HIV Replication Under High-Level Cabotegravir Is Associated with the Appearance of 3′-PPT Mutations, Circular DNA Transcription and Recombination
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Cultures Under Cabotegravir Treatment
2.2. HIV Genome Amplification
2.3. Sequencing of HIV Integrase Gene and LTRs
2.4. Strategies to Detect Integrated HIV
2.5. In Vitro Culture Fitness Competition in the Absence of CAB
3. Results
3.1. Assessment of In Vitro Int Resistance with RAL and CAB Treatment
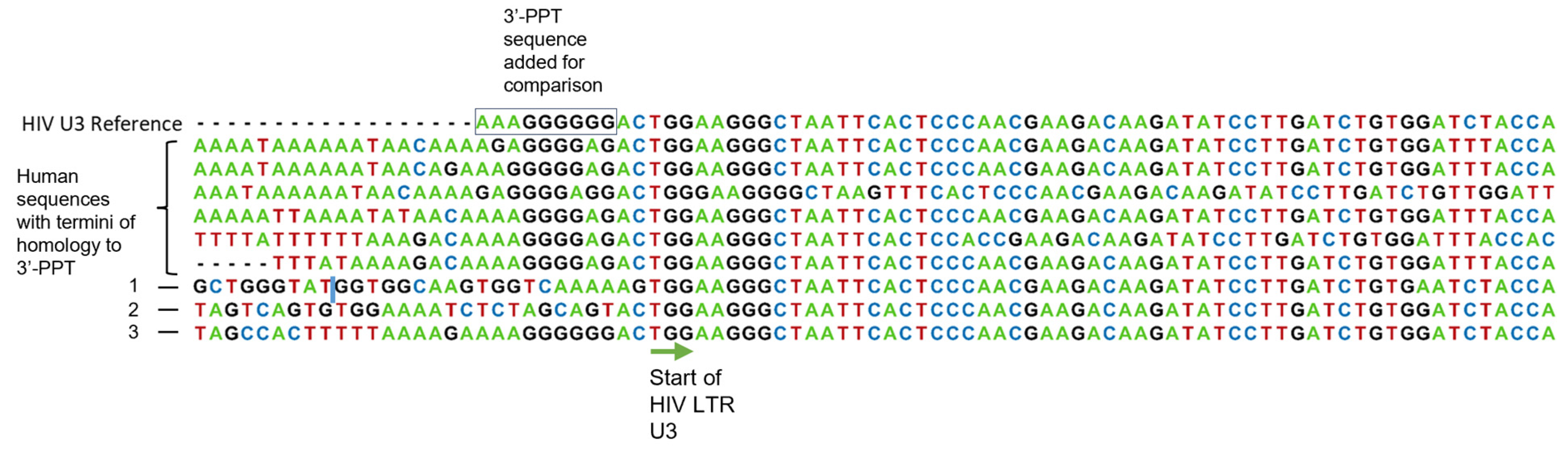
3.2. Appearance of 3′-PPT Mutations In Vitro Under Steady-State Therapeutic Levels of CAB
3.2.1. HIV RNA
3.2.2. HIV DNA
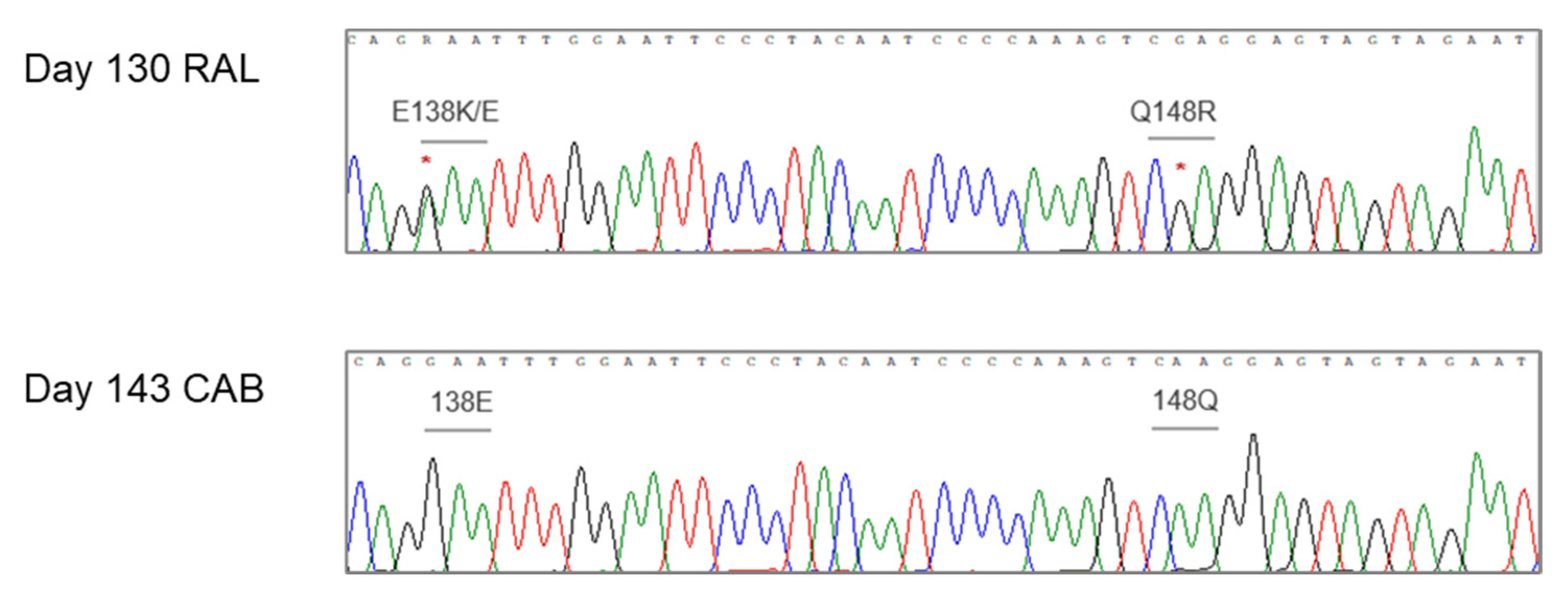
3.3. Evidence of the Origin of 3′-PPT Mutations
