Generation and Characterization of a Replication-Competent Human Adenovirus Type 55 Encoding EGFP
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Cells
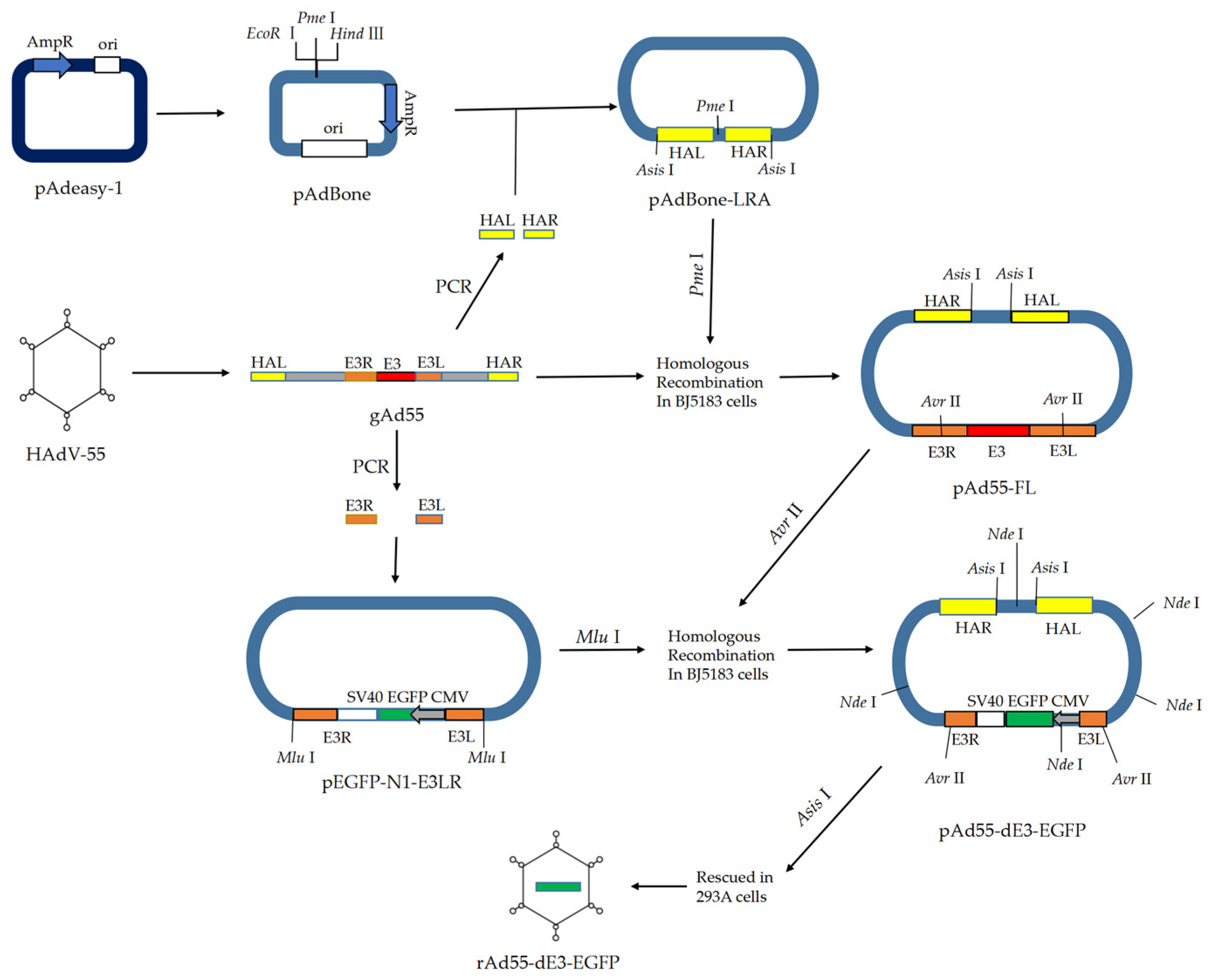
2.2. Construction of Full-Length Infectious cDNA Clone of HAdV-55
2.3. Constructure of the E3-Defective Replication-Competent EGFP Expression Vector pAd55-dE3-EGFP
2.4. Virus Rescue
2.5. Virus Growth Kinetics in A549 Cells
2.6. Stability Testing of rAd55-dE3-EGFP Containing eGFP and Viral Genome in A549
2.7. Indirect Immunofluorescence and Fluorescence Focus Assay
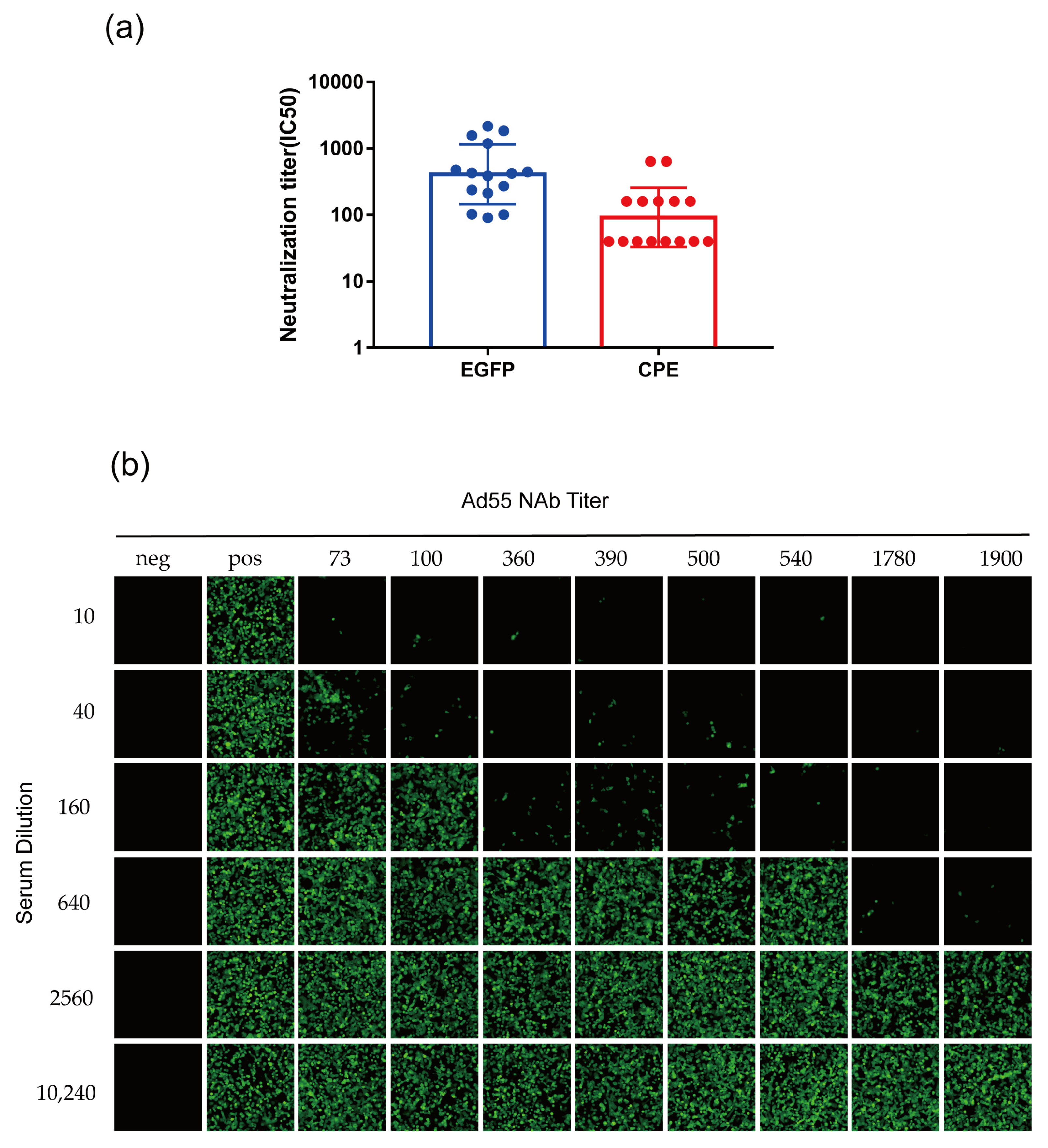
2.8. Microneutralization Assay
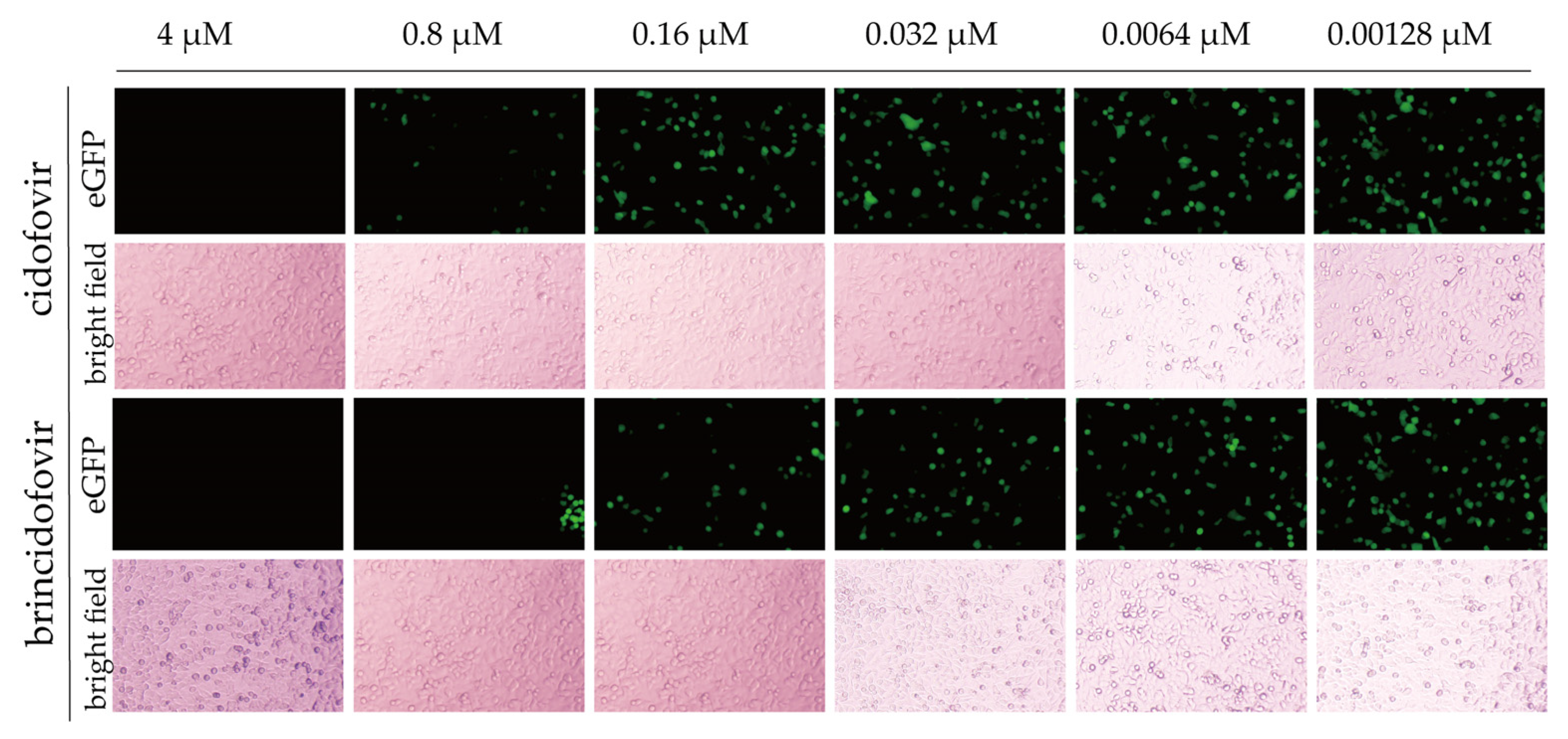
2.9. Antiviral Drug Screening
2.10. Electron Microscopic Observation
2.11. Mouse Experiment
