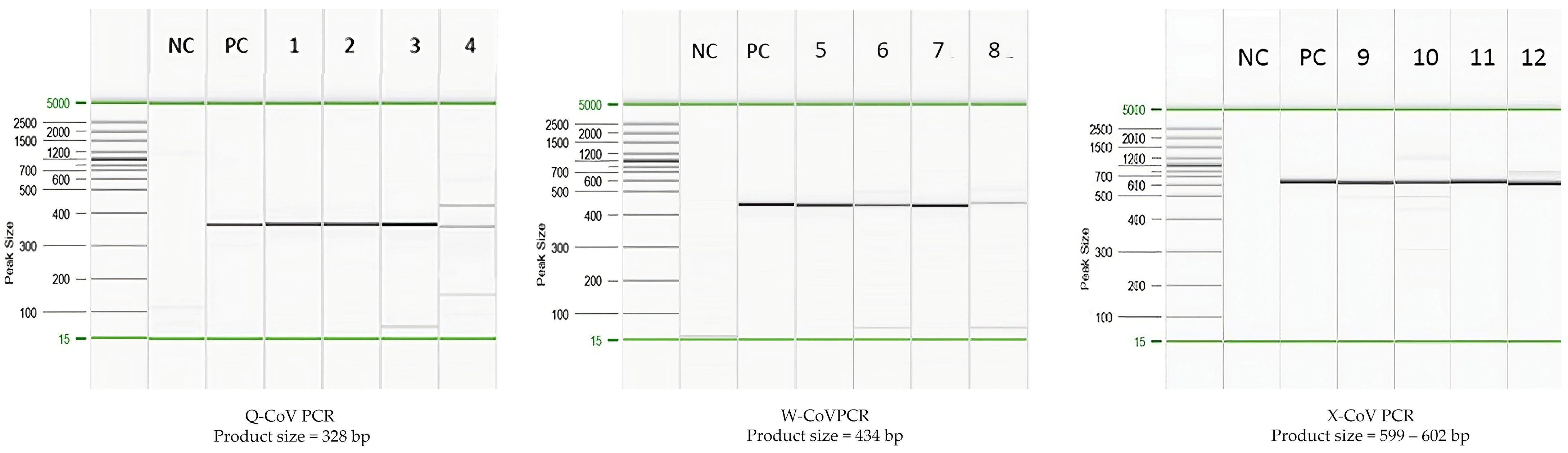

| Virus Name | CoV Genus | Host/Source | Sample Type | Confirmed Assay | Number |
|---|---|---|---|---|---|
| HCoV-229E | α CoV | Human | NPS * | Real-time PCR | 5 |
| HCoV-NL63 | α CoV | Human | NPS | Real-time PCR | 3 |
| Bat Alphacoronavirus HKU10 | α CoV | Bat | Rectal swab | Q-CoV PCR and sequencing | 14 |
| Bat Alphacoronavirus RsYN14 | α CoV | Bat | Rectal swab | Q-CoV PCR and sequencing | 2 |
| HCoV- HKU1 | β CoV | Human | NPS | Real-time PCR | 1 |
| HCoV-OC43 | β CoV | Human | NPS | Real-time PCR | 7 |
| SARS-CoV-2 | β CoV | Human | NPS | Real-time PCR | 7 |
| SARS-CoV-2 | β CoV | Hospital, aircraft | Wastewater | Real-time PCR | 27 |
| Bat Sarbecovirus isolate Ra22QT77 | β CoV | Bat | Rectal swab | Q-CoV PCR and sequencing | 12 |
| Anser fabalis coronavirus NCN2 | γ CoV | Avian | Oral swab Rectal swab | X-CoV PCR and sequencing | 3 |
| Avian coronavirus Glaucous-winged gull CIR-66002 | γ CoV | Avian | Oral swab Rectal swab | X-CoV PCR and sequencing | 1 |
| Avian coronavirus isolate CoV-9518-2016 | γ CoV | Avian | Oral swab Rectal swab | X-CoV PCR and sequencing | 2 |
| Pigeon-dominant Coronavirus | γ CoV | Avian | Oral swab | X-CoV PCR and sequencing | 1 |
| Night-heron coronavirus HKU19 | δ CoV | Avian | Rectal swab | X-CoV PCR and sequencing | 1 |
| Magpie-robin coronavirus HKU18 | δ CoV | Avian | Oral swab Rectal swab | X-CoV PCR and sequencing | 2 |
| unclassified Deltacoronavirus | δ CoV | Avian | Rectal swab | X-CoV PCR and sequencing | 7 |
| Number of PCR Positive Samples Using Three PCR Assays in Four Sample Sources | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CoV Genus | Human | Water | Bat | Avian | ||||||||
| Q | W | X | Q | W | X | Q | W | X | Q | W | X | |
| α | 6 | 4 | 8 | 7 | 2 | 2 | 16 | 16 | 16 | 0 | 0 | 0 |
| β | 15 | 5 | 13 | 23 | 4 | 25 | 12 | 0 | 10 | 0 | 0 | 0 |
| γ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 7 |
| δ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| Negative no. | 29 | 41 | 29 | 16 | 40 | 19 | 0 | 12 | 2 | 16 | 13 | 0 |
| Positive no. | 21 | 9 | 21 | 30 | 6 | 27 | 28 | 16 | 26 | 1 | 4 | 17 |