3.4. Recombination of 2-LTR Circles with Human DNA
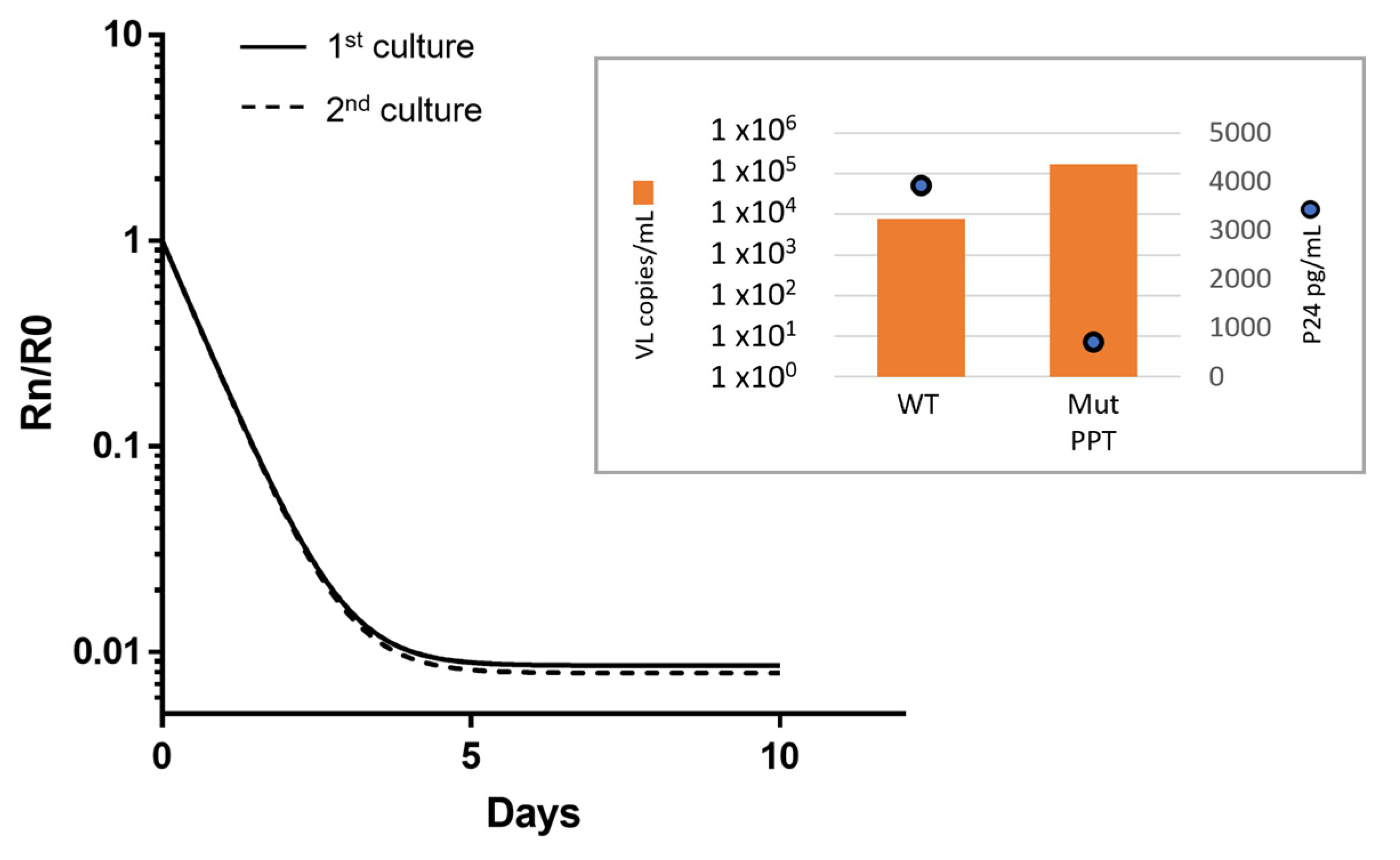
3.5. Fitness of the CAB-Selected 3′-PPT Mutant Virus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Craigie, R.; Bushman, F.D. HIV DNA integration. Cold Spring Harb. Perspect. Med. 2012, 2, a006890. [Google Scholar] [CrossRef] [PubMed]
- Passos, D.O.; Li, M.; Yang, R.; Rebensburg, S.V.; Ghirlando, R.; Jeon, Y.; Shkriabai, N.; Kvaratskhelia, M.; Craigie, R.; Lyumkis, D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 2017, 355, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Passos, D.O.; Li, M.; Jóźwik, I.K.; Zhao, X.Z.; Santos-Martins, D.; Yang, R.; Smith, S.J.; Jeon, Y.; Forli, S.; Hughes, S.H.; et al. Structural basis for strand-transfer inhibitor binding to HIV intasomes. Science 2020, 367, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.N.; Cherepanov, P. Retroviral intasomes arising. Curr. Opin. Struct. Biol. 2017, 47, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Mizuuchi, K.; Craigie, R. HIV-1 DNA integration: Mecshanism of viral DNA cleavage and DNA strand transfer. Cell 1991, 67, 1211–1221. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Available online: https://www.fda.gov/drugs/human-immunodeficiency-virus-hiv/fda-approves-cabenuva-and-vocabria-treatment-hiv-1-infection (accessed on 1 August 2024).
- US Food and Drug Administration. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-injectable-treatment-hiv-pre-exposure-prevention (accessed on 1 August 2024).
