The ISG15-Protease USP18 Is a Pleiotropic Enhancer of HIV-1 Replication
Abstract
1. Introduction
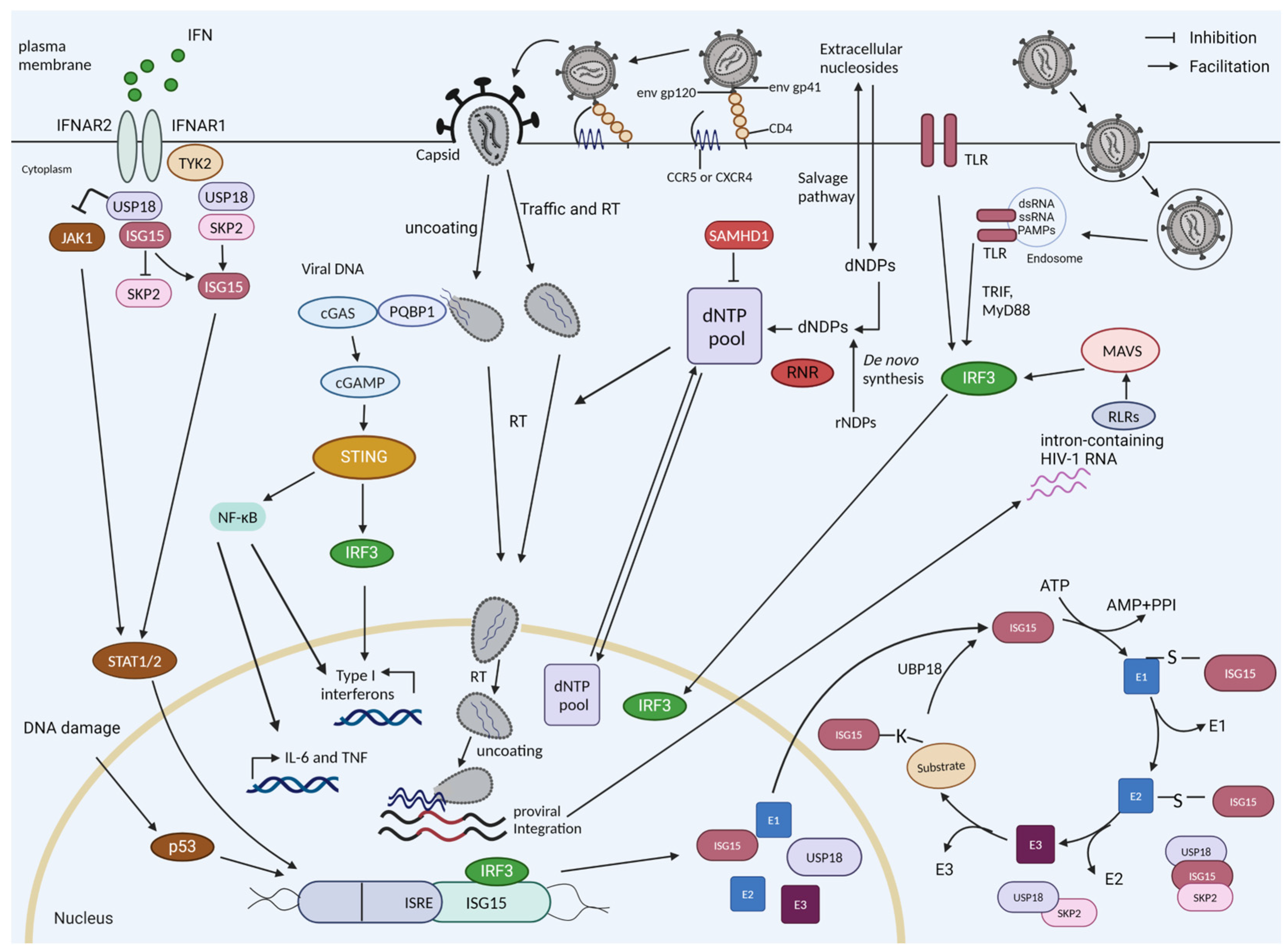
1.1. Interferon and Interferon Induction by HIV-1
1.2. ISG15
1.3. USP18
1.4. P53
1.5. Restriction by SAMHD1
2. USP18 Expression Enhances HIV-1 Replication
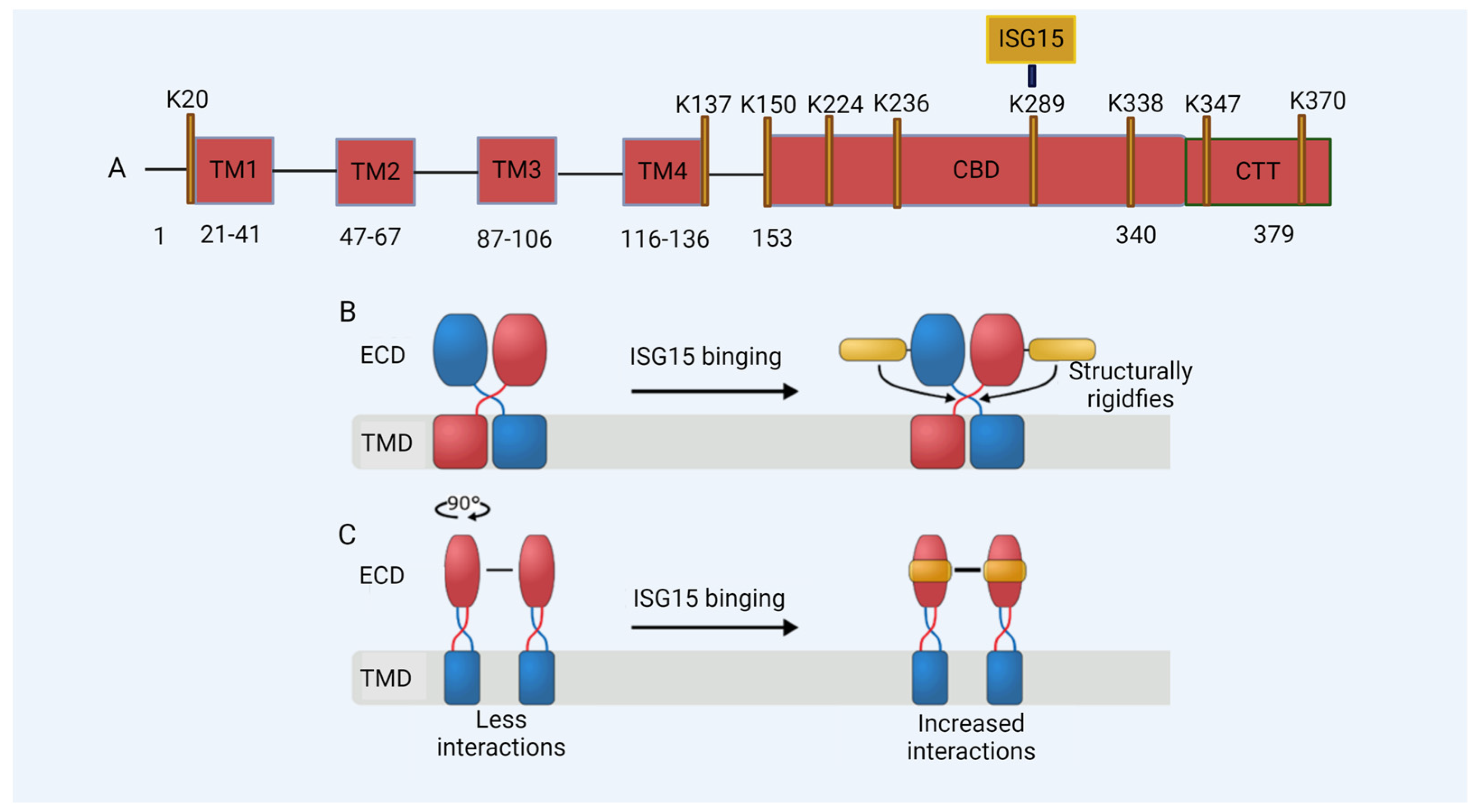
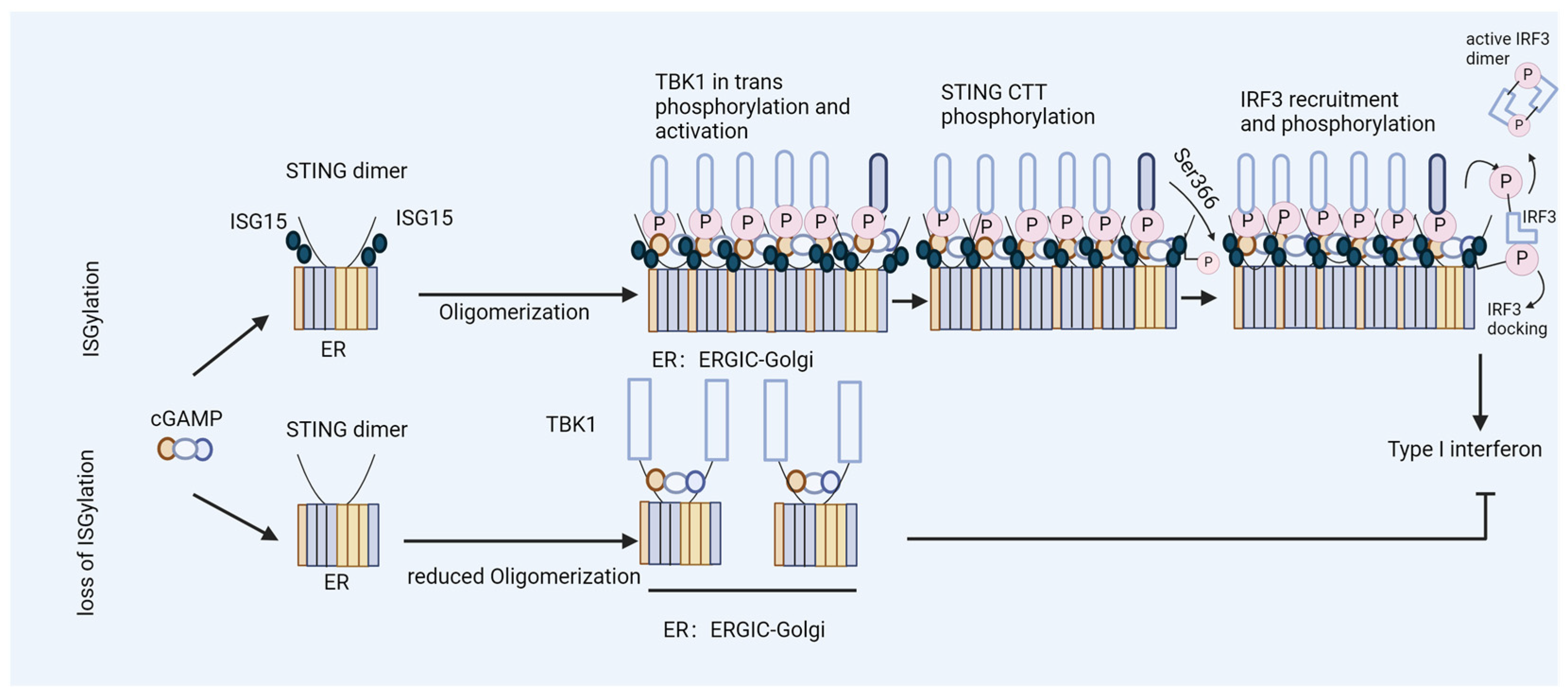
3. ISG15 Is a Regulator of HIV-1 DNA Sensing
4. Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coffin, J.; Swanstrom, R. HIV pathogenesis: Dynamics and genetics of viral populations and infected cells. Cold Spring Harb. Perspect. Med. 2013, 3, a012526. [Google Scholar] [CrossRef] [PubMed]
- Swanstrom, R.; Coffin, J. HIV-1 pathogenesis: The virus. Cold Spring Harb. Perspect. Med. 2012, 2, a007443. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.R.; Olagnier, D.; Hiscott, J. Innate immune sensing of HIV-1 infection. Curr. Opin. HIV AIDS 2015, 10, 96–102. [Google Scholar] [CrossRef]
- Sumner, R.P.; Thorne, L.G.; Fink, D.L.; Khan, H.; Milne, R.S.; Towers, G.J. Are Evolution and the Intracellular Innate Immune System Key Determinants in HIV Transmission? Front. Immunol. 2017, 8, 1246. [Google Scholar] [CrossRef] [PubMed]
- Hertoghs, N.; Geijtenbeek, T.B.H.; Ribeiro, C.M.S. Interplay between HIV-1 innate sensing and restriction in mucosal dendritic cells: Balancing defense and viral transmission. Curr. Opin. Virol. 2017, 22, 112–119. [Google Scholar] [CrossRef]
- Abbink, T.E.; Berkhout, B. HIV-1 reverse transcription initiation: A potential target for novel antivirals? Virus Res. 2008, 134, 4–18. [Google Scholar] [CrossRef]
- Yin, X.; Langer, S.; Zhang, Z.; Herbert, K.M.; Yoh, S.; König, R.; Chanda, S.K. Sensor Sensibility-HIV-1 and the Innate Immune Response. Cells 2020, 9, 254. [Google Scholar] [CrossRef]
- Altfeld, M.; Gale, M., Jr. Innate immunity against HIV-1 infection. Nat. Immunol. 2015, 16, 554–562. [Google Scholar] [CrossRef]
- Gao, D.; Wu, J.; Wu, Y.T.; Du, F.; Aroh, C.; Yan, N.; Sun, L.; Chen, Z.J. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013, 341, 903–906. [Google Scholar] [CrossRef]
- Jakobsen, M.R.; Bak, R.O.; Andersen, A.; Berg, R.K.; Jensen, S.B.; Tengchuan, J.; Laustsen, A.; Hansen, K.; Ostergaard, L.; Fitzgerald, K.A.; et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. USA 2013, 110, E4571–E4580. [Google Scholar] [CrossRef]
- Bode, C.; Fox, M.; Tewary, P.; Steinhagen, A.; Ellerkmann, R.K.; Klinman, D.; Baumgarten, G.; Hornung, V.; Steinhagen, F. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur. J. Immunol. 2016, 46, 1615–1621. [Google Scholar] [CrossRef]
- Meås, H.Z.; Haug, M.; Beckwith, M.S.; Louet, C.; Ryan, L.; Hu, Z.; Landskron, J.; Nordbø, S.A.; Taskén, K.; Yin, H.; et al. Sensing of HIV-1 by TLR8 activates human T cells and reverses latency. Nat. Commun. 2020, 11, 147. [Google Scholar] [CrossRef]
- Kayesh, M.E.H.; Kohara, M.; Tsukiyama-Kohara, K. Toll-like Receptor Response to Human Immunodeficiency Virus Type 1 or Co-Infection with Hepatitis B or C Virus: An Overview. Int. J. Mol. Sci. 2023, 24, 9624. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Miller, C.M.; Ettinger, C.R.; Belkina, A.C.; Snyder-Cappione, J.E.; Gummuluru, S. HIV-1 intron-containing RNA expression induces innate immune activation and T cell dysfunction. Nat. Commun. 2018, 9, 3450. [Google Scholar] [CrossRef]
- McCauley, S.M.; Kim, K.; Nowosielska, A.; Dauphin, A.; Yurkovetskiy, L.; Diehl, W.E.; Luban, J. Intron-containing RNA from the HIV-1 provirus activates type I interferon and inflammatory cytokines. Nat. Commun. 2018, 9, 5305. [Google Scholar] [CrossRef]
- Elsner, C.; Ponnurangam, A.; Kazmierski, J.; Zillinger, T.; Jansen, J.; Todt, D.; Döhner, K.; Xu, S.; Ducroux, A.; Kriedemann, N.; et al. Absence of cGAS-mediated type I IFN responses in HIV-1-infected T cells. Proc. Natl. Acad. Sci. USA 2020, 117, 19475–19486. [Google Scholar] [CrossRef] [PubMed]
- Cingöz, O.; Goff, S.P. HIV-1 Is a Poor Inducer of Innate Immune Responses. mBio 2019, 10, e02834-18. [Google Scholar] [CrossRef] [PubMed]
- Vermeire, J.; Roesch, F.; Sauter, D.; Rua, R.; Hotter, D.; Van Nuffel, A.; Vanderstraeten, H.; Naessens, E.; Iannucci, V.; Landi, A.; et al. HIV Triggers a cGAS-Dependent, Vpu- and Vpr-Regulated Type I Interferon Response in CD4(+) T Cells. Cell Rep. 2016, 17, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Zuliani-Alvarez, L.; Govasli, M.L.; Rasaiyaah, J.; Monit, C.; Perry, S.O.; Sumner, R.P.; McAlpine-Scott, S.; Dickson, C.; Rifat Faysal, K.M.; Hilditch, L.; et al. Evasion of cGAS and TRIM5 defines pandemic HIV. Nat. Microbiol. 2022, 7, 1762–1776. [Google Scholar] [CrossRef]