2.12. Statistical Analysis
3. Results
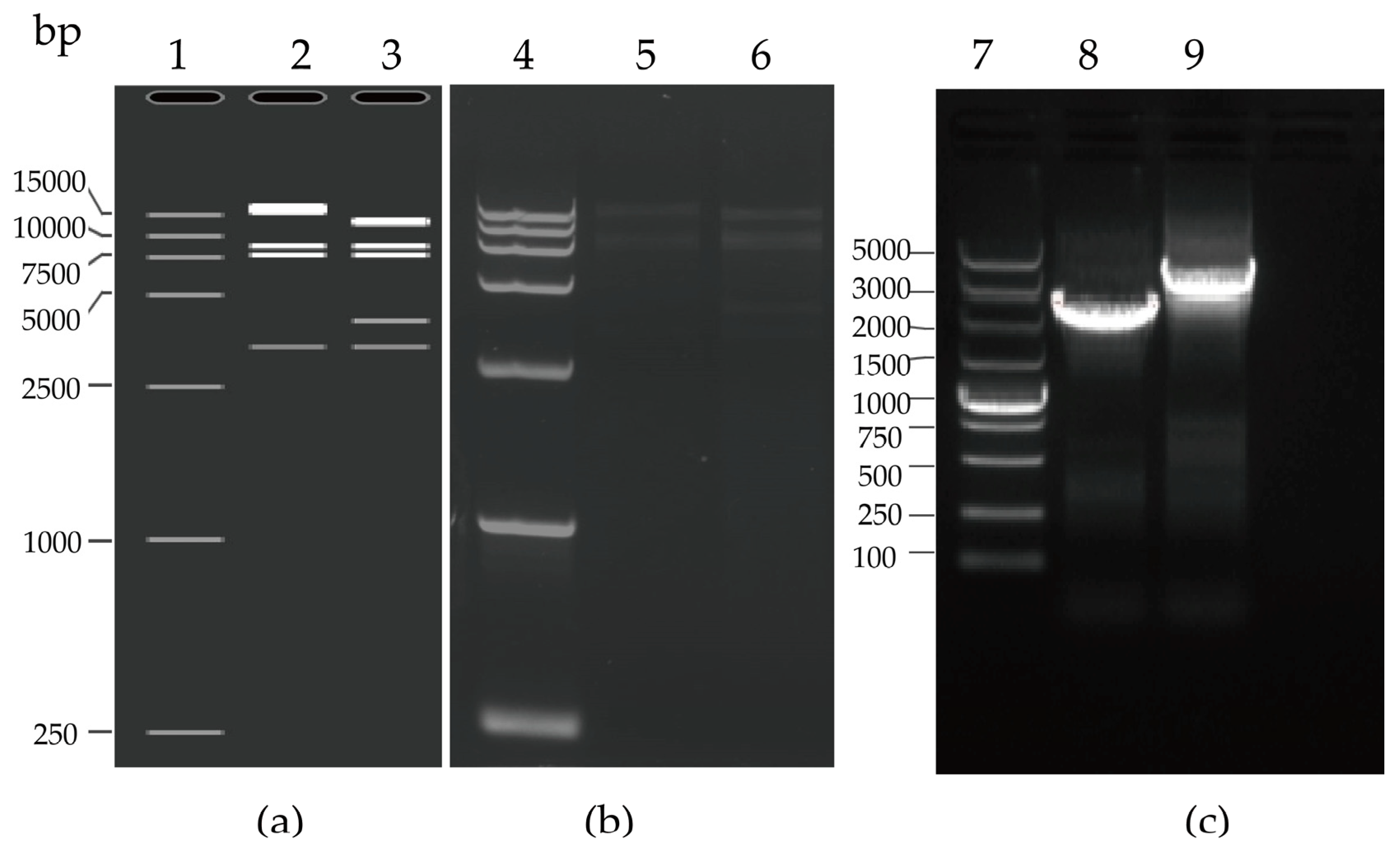
3.1. Generation of the Infectious Clones pAd55-FL/pAd55-dE3-EGFP
3.2. rAd55 and rAd55-dE3-EGFP Rescue
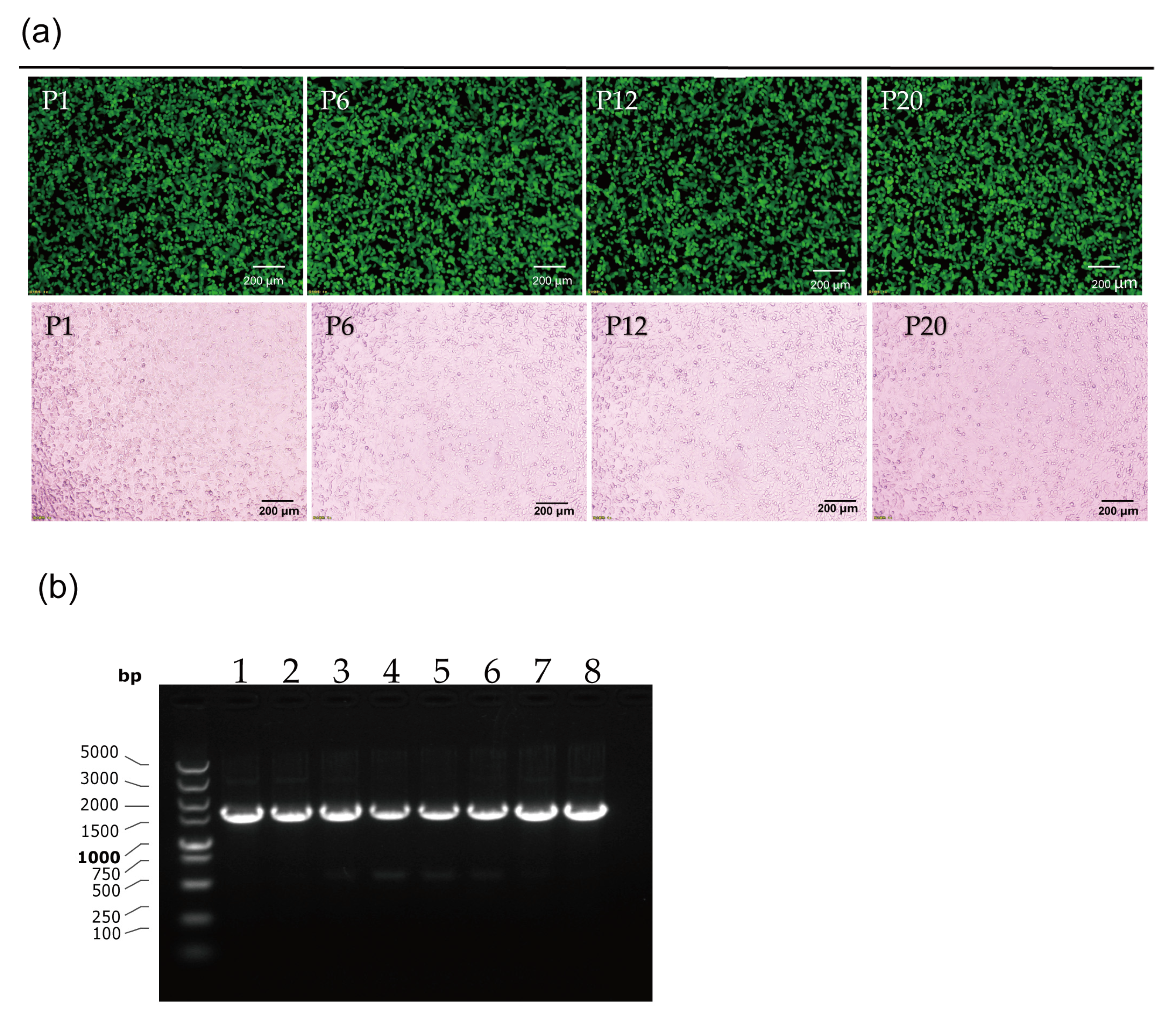
3.3. Stability Testing of rAd55-dE3-EGFP
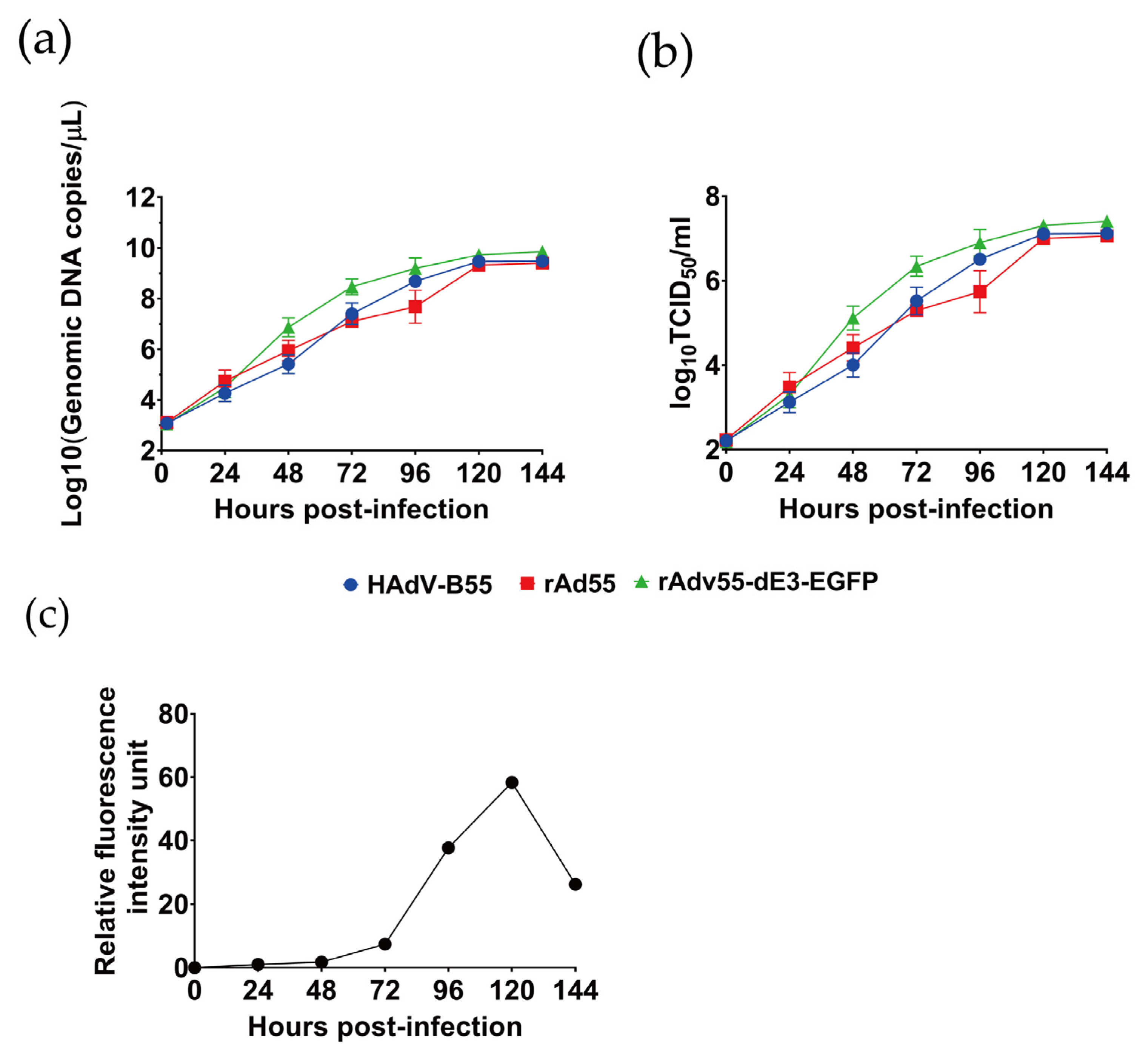
3.4. Growth Kinetics Characteristics of rAd55 and rAd55-dE3-EGFP
3.5. Indirect Immunofluorescence Assay
3.6. Development of rAd55-dE3-EGFP-Based MN Assays
3.7. Antiviral Screening Experiment
3.8. Electron Microscopic Observation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P., 3rd; Kajon, A.E. Adenovirus: Epidemiology, global spread of novel types, and approach to treatment. Semin. Respir. Crit. Care Med. 2021, 42, 800–821. [Google Scholar] [CrossRef] [PubMed]