- Malet, I.; Subra, F.; Charpentier, C.; Collin, G.; Descamps, D.; Calvez, V.; Marcelin, A.G.; Delelis, O. Mutations Located outside the Integrase Gene Can Confer Resistance to HIV-1 Integrase Strand Transfer Inhibitors. mBio 2017, 8, e00922-17. [Google Scholar] [CrossRef]
- Wijting, I.E.A.; Lungu, C.; Rijnders, B.J.A.; van der Ende, M.E.; Pham, H.T.; Mesplede, T.; Pas, S.D.; Voermans, J.J.C.; Schuurman, R.; van de Vijver, D.A.M.C.; et al. HIV-1 Resistance Dynamics in Patients with Virologic Failure to Dolutegravir Maintenance Monotherapy. J. Infect. Dis. 2018, 218, 688–697. [Google Scholar] [CrossRef]
- Hassounah, S.A.; Alikhani, A.; Oliveira, M.; Bharaj, S.; Ibanescu, R.I.; Osman, N.; Xu, H.T.; Brenner, B.G.; Mesplède, T.; Wainberg, M.A. Antiviral Activity of Bictegravir and Cabotegravir against Integrase Inhibitor-Resistant SIVmac239 and HIV-1. Antimicrob. Agents Chemother. 2017, 61, e01695-17. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Integrated Review. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2021/212887Orig1s000,212888Orig1s000IntegratedR.pdf (accessed on 1 August 2024).
- Whitcomb, J.M.; Kumar, R.; Hughes, S.H. Sequence of the circle junction of human immunodeficiency virus type 1: Implications for reverse transcription and integration. J. Virol. 1990, 64, 4903–4906, Erratum in J. Virol. 1990, 64, 6360. [Google Scholar] [CrossRef]
- Holland, J.J.; de la Torre, J.C.; Clarke, D.K.; Duarte, E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 1991, 65, 2960–2967. [Google Scholar] [CrossRef]
- Cong, M.E.; Heneine, W.; García-Lerma, J.G. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 2007, 81, 3037–3041. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jurriaans, S.; de Ronde, A.; Dekker, J.; Goudsmit, J.; Cornelissen, M. Analysis of human immunodeficiency virus type 1 LTR-LTR junctions in peripheral blood mononuclear cells of infected individuals. J. Gen. Virol. 1992, 73, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Delelis, O.; Parissi, V.; Leh, H.; Mbemba, G.; Petit, C.; Sonigo, P.; Deprez, E.; Mouscadet, J.F. Efficient and specific internal cleavage of a retroviral palindromic DNA sequence by tetrameric HIV-1 integrase. PLoS ONE 2007, 2, e608. [Google Scholar] [CrossRef] [PubMed]