- Sumner, R.P.; Harrison, L.; Touizer, E.; Peacock, T.P.; Spencer, M.; Zuliani-Alvarez, L.; Towers, G.J. Disrupting HIV-1 capsid formation causes cGAS sensing of viral DNA. EMBO J. 2020, 39, e103958. [Google Scholar] [CrossRef]
- Rasaiyaah, J.; Tan, C.P.; Fletcher, A.J.; Price, A.J.; Blondeau, C.; Hilditch, L.; Jacques, D.A.; Selwood, D.L.; James, L.C.; Noursadeghi, M.; et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 2013, 503, 402–405. [Google Scholar] [CrossRef]
- Papa, G.; Albecka, A.; Mallery, D.; Vaysburd, M.; Renner, N.; James, L.C. IP6-stabilised HIV capsids evade cGAS/STING-mediated host immune sensing. EMBO Rep. 2023, 24, e56275. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, X.; Satoh, T.; Gentili, M.; Cerboni, S.; Conrad, C.; Hurbain, I.; El Marjou, A.; Lacabaratz, C.; Lelièvre, J.D.; Manel, N. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 2013, 39, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Maelfait, J.; Seiradake, E.; Rehwinkel, J. Keeping your armour intact: How HIV-1 evades detection by the innate immune system: HIV-1 capsid controls detection of reverse transcription products by the cytosolic DNA sensor cGAS. Bioessays 2014, 36, 649–657. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Saito, A.; Halambage, U.D.; Ferhadian, D.; Fischer, D.K.; Francis, A.C.; Melikyan, G.B.; Ambrose, Z.; Aiken, C.; Yamashita, M. A Novel Phenotype Links HIV-1 Capsid Stability to cGAS-Mediated DNA Sensing. J. Virol. 2019, 93, e00706-19. [Google Scholar] [CrossRef]
- Yoh, S.M.; Mamede, J.I.; Lau, D.; Ahn, N.; Sánchez-Aparicio, M.T.; Temple, J.; Tuckwell, A.; Fuchs, N.V.; Cianci, G.C.; Riva, L.; et al. Recognition of HIV-1 capsid by PQBP1 licenses an innate immune sensing of nascent HIV-1 DNA. Mol. Cell 2022, 82, 2871–2884.e6. [Google Scholar] [CrossRef] [PubMed]
- Yoh, S.M.; Schneider, M.; Seifried, J.; Soonthornvacharin, S.; Akleh, R.E.; Olivieri, K.C.; De Jesus, P.D.; Ruan, C.; de Castro, E.; Ruiz, P.A.; et al. PQBP1 Is a Proximal Sensor of the cGAS-Dependent Innate Response to HIV-1. Cell 2015, 161, 1293–1305. [Google Scholar] [CrossRef]
- Lahaye, X.; Gentili, M.; Silvin, A.; Conrad, C.; Picard, L.; Jouve, M.; Zueva, E.; Maurin, M.; Nadalin, F.; Knott, G.J.; et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell 2018, 175, 488–501.e22. [Google Scholar] [CrossRef]
- Xu, S.; Ducroux, A.; Ponnurangam, A.; Vieyres, G.; Franz, S.; Müsken, M.; Zillinger, T.; Malassa, A.; Ewald, E.; Hornung, V.; et al. cGAS-Mediated Innate Immunity Spreads Intercellularly through HIV-1 Env-Induced Membrane Fusion Sites. Cell Host Microbe 2016, 20, 443–457. [Google Scholar] [CrossRef]
- Doitsh, G.; Galloway, N.L.; Geng, X.; Yang, Z.; Monroe, K.M.; Zepeda, O.; Hunt, P.W.; Hatano, H.; Sowinski, S.; Muñoz-Arias, I.; et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 2014, 505, 509–514. [Google Scholar] [CrossRef]
- Doyle, T.; Goujon, C.; Malim, M.H. HIV-1 and interferons: Who’s interfering with whom? Nat. Rev. Microbiol. 2015, 13, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Ruiz, A.; Moris, A.; Prado, J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018, 9, 2876. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.C.; Wei, M.C.; Li, F.X.; Shi, M.; Gan, H.; Tang, Z.; Dong, H.P.; Liuyu, T.; Gao, P.; Zhong, B.; et al. The E3 ubiquitin ligase ARIH1 promotes antiviral immunity and autoimmunity by inducing mono-ISGylation and oligomerization of cGAS. Nat. Commun. 2022, 13, 5973. [Google Scholar] [CrossRef]
- Zhao, C.; Sridharan, H.; Chen, R.; Baker, D.P.; Wang, S.; Krug, R.M. Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat. Commun. 2016, 7, 12754. [Google Scholar] [CrossRef]
- Chu, L.; Qian, L.; Chen, Y.; Duan, S.; Ding, M.; Sun, W.; Meng, W.; Zhu, J.; Wang, Q.; Hao, H.; et al. HERC5-catalyzed ISGylation potentiates cGAS-mediated innate immunity. Cell Rep. 2024, 43, 113870. [Google Scholar] [CrossRef] [PubMed]
- François-Newton, V.; Magno de Freitas Almeida, G.; Payelle-Brogard, B.; Monneron, D.; Pichard-Garcia, L.; Piehler, J.; Pellegrini, S.; Uzé, G. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS ONE 2011, 6, e22200. [Google Scholar] [CrossRef]
- Honke, N.; Shaabani, N.; Zhang, D.E.; Hardt, C.; Lang, K.S. Multiple functions of USP18. Cell Death Dis. 2016, 7, e2444. [Google Scholar] [CrossRef]
- Yang, Z.; Xian, H.; Hu, J.; Tian, S.; Qin, Y.; Wang, R.F.; Cui, J. USP18 negatively regulates NF-κB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. Sci. Rep. 2015, 5, 12738. [Google Scholar] [CrossRef]