- Benkő, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kaján, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV Virus Taxonomy Profile: Adenoviridae 2022. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef] [PubMed]
- Pscheidt, V.M.; Gregianini, T.S.; Martins, L.G.; Veiga, A. Epidemiology of human adenovirus associated with respiratory infection in southern Brazil. Rev. Med. Virol. 2021, 31, e2189. [Google Scholar] [CrossRef]
- Wang, X.; Wang, D.; Umar, S.; Qin, S.; Ling, Q.; Gray, G.C.; Liu, Y. Molecular typing of human adenoviruses among hospitalized patients with respiratory tract infections in a tertiary Hospital in Guangzhou, China between 2017 and 2019. BMC Infect. Dis. 2021, 21, 748. [Google Scholar] [CrossRef]
- Lynch, J.P., 3rd; Kajon, A.E. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention. Semin. Respir. Crit. Care Med. 2016, 37, 586–602. [Google Scholar] [CrossRef]
- Mao, N.Y.; Zhu, Z.; Zhang, Y.; Xu, W.B. Current status of human adenovirus infection in China. World J. Pediatr. 2022, 18, 533–537. [Google Scholar] [CrossRef]
- Liu, J.; Nian, Q.G.; Zhang, Y.; Xu, L.J.; Hu, Y.; Li, J.; Deng, Y.Q.; Zhu, S.Y.; Wu, X.Y.; Qin, E.D.; et al. In vitro characterization of human adenovirus type 55 in comparison with its parental adenoviruses, types 11 and 14. PLoS ONE 2014, 9, e100665. [Google Scholar] [CrossRef]
- Liu, H.; Li, Q.; Xiang, Y.; Li, H.; Liu, K.; Du, X.; Yang, C.; Liu, H.; Shi, M.; Hu, X.; et al. An outbreak of acute respiratory disease caused by HAdV-55 in Beijing, China, 2020. J. Med. Virol. 2022, 94, 6111–6115. [Google Scholar] [CrossRef]