- Delelis, O.; Carayon, K.; Saïb, A.; Deprez, E.; Mouscadet, J.F. Integrase and integration: Biochemical activities of HIV-1 integrase. Retrovirology 2008, 5, 114. [Google Scholar] [CrossRef]
- Dekker, J.G.; Klaver, B.; Berkhout, B.; Das, A.T. Reverse Transcription of HIV-1 2-LTR circle transcripts does frequently cause 3′-polypurine tract mutations implicated in dolutegravir resistance. J. Virol. 2024, 98, e0033524. [Google Scholar] [CrossRef]
- Kirk, P.D.; Huvet, M.; Melamed, A.; Maertens, G.N.; Bangham, C.R. Retroviruses integrate into a shared, non-palindromic DNA motif. Nat. Microbiol. 2016, 2, 16212. [Google Scholar] [CrossRef]
- Julias, J.G.; McWilliams, M.J.; Sarafianos, S.G.; Alvord, W.G.; Arnold, E.; Hughes, S.H. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 2004, 78, 13315–13324. [Google Scholar] [CrossRef]
- McWilliams, M.J.; Julias, J.G.; Hughes, S.H. Mutations in the human immunodeficiency virus type 1 polypurine tract (PPT) reduce the rate of PPT cleavage and plus-strand DNA synthesis. J. Virol. 2008, 82, 5104–5108. [Google Scholar] [CrossRef]
- Richetta, C.; Subra, F.; Malet, I.; Leh, H.; Charpentier, C.; Corona, A.; Collin, G.; Descamps, D.; Deprez, E.; Parissi, V.; et al. Mutations in the 3′-PPT Lead to HIV-1 Replication without Integration. J. Virol. 2022, 96, e0067622. [Google Scholar] [CrossRef]
- Dekker, J.G.; Klaver, B.; Berkhout, B.; Das, A.T. HIV-1 3′-Polypurine Tract Mutations Confer Dolutegravir Resistance by Switching to an Integration-Independent Replication Mechanism via 1-LTR Circles. J. Virol. 2023, 97, e0036123. [Google Scholar] [CrossRef]
- Malbec, M.; Sourisseau, M.; Guivel-Benhassine, F.; Porrot, F.; Blanchet, F.; Schwartz, O.; Casartelli, N. HIV-1 Nef promotes the localization of Gag to the cell membrane and facilitates viral cell-to-cell transfer. Retrovirology 2013, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Richetta, C.; Thierry, S.; Thierry, E.; Lesbats, P.; Lapaillerie, D.; Munir, S.; Subra, F.; Leh, H.; Deprez, E.; Parissi, V.; et al. Two-long terminal repeat (LTR) DNA circles are a substrate for HIV-1 integrase. J. Biol. Chem. 2019, 294, 8286–8295. [Google Scholar] [CrossRef] [PubMed]