- Basters, A.; Knobeloch, K.P.; Fritz, G. USP18—A multifunctional component in the interferon response. Biosci. Rep. 2018, 38, BSR20180250. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; El-Diwany, R.; Cohen, L.K.; Rousseau, K.E.; Williams, C.Y.K.; Veenhuis, R.T.; Mehta, S.H.; Blankson, J.N.; Thomas, D.L.; Cox, A.L.; et al. People with HIV-1 demonstrate type 1 interferon refractoriness associated with upregulated USP18. J. Virol. 2021, 95, e01777-20. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Cash, M.N.; Santostefano, K.E.; Nakanishi, M.; Terada, N.; Wallet, M.A. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV-1 infection in macrophages. J. Leukoc. Biol. 2018, 103, 1225–1240. [Google Scholar] [CrossRef]
- Lin, C. Impact of USP18 and ISG15 on Sensing of HIV-1 Infection. Doctoral Thesis, University and State Library of the Heinrich Heine University Düsseldorf, Dusseldorf, Germany, 2023. [Google Scholar]
- Osei Kuffour, E. The Unending War between HIV and the Intrinsic Innate and Adaptive Immune System: Dissecting the Role of USP18 (UBP43) and H. Pylori Coinfection. Doctoral Thesis, Universitäts- und Landesbibliothek der Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany, 2019. [Google Scholar]
- Lin, C.; Kuffour, E.O.; Fuchs, N.V.; Gertzen, C.G.W.; Kaiser, J.; Hirschenberger, M.; Tang, X.; Xu, H.C.; Michel, O.; Tao, R.; et al. Regulation of STING activity in DNA sensing by ISG15 modification. Cell Rep. 2023, 42, 113277. [Google Scholar] [CrossRef]
- Desai, S.D.; Haas, A.L.; Wood, L.M.; Tsai, Y.C.; Pestka, S.; Rubin, E.H.; Saleem, A.; Nur, E.K.A.; Liu, L.F. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006, 66, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Haas, A.L.; Ahrens, P.; Bright, P.M.; Ankel, H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987, 262, 11315–11323. [Google Scholar] [CrossRef]
- Narasimhan, J.; Wang, M.; Fu, Z.; Klein, J.M.; Haas, A.L.; Kim, J.J. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 2005, 280, 27356–27365. [Google Scholar] [CrossRef] [PubMed]
- Durfee, L.A.; Huibregtse, J.M. The ISG15 conjugation system. Methods Mol. Biol. 2012, 832, 141–149. [Google Scholar] [PubMed]
- Malakhov, M.P.; Malakhova, O.A.; Kim, K.I.; Ritchie, K.J.; Zhang, D.E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2002, 277, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Pinto-Fernández, A.; Flierman, D.; Akkermans, J.; O’Brien, D.P.; Greenwood, H.; Scott, H.C.; Fritz, G.; Knobeloch, K.P.; Neefjes, J.; et al. USP16 is an ISG15 cross-reactive deubiquitinase that targets pro-ISG15 and ISGylated proteins involved in metabolism. Proc. Natl. Acad. Sci. USA 2023, 120, e2315163120. [Google Scholar] [CrossRef] [PubMed]
- Malakhova, O.; Malakhov, M.; Hetherington, C.; Zhang, D.E. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J. Biol. Chem. 2002, 277, 14703–14711. [Google Scholar] [CrossRef] [PubMed]
- Pitha-Rowe, I.; Hassel, B.A.; Dmitrovsky, E. Involvement of UBE1L in ISG15 conjugation during retinoid-induced differentiation of acute promyelocytic leukemia. J. Biol. Chem. 2004, 279, 18178–18187. [Google Scholar] [CrossRef]
- Radoshevich, L.; Impens, F.; Ribet, D.; Quereda, J.J.; Nam Tham, T.; Nahori, M.A.; Bierne, H.; Dussurget, O.; Pizarro-Cerdá, J.; Knobeloch, K.P.; et al. ISG15 counteracts Listeria monocytogenes infection. eLife 2015, 4, e06848. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Jo, M.G.; Yoo, H.M.; Hong, S.H.; Park, J.M.; Ka, S.H.; Oh, K.H.; Seol, J.H.; Jung, Y.K.; Chung, C.H. Chemosensitivity is controlled by p63 modification with ubiquitin-like protein ISG15. J. Clin. Investig. 2012, 122, 2622–2636. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Yang, S.W.; Yu, K.R.; Ka, S.H.; Lee, S.W.; Seol, J.H.; Jeon, Y.J.; Chung, C.H. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell 2014, 54, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Crimmins, G.T.; Herskovits, A.A.; Rehder, K.; Sivick, K.E.; Lauer, P.; Dubensky, T.W., Jr.; Portnoy, D.A. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc. Natl. Acad. Sci. USA 2008, 105, 10191–10196. [Google Scholar] [CrossRef] [PubMed]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Werneke, S.W.; Schilte, C.; Rohatgi, A.; Monte, K.J.; Michault, A.; Arenzana-Seisdedos, F.; Vanlandingham, D.L.; Higgs, S.; Fontanet, A.; Albert, M.L.; et al. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 2011, 7, e1002322. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Struckhoff, J.J.; Schneider, J.; Martinez-Sobrido, L.; Wolff, T.; García-Sastre, A.; Zhang, D.E.; Lenschow, D.J. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J. Virol. 2009, 83, 1147–1151. [Google Scholar] [CrossRef]