| Total | 50 | 50 | 50 | 46 | 46 | 46 | 28 | 28 | 28 | 17 | 17 | 17 |
| % positive | 42.00 | 18.00 | 42.00 | 65.22 | 13.04 | 58.70 | 100.00 | 57.14 | 92.86 | 5.88 | 23.53 | 100.00 |
| 95% CI | 79.79–100% | 19.18–59.08% | 79.79–100% | GS | 5.69–34.31% | 79.26–100% | GS | 38.83–75.47% | 83.32–100% | 0.00–17.07% | 3.34–43.69% | GS |
| CoV Genus | Number (%) Positive/Negative Detected Using Three Assays | ||
|---|---|---|---|
| Q-CoV PCR | W-CoV PCR | X-CoV PCR | |
| α | 29 (20.57) | 22 (15.60) | 26 (18.44) |
| β | 50 (35.46) | 9 (6.38) | 48 (34.04) |
| γ | 1 (0.71) | 4 (2.84) | 7 (4.96) |
| δ | 0 (0.00) | 0 (0.00) | 10 (7.09) |
| Negative | 61 (43.26) | 106 (2.84) | 50 (35.46) |
| Total | 141 | 141 | 141 |
| Positive | 80 | 35 | 91 |
| % positive | 56.74 | 24.82 | 64.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wacharapluesadee, S.; Thippamom, N.; Hirunpatrawong, P.; Rattanatumhi, K.; Sterling, S.L.; Khunnawutmanotham, W.; Noradechanon, K.; Maneeorn, P.; Buathong, R.; Paitoonpong, L.; et al. Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens. Viruses 2024, 16, 534. https://doi.org/10.3390/v16040534
Wacharapluesadee S, Thippamom N, Hirunpatrawong P, Rattanatumhi K, Sterling SL, Khunnawutmanotham W, Noradechanon K, Maneeorn P, Buathong R, Paitoonpong L, et al. Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens. Viruses. 2024; 16(4):534. https://doi.org/10.3390/v16040534
Chicago/Turabian StyleWacharapluesadee, Supaporn, Nattakarn Thippamom, Piyapha Hirunpatrawong, Khwankamon Rattanatumhi, Spencer L. Sterling, Wiparat Khunnawutmanotham, Kirana Noradechanon, Patarapol Maneeorn, Rome Buathong, Leilani Paitoonpong, and et al. 2024. "Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens" Viruses 16, no. 4: 534. https://doi.org/10.3390/v16040534
APA StyleWacharapluesadee, S., Thippamom, N., Hirunpatrawong, P., Rattanatumhi, K., Sterling, S. L., Khunnawutmanotham, W., Noradechanon, K., Maneeorn, P., Buathong, R., Paitoonpong, L., & Putcharoen, O. (2024). Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens. Viruses, 16(4), 534. https://doi.org/10.3390/v16040534