- Post, K.; Kankia, B.; Gopalakrishnan, S.; Yang, V.; Cramer, E.; Saladores, P.; Gorelick, R.J.; Guo, J.; Musier-Forsyth, K.; Levin, J.G. Fidelity of plus-strand priming requires the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2009, 37, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Else, L.J.; Taylor, S.; Back, D.J.; Khoo, S.H. Pharmacokinetics of antiretroviral drugs in anatomical sanctuary sites: The male and female genital tract. Antivir. Ther. 2011, 16, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Greener, B.N.; Patterson, K.B.; Prince, H.M.; Sykes, C.S.; Adams, J.L.; Dumond, J.B.; Shaheen, N.J.; Madanick, R.D.; Dellon, E.S.; Cohen, M.S.; et al. Dolutegravir pharmacokinetics in the genital tract and colorectum of HIV-negative men after single and multiple dosing. J. Acquir. Immune Defic. Syndr. 2013, 64, 39–44. [Google Scholar] [CrossRef]
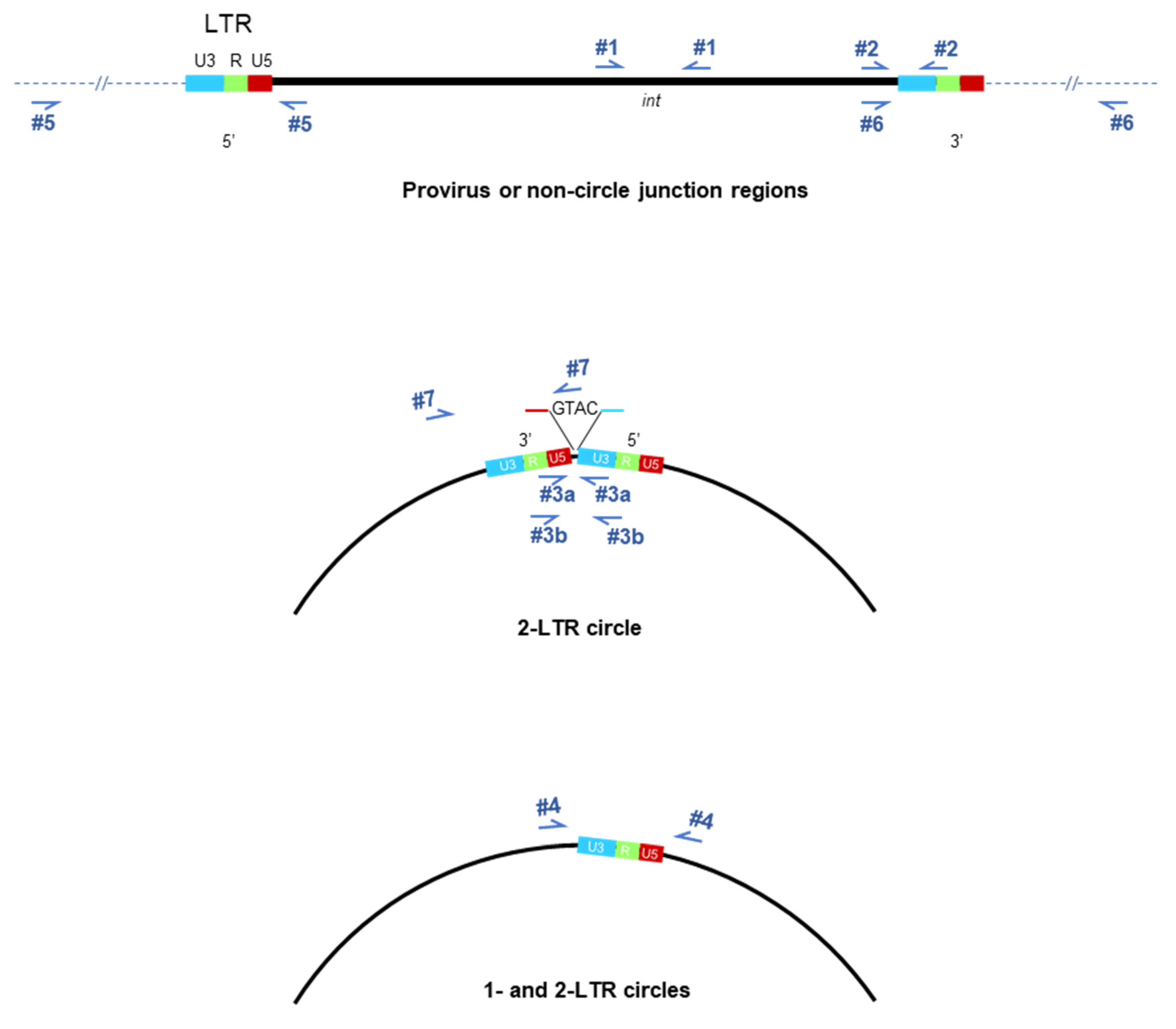

| (RT)-PCR Primer Pair | Genomic Region | Oligonucleotide Sequence 5′-3′ | Nt Positions in HIV-1IIIB Genomic DNA |
|---|---|---|---|
| 1 | int | F: TCTAGCTTTGCAGGATTCGG R: CAGTCTCTTTCTCCTGTATG | 4034–4053 5293–5312 |
| 2 | Nef-3′LTR R | F: CTAACGCTGATTGTGCCTGGC R: TGTACAGCCAAAAAGCAGCTGC | 8981–9001 9550–9570 |
| 3a | 2-LTR cross junction (short) | F: ATCTGAGCCTGGGAGCTCTC R: GTACTAGCTTGTAGCACCATC | 9593–9612 130–150 |
| 3b | 2-LTR cross junction (long) | F:TCAAGTAGTGTGTGCCCGTCTGT R: GCAGCTGCTTATATGTAGGATCTGAG | 9667–9689 414–439 |
| 4 | 1 and 2 LTR circle | F:CTAACGCTGATTGTGCCTGGC R:TAATAYTGACGCTCTCGCACCCAT | 8981–9001 790–813 |
| 5 | HuAluF HuAluR To gag | CTCCCAAAGTGCTGGGATTACA TGTAATCCCAGCACTTTGGGAG TAATAYTGACGCTCTCGCACCCAT | N/A N/A 790–813 |
| 6 | HuAluF HuAluR To nef | CTCCCAAAGTGCTGGGATTACA TGTAATCCCAGCACTTTGGGAG CTAACGCTGATTGTGCCTGGC | N/A N/A 8981–9001 |
| 7 | Nef-2-LTR junction | F:CTAACGCTGATTGTGCCTGGC R:GAGTGAATTAGCACTTCCAGTAC | 8981–9001 Junction (−2)–19 |