- Dos Santos, P.F.; Mansur, D.S. Beyond ISGlylation: Functions of Free Intracellular and Extracellular ISG15. J. Interferon Cytokine Res. 2017, 37, 246–253. [Google Scholar] [CrossRef]
- Sun, L.; Wang, X.; Zhou, Y.; Zhou, R.H.; Ho, W.Z.; Li, J.L. Exosomes contribute to the transmission of anti-HIV activity from TLR3-activated brain microvascular endothelial cells to macrophages. Antivir. Res. 2016, 134, 167–171. [Google Scholar] [CrossRef]
- Munnur, D.; Teo, Q.; Eggermont, D.; Lee, H.H.Y.; Thery, F.; Ho, J.; van Leur, S.W.; Ng, W.W.S.; Siu, L.Y.L.; Beling, A.; et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021, 22, 1416–1427. [Google Scholar] [CrossRef]
- Swaim, C.D.; Scott, A.F.; Canadeo, L.A.; Huibregtse, J.M. Extracellular ISG15 Signals Cytokine Secretion through the LFA-1 Integrin Receptor. Mol. Cell 2017, 68, 581–590.e5. [Google Scholar] [CrossRef] [PubMed]
- Recht, M.; Borden, E.C.; Knight, E., Jr. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J. Immunol. 1991, 147, 2617–2623. [Google Scholar] [CrossRef]
- D’Cunha, J.; Knight, E., Jr.; Haas, A.L.; Truitt, R.L.; Borden, E.C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 1996, 93, 211–215.e5. [Google Scholar] [CrossRef]
- Villarreal, D.O.; Wise, M.C.; Siefert, R.J.; Yan, J.; Wood, L.M.; Weiner, D.B. Ubiquitin-like Molecule ISG15 Acts as an Immune Adjuvant to Enhance Antigen-specific CD8 T-cell Tumor Immunity. Mol. Ther. 2015, 23, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Padovan, E.; Terracciano, L.; Certa, U.; Jacobs, B.; Reschner, A.; Bolli, M.; Spagnoli, G.C.; Borden, E.C.; Heberer, M. Interferon stimulated gene 15 constitutively produced by melanoma cells induces e-cadherin expression on human dendritic cells. Cancer Res. 2002, 62, 3453–3458. [Google Scholar]
- Owhashi, M.; Taoka, Y.; Ishii, K.; Nakazawa, S.; Uemura, H.; Kambara, H. Identification of a ubiquitin family protein as a novel neutrophil chemotactic factor. Biochem. Biophys. Res. Commun. 2003, 309, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Kespohl, M.; Bredow, C.; Klingel, K.; Voß, M.; Paeschke, A.; Zickler, M.; Poller, W.; Kaya, Z.; Eckstein, J.; Fechner, H.; et al. Protein modification with ISG15 blocks coxsackievirus pathology by antiviral and metabolic reprogramming. Sci. Adv. 2020, 6, eaay1109. [Google Scholar] [CrossRef]
- Du, Y.; Duan, T.; Feng, Y.; Liu, Q.; Lin, M.; Cui, J.; Wang, R.F. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018, 37, 351–366. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Nakashima, H.; Nguyen, T.; Goins, W.F.; Chiocca, E.A. Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J. Biol. Chem. 2015, 290, 1485–1495. [Google Scholar] [CrossRef]
- Yasuda, J.; Nakao, M.; Kawaoka, Y.; Shida, H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 2003, 77, 9987–9992. [Google Scholar] [CrossRef]
- Okumura, A.; Pitha, P.M.; Harty, R.N. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. USA 2008, 105, 3974–3979. [Google Scholar] [CrossRef]
- Tokarz, S.; Berset, C.; La Rue, J.; Friedman, K.; Nakayama, K.; Nakayama, K.; Zhang, D.E.; Lanker, S. The ISG15 isopeptidase UBP43 is regulated by proteolysis via the SCFSkp2 ubiquitin ligase. J. Biol. Chem. 2004, 279, 46424–46430. [Google Scholar] [CrossRef]
- Yeh, Y.H.; Yang, Y.C.; Hsieh, M.Y.; Yeh, Y.C.; Li, T.K. A negative feedback of the HIF-1α pathway via interferon-stimulated gene 15 and ISGylation. Clin. Cancer Res. 2013, 19, 5927–5939. [Google Scholar] [CrossRef]
- Zhang, X.; Bogunovic, D.; Payelle-Brogard, B.; Francois-Newton, V.; Speer, S.D.; Yuan, C.; Volpi, S.; Li, Z.; Sanal, O.; Mansouri, D.; et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 2015, 517, 89–93. [Google Scholar] [CrossRef]
- Speer, S.D.; Li, Z.; Buta, S.; Payelle-Brogard, B.; Qian, L.; Vigant, F.; Rubino, E.; Gardner, T.J.; Wedeking, T.; Hermann, M.; et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat. Commun. 2016, 7, 11496. [Google Scholar] [PubMed]
- Kang, D.; Jiang, H.; Wu, Q.; Pestka, S.; Fisher, P.B. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene 2001, 267, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Q.; Ilaria, R., Jr.; Kingsley, P.D.; Iwama, A.; van Etten, R.A.; Palis, J.; Zhang, D.E. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 1999, 19, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shin, J.; Molitor, T.W.; Schook, L.B.; Rutherford, M.S. Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology 1999, 262, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Zhong, B.; Blonska, M.; Gorjestani, S.; Yan, M.; Tian, Q.; Zhang, D.E.; Lin, X.; Dong, C. USP18 inhibits NF-κB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. J. Exp. Med. 2013, 210, 1575–1590. [Google Scholar] [CrossRef]
- Cong, X.L.; Lo, M.C.; Reuter, B.A.; Yan, M.; Fan, J.B.; Zhang, D.E. Usp18 promotes conventional CD11b+ dendritic cell development. J. Immunol. 2012, 188, 4776–4781. [Google Scholar] [CrossRef] [PubMed]
- Colonne, P.M.; Sahni, A.; Sahni, S.K. Rickettsia conorii infection stimulates the expression of ISG15 and ISG15 protease UBP43 in human microvascular endothelial cells. Biochem. Biophys. Res. Commun. 2011, 416, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lei, Q.S.; Zhang, S.J.; Kong, L.N.; Qin, B. Suppression of USP18 Potentiates the Anti-HBV Activity of Interferon Alpha in HepG2.2.15 Cells via JAK/STAT Signaling. PLoS ONE 2016, 11, e0156496. [Google Scholar]
- Ye, H.; Duan, X.; Yao, M.; Kang, L.; Li, Y.; Li, S.; Li, B.; Chen, L. USP18 Mediates Interferon Resistance of Dengue Virus Infection. Front. Microbiol. 2021, 12, 682380. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Feng, S.; Zhang, J.; Qiu, W.; Qian, M.; Wang, Y. USP18 deubiquitinates and stabilizes Twist1 to promote epithelial-mesenchymal transition in glioblastoma cells. Am. J. Cancer Res. 2020, 10, 1156–1169. [Google Scholar]
- Huang, F.; Zheng, C.; Huang, L.; Lin, C.; Wang, J. USP18 directly regulates Snail1 protein through ubiquitination pathway in colorectal cancer. Cancer Cell Int. 2020, 20, 346. [Google Scholar] [CrossRef]
- Liu, W.; Ge, X.; Zhou, Z.; Jiang, D.; Rong, Y.; Wang, J.; Ji, C.; Fan, J.; Yin, G.; Cai, W. Deubiquitinase USP18 regulates reactive astrogliosis by stabilizing SOX9. Glia 2021, 69, 1782–1798. [Google Scholar] [CrossRef]
- Song, C.; Peng, J.; Wei, Y.; Shao, J.; Chen, X.; Zhang, X.; Xu, J. USP18 promotes tumor metastasis in esophageal squamous cell carcinomas via deubiquitinating ZEB1. Exp. Cell Res. 2021, 409, 112884. [Google Scholar] [CrossRef]
- Goldmann, T.; Zeller, N.; Raasch, J.; Kierdorf, K.; Frenzel, K.; Ketscher, L.; Basters, A.; Staszewski, O.; Brendecke, S.M.; Spiess, A.; et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015, 34, 1612–1629. [Google Scholar] [CrossRef]
- Meuwissen, M.E.; Schot, R.; Buta, S.; Oudesluijs, G.; Tinschert, S.; Speer, S.D.; Li, Z.; van Unen, L.; Heijsman, D.; Goldmann, T.; et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 2016, 213, 1163–1174. [Google Scholar] [CrossRef]
- Arimoto, K.I.; Löchte, S.; Stoner, S.A.; Burkart, C.; Zhang, Y.; Miyauchi, S.; Wilmes, S.; Fan, J.B.; Heinisch, J.J.; Li, Z.; et al. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat. Struct. Mol. Biol. 2017, 24, 279–289. [Google Scholar] [CrossRef]
- Miyauchi, S.; Arimoto, K.I.; Liu, M.; Zhang, Y.; Zhang, D.E. Reprogramming of tumor-associated macrophages via NEDD4-mediated CSF1R degradation by targeting USP18. Cell Rep. 2023, 42, 113560. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Han, L.; Zhao, Z.; Liu, H.; Zhang, L.; Ma, C.; Yi, F.; Liu, B.; Zheng, Y.; Gao, C. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat. Commun. 2021, 12, 2970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, M.X.; Zhang, Q.; Zhu, G.F.; Yuan, L.; Zhang, D.E.; Zhu, Q.; Yao, J.; Shu, H.B.; Zhong, B. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016, 26, 1302–1319. [Google Scholar] [CrossRef]
- Arimoto, K.I.; Miyauchi, S.; Troutman, T.D.; Zhang, Y.; Liu, M.; Stoner, S.A.; Davis, A.G.; Fan, J.B.; Huang, Y.J.; Yan, M.; et al. Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis. Nat. Commun. 2023, 14, 251. [Google Scholar] [CrossRef]
- Olie, C.S.; Pinto-Fernández, A.; Damianou, A.; Vendrell, I.; Mei, H.; den Hamer, B.; van der Wal, E.; de Greef, J.C.; Raz, V.; Kessler, B.M. USP18 is an essential regulator of muscle cell differentiation and maturation. Cell Death Dis. 2023, 14, 231. [Google Scholar] [CrossRef]
- Osei Kuffour, E.; König, R.; Häussinger, D.; Schulz, W.A.; Münk, C. ISG15 Deficiency Enhances HIV-1 Infection by Accumulating Misfolded p53. mBio 2019, 10, e01342-19. [Google Scholar] [CrossRef]
- Baker, S.J.; Fearon, E.R.; Nigro, J.M.; Hamilton, S.R.; Preisinger, A.C.; Jessup, J.M.; vanTuinen, P.; Ledbetter, D.H.; Barker, D.F.; Nakamura, Y.; et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989, 244, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. p53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef]