| 8 | Viral load | F:TGCTTAAGCCTCAATAAAGCTTGCCTTGA R:TCTGAGGGATCTCTAGTTACCAG | 515–543 581–603 |
| (A) AGCAGT variant | ||||||
| Culture 1 | Culture 2 | |||||
| Day | Nef-U3 (#2) | Nef-gag (#3a) | Nef-junct * (#7) | Nef-U3 (#2) | Nef-gag (#3a) | Nef-junct * (#7) |
| 14 | NS | NS | NS | 8 | <2% | NS |
| 30 | NS | NS | NS | 26.6 | <2% | NS |
| 98 | 10.1 | 34.2 | 13.9 | 62.3 | 75.6 | 52.9 |
| 112 | 5.9 | 13.3 | 1.7 | 78 | 66 | 55.7 |
| 120 | 3.1 | 11.2 | 3.3 | 53.1 | 50.9 | 35.6 |
| 127 | 3.5 | 9.3 | 1.7 | 42.7 | 51.4 | 24.2 |
| (B) GGAGCA variant | ||||||
| Culture 1 | ||||||
| Day | Nef-U3 (#2) | Nef-gag (#3a) | Nef-junct * (#7) | |||
| 98 | 78.2 | 51.9 | 13 | |||
| 112 | 90 | 81.6 | 17 | |||
| 120 | 93.7 | 84.9 | 27 | |||
| 127 | 93.8 | 82.6 | 16.5 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Lipscomb, J.T.; Tino, A.S.; Cong, M.-e.; Ruone, S.; Bentz, M.L.; Sheth, M.; Garcia-Lerma, G.; Johnson, J.A. HIV Replication Under High-Level Cabotegravir Is Associated with the Appearance of 3′-PPT Mutations, Circular DNA Transcription and Recombination. Viruses 2024, 16, 1874. https://doi.org/10.3390/v16121874
Wei X, Lipscomb JT, Tino AS, Cong M-e, Ruone S, Bentz ML, Sheth M, Garcia-Lerma G, Johnson JA. HIV Replication Under High-Level Cabotegravir Is Associated with the Appearance of 3′-PPT Mutations, Circular DNA Transcription and Recombination. Viruses. 2024; 16(12):1874. https://doi.org/10.3390/v16121874
Chicago/Turabian StyleWei, Xierong, Jonathan T. Lipscomb, Ariana Santos Tino, Mian-er Cong, Susan Ruone, Meghan L. Bentz, Mili Sheth, Gerardo Garcia-Lerma, and Jeffrey A. Johnson. 2024. "HIV Replication Under High-Level Cabotegravir Is Associated with the Appearance of 3′-PPT Mutations, Circular DNA Transcription and Recombination" Viruses 16, no. 12: 1874. https://doi.org/10.3390/v16121874
APA StyleWei, X., Lipscomb, J. T., Tino, A. S., Cong, M.-e., Ruone, S., Bentz, M. L., Sheth, M., Garcia-Lerma, G., & Johnson, J. A. (2024). HIV Replication Under High-Level Cabotegravir Is Associated with the Appearance of 3′-PPT Mutations, Circular DNA Transcription and Recombination. Viruses, 16(12), 1874. https://doi.org/10.3390/v16121874