- Lane, D.P.; Crawford, L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979, 278, 261–263. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Mandinova, A.; Aaronson, S.A.; Lee, S.W. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat. Rev. Immunol. 2016, 16, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Bullock, A.N.; Fersht, A.R. Rescuing the function of mutant p53. Nat. Rev. Cancer 2001, 1, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Oren, M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003, 10, 431–442. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. p53 signaling in cancer progression and therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Oren, M.; Rotter, V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a001107. [Google Scholar] [CrossRef]
- Brosh, R.; Rotter, V. When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer 2009, 9, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Wee, S.; Gunaratne, J.; Lane, D.P.; Bulavin, D.V. Isg15 controls p53 stability and functions. Cell Cycle 2014, 13, 2200–2210. [Google Scholar] [CrossRef]
- Huang, Y.F.; Bulavin, D.V. Oncogene-mediated regulation of p53 ISGylation and functions. Oncotarget 2014, 5, 5808–5818. [Google Scholar] [CrossRef]
- Park, J.H.; Yang, S.W.; Park, J.M.; Ka, S.H.; Kim, J.H.; Kong, Y.Y.; Jeon, Y.J.; Seol, J.H.; Chung, C.H. Positive feedback regulation of p53 transactivity by DNA damage-induced ISG15 modification. Nat. Commun. 2016, 7, 12513. [Google Scholar] [CrossRef]
- Laptenko, O.; Prives, C. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 2006, 13, 951–961. [Google Scholar] [CrossRef]
- Kim, E.; Deppert, W. The versatile interactions of p53 with DNA: When flexibility serves specificity. Cell Death Differ. 2006, 13, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Zhu, W.G. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 2012, 8, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Shiff, I.; Freed-Pastor, W.; Zupnick, A.; Mattia, M.; Freulich, E.; Shamir, I.; Kadouri, N.; Kahan, T.; Manfredi, J.; et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell 2015, 57, 1034–1046. [Google Scholar] [CrossRef] [PubMed]
- el-Deiry, W.S.; Harper, J.W.; O’Connor, P.M.; Velculescu, V.E.; Canman, C.E.; Jackman, J.; Pietenpol, J.A.; Burrell, M.; Hill, D.E.; Wang, Y.; et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994, 54, 1169–1174. [Google Scholar]
- He, G.; Siddik, Z.H.; Huang, Z.; Wang, R.; Koomen, J.; Kobayashi, R.; Khokhar, A.R.; Kuang, J. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene 2005, 24, 2929–2943. [Google Scholar] [CrossRef]
- Krześniak, M.; Zajkowicz, A.; Gdowicz-Kłosok, A.; Głowala-Kosińska, M.; Łasut-Szyszka, B.; Rusin, M. Synergistic activation of p53 by actinomycin D and nutlin-3a is associated with the upregulation of crucial regulators and effectors of innate immunity. Cell. Signal. 2020, 69, 109552. [Google Scholar] [CrossRef] [PubMed]
- Textor, S.; Fiegler, N.; Arnold, A.; Porgador, A.; Hofmann, T.G.; Cerwenka, A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011, 71, 5998–6009. [Google Scholar] [CrossRef]
- Kadaja-Saarepuu, L.; Lõoke, M.; Balikova, A.; Maimets, T. Tumor suppressor p53 down-regulates expression of human leukocyte marker CD43 in non-hematopoietic tumor cells. Int. J. Oncol. 2012, 40, 567–576. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Pazos, M.; Delgado, I.; Murk, W.; Mungamuri, S.K.; Lee, S.W.; García-Sastre, A.; Moran, T.M.; Aaronson, S.A. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J. Immunol. 2011, 187, 6428–6436. [Google Scholar] [CrossRef]
- Yamanishi, Y.; Boyle, D.L.; Rosengren, S.; Green, D.R.; Zvaifler, N.J.; Firestein, G.S. Regional analysis of p53 mutations in rheumatoid arthritis synovium. Proc. Natl. Acad. Sci. USA 2002, 99, 10025–10030. [Google Scholar] [CrossRef]
- Kawashima, H.; Takatori, H.; Suzuki, K.; Iwata, A.; Yokota, M.; Suto, A.; Minamino, T.; Hirose, K.; Nakajima, H. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. J. Immunol. 2013, 191, 3614–3623. [Google Scholar] [CrossRef]
- Osei Kuffour, E.; Schott, K.; Jaguva Vasudevan, A.A.; Holler, J.; Schulz, W.A.; Lang, P.A.; Lang, K.S.; Kim, B.; Häussinger, D.; König, R.; et al. USP18 (UBP43) Abrogates p21-Mediated Inhibition of HIV-1. J. Virol. 2018, 92, e00592-18. [Google Scholar] [CrossRef] [PubMed]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Baldauf, H.M.; Pan, X.; Erikson, E.; Schmidt, S.; Daddacha, W.; Burggraf, M.; Schenkova, K.; Ambiel, I.; Wabnitz, G.; Gramberg, T.; et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012, 18, 1682–1687. [Google Scholar] [CrossRef] [PubMed]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 2011, 474, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Descours, B.; Cribier, A.; Chable-Bessia, C.; Ayinde, D.; Rice, G.; Crow, Y.; Yatim, A.; Schwartz, O.; Laguette, N.; Benkirane, M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 2012, 9, 87. [Google Scholar] [CrossRef]
- Berger, A.; Sommer, A.F.; Zwarg, J.; Hamdorf, M.; Welzel, K.; Esly, N.; Panitz, S.; Reuter, A.; Ramos, I.; Jatiani, A.; et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011, 7, e1002425. [Google Scholar] [CrossRef]
- Volcic, M.; Wiesmüller, L.; Kirchhoff, F. Small but Highly Versatile: The Viral Accessory Protein Vpu. Annu. Rev. Virol. 2023, 10, 243–259. [Google Scholar] [CrossRef]
- Majer, C.; Schüssler, J.M.; König, R. Intertwined: SAMHD1 cellular functions, restriction, and viral evasion strategies. Med. Microbiol. Immunol. 2019, 208, 513–529. [Google Scholar] [CrossRef]
- Powell, R.D.; Holland, P.J.; Hollis, T.; Perrino, F.W. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 2011, 286, 43596–43600. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef] [PubMed]
- St Gelais, C.; de Silva, S.; Amie, S.M.; Coleman, C.M.; Hoy, H.; Hollenbaugh, J.A.; Kim, B.; Wu, L. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 2012, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Welbourn, S.; Strebel, K. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology 2016, 488, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Schüssler, M.; Schott, K.; Fuchs, N.V.; Oo, A.; Zahadi, M.; Rauch, P.; Kim, B.; König, R. Gene editing of SAMHD1 in macrophage-like cells reveals complex relationships between SAMHD1 phospho-regulation, HIV-1 restriction, and cellular dNTP levels. mBio 2023, 14, e0225223. [Google Scholar] [CrossRef]
- Beloglazova, N.; Flick, R.; Tchigvintsev, A.; Brown, G.; Popovic, A.; Nocek, B.; Yakunin, A.F. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 2013, 288, 8101–8110. [Google Scholar] [CrossRef]
- Maharana, S.; Kretschmer, S.; Hunger, S.; Yan, X.; Kuster, D.; Traikov, S.; Zillinger, T.; Gentzel, M.; Elangovan, S.; Dasgupta, P.; et al. SAMHD1 controls innate immunity by regulating condensation of immunogenic self RNA. Mol. Cell 2022, 82, 3712–3728.e10. [Google Scholar] [CrossRef]
- Schmidt, S.; Schenkova, K.; Adam, T.; Erikson, E.; Lehmann-Koch, J.; Sertel, S.; Verhasselt, B.; Fackler, O.T.; Lasitschka, F.; Keppler, O.T. SAMHD1’s protein expression profile in humans. J. Leukoc. Biol. 2015, 98, 5–14. [Google Scholar] [CrossRef]
- Yang, S.; Zhan, Y.; Zhou, Y.; Jiang, Y.; Zheng, X.; Yu, L.; Tong, W.; Gao, F.; Li, L.; Huang, Q.; et al. Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci. Rep. 2016, 6, 29665. [Google Scholar] [CrossRef]
- Riess, M.; Fuchs, N.V.; Idica, A.; Hamdorf, M.; Flory, E.; Pedersen, I.M.; König, R. Interferons Induce Expression of SAMHD1 in Monocytes through Down-regulation of miR-181a and miR-30a. J. Biol. Chem. 2017, 292, 264–277. [Google Scholar] [CrossRef]
- Cribier, A.; Descours, B.; Valadão, A.L.; Laguette, N.; Benkirane, M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013, 3, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- White, T.E.; Brandariz-Nuñez, A.; Valle-Casuso, J.C.; Amie, S.; Nguyen, L.A.; Kim, B.; Tuzova, M.; Diaz-Griffero, F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 2013, 13, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Schott, K.; Fuchs, N.V.; Derua, R.; Mahboubi, B.; Schnellbächer, E.; Seifried, J.; Tondera, C.; Schmitz, H.; Shepard, C.; Brandariz-Nuñez, A.; et al. Dephosphorylation of the HIV-1 restriction factor SAMHD1 is mediated by PP2A-B55α holoenzymes during mitotic exit. Nat. Commun. 2018, 9, 2227. [Google Scholar] [CrossRef] [PubMed]
- Mlcochova, P.; Sutherland, K.A.; Watters, S.A.; Bertoli, C.; de Bruin, R.A.; Rehwinkel, J.; Neil, S.J.; Lenzi, G.M.; Kim, B.; Khwaja, A.; et al. A G1-like state allows HIV-1 to bypass SAMHD1 restriction in macrophages. EMBO J. 2017, 36, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Allouch, A.; David, A.; Amie, S.M.; Lahouassa, H.; Chartier, L.; Margottin-Goguet, F.; Barré-Sinoussi, F.; Kim, B.; Sáez-Cirión, A.; Pancino, G. Reply to Pauls et al.: p21 is a master regulator of HIV replication in macrophages through dNTP synthesis block. Proc. Natl. Acad. Sci. USA 2014, 111, E1325–E1326. [Google Scholar] [CrossRef]
- St Gelais, C.; de Silva, S.; Hach, J.C.; White, T.E.; Diaz-Griffero, F.; Yount, J.S.; Wu, L. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 2014, 88, 5834–5844. [Google Scholar] [CrossRef]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef]
- Badia, R.; Pujantell, M.; Riveira-Muñoz, E.; Puig, T.; Torres-Torronteras, J.; Martí, R.; Clotet, B.; Ampudia, R.M.; Vives-Pi, M.; Esté, J.A.; et al. The G1/S Specific Cyclin D2 Is a Regulator of HIV-1 Restriction in Non-proliferating Cells. PLoS Pathog. 2016, 12, e1005829. [Google Scholar] [CrossRef]
- Li, J.; Guo, M.; Chen, L.; Chen, Z.; Fu, Y.; Chen, Y. p53 amyloid aggregation in cancer: Function, mechanism, and therapy. Exp. Hematol. Oncol. 2022, 11, 66. [Google Scholar] [CrossRef]
- Shi, H.X.; Yang, K.; Liu, X.; Liu, X.Y.; Wei, B.; Shan, Y.F.; Zhu, L.H.; Wang, C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 2010, 30, 2424–2436. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lee, J.H.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; van Gent, M.; Riedl, W.; Davis-Gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Hwang, S.Y.; Imaizumi, T.; Yoo, J.Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 2008, 82, 1474–1483. [Google Scholar] [CrossRef] [PubMed]
- Malakhov, M.P.; Kim, K.I.; Malakhova, O.A.; Jacobs, B.S.; Borden, E.C.; Zhang, D.E. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2003, 278, 16608–16613. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Murugaiah, V.; Yasmin, H.; Pandit, H.; Ganguly, K.; Subedi, R.; Al-Mozaini, M.; Madan, T.; Kishore, U. Innate Immune Response Against HIV-1. Adv. Exp. Med. Biol. 2021, 1313, 23–58. [Google Scholar]
- Kang, J.; Wu, J.; Liu, Q.; Wu, X.; Zhao, Y.; Ren, J. Post-Translational Modifications of STING: A Potential Therapeutic Target. Front. Immunol. 2022, 13, 888147. [Google Scholar] [CrossRef]
- Hong, Z.; Ma, T.; Liu, X.; Wang, C. cGAS-STING pathway: Post-translational modifications and functions in sterile inflammatory diseases. FEBS J. 2022, 289, 6187–6208. [Google Scholar] [CrossRef]
- Qian, G.; Zhang, Y.; Liu, Y.; Li, M.; Xin, B.; Jiang, W.; Han, W.; Wang, Y.; Tang, X.; Li, L.; et al. Glutamylation of an HIV-1 protein inhibits the immune response by hijacking STING. Cell Rep. 2023, 42, 112442. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, G.; Zhu, L.; Zhao, Z.; Liu, Y.; Han, W.; Zhang, X.; Zhang, Y.; Xiong, T.; Zeng, H.; et al. HIV-1 Vif suppresses antiviral immunity by targeting STING. Cell Mol. Immunol. 2022, 19, 108–121. [Google Scholar] [CrossRef]
- Fremond, M.L.; Crow, Y.J. STING-Mediated Lung Inflammation and Beyond. J. Clin. Immunol. 2021, 41, 501–514. [Google Scholar] [CrossRef]
- Jurczyszak, D.; Manganaro, L.; Buta, S.; Gruber, C.; Martin-Fernandez, M.; Taft, J.; Patel, R.S.; Cipolla, M.; Alshammary, H.; Mulder, L.C.F.; et al. ISG15 deficiency restricts HIV-1 infection. PLoS Pathog. 2022, 18, e1010405. [Google Scholar] [CrossRef]
- Pincetic, A.; Kuang, Z.; Seo, E.J.; Leis, J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J. Virol. 2010, 84, 4725–4736. [Google Scholar] [CrossRef] [PubMed]
- Okumura, A.; Lu, G.; Pitha-Rowe, I.; Pitha, P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. USA 2006, 103, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Woods, M.W.; Kelly, J.N.; Hattlmann, C.J.; Tong, J.G.; Xu, L.S.; Coleman, M.D.; Quest, G.R.; Smiley, J.R.; Barr, S.D. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology 2011, 8, 95. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Kuffour, E.O.; Li, T.; Gertzen, C.G.W.; Kaiser, J.; Luedde, T.; König, R.; Gohlke, H.; Münk, C. The ISG15-Protease USP18 Is a Pleiotropic Enhancer of HIV-1 Replication. Viruses 2024, 16, 485. https://doi.org/10.3390/v16040485
Lin C, Kuffour EO, Li T, Gertzen CGW, Kaiser J, Luedde T, König R, Gohlke H, Münk C. The ISG15-Protease USP18 Is a Pleiotropic Enhancer of HIV-1 Replication. Viruses. 2024; 16(4):485. https://doi.org/10.3390/v16040485
Chicago/Turabian StyleLin, Chaohui, Edmund Osei Kuffour, Taolan Li, Christoph G. W. Gertzen, Jesko Kaiser, Tom Luedde, Renate König, Holger Gohlke, and Carsten Münk. 2024. "The ISG15-Protease USP18 Is a Pleiotropic Enhancer of HIV-1 Replication" Viruses 16, no. 4: 485. https://doi.org/10.3390/v16040485
APA StyleLin, C., Kuffour, E. O., Li, T., Gertzen, C. G. W., Kaiser, J., Luedde, T., König, R., Gohlke, H., & Münk, C. (2024). The ISG15-Protease USP18 Is a Pleiotropic Enhancer of HIV-1 Replication. Viruses, 16(4), 485. https://doi.org/10.3390/v16040485