- Cao, B.; Huang, G.H.; Pu, Z.H.; Qu, J.X.; Yu, X.M.; Zhu, Z.; Dong, J.P.; Gao, Y.; Zhang, Y.X.; Li, X.H.; et al. Emergence of community-acquired adenovirus type 55 as a cause of community-onset pneumonia. Chest 2014, 145, 79–86. [Google Scholar] [CrossRef]
- Walsh, M.P.; Seto, J.; Jones, M.S.; Chodosh, J.; Xu, W.; Seto, D. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 2010, 48, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhao, X.; Tang, F.; Huang, D.; Pei, G.; Zhang, X.; Jiang, B.; Lu, Q.; Liu, W.; Tong, Y. Outbreaks of acute respiratory disease associated with human adenovirus infection in closed camps, China, December 2011-March 2014. China CDC Wkly. 2021, 3, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.B.; Tong, Y.G.; Wo, Y.; Wang, H.Y.; Liu, E.M.; Gray, G.C.; Liu, W.; Cao, W.C. Epidemiology of human adenovirus and molecular characterization of human adenovirus 55 in China, 2009–2012. Influenza Other Respir. Viruses 2014, 8, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-C.; Xu, Q.; Li, T.-T.; Wang, T.; Jiang, B.-G.; Lv, C.-L.; Zhang, X.-A.; Liu, W.; Fang, L.-Q. Prevalence of human infection with respiratory adenovirus in China: A systematic review and meta-analysis. PLOS Negl. Trop. Dis. 2023, 17, e0011151. [Google Scholar] [CrossRef] [PubMed]
- Binder, A.M.; Biggs, H.M.; Haynes, A.K.; Chommanard, C.; Lu, X.; Erdman, D.D.; Watson, J.T.; Gerber, S.I. Human adenovirus surveillance—United States, 2003–2016. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 1039–1042. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, Y.; Wu, H.; Wang, Z.; Zhan, Y.; Feng, X.; Geng, R.; Wu, Y.; Kong, W.; Yu, X. Seroprevalence of neutralizing antibodies to human adenovirus type 5 in healthy adults in China. J. Med. Virol. 2012, 84, 1408–1414. [Google Scholar] [CrossRef]
- Malasig, M.D.; Goswami, P.R.; Crawford-Miksza, L.K.; Schnurr, D.P.; Gray, G.C. Simplified microneutralization test for serotyping adenovirus isolates. J. Clin. Microbiol. 2001, 39, 2984–2986. [Google Scholar] [CrossRef]
- Crawford-Miksza, L.K.; Schnurr, D.P. Quantitative colorimetric microneutralization assay for characterization of adenoviruses. J. Clin. Microbiol. 1994, 32, 2331–2334. [Google Scholar] [CrossRef]
- Pauwels, R.; Balzarini, J.; Baba, M.; Snoeck, R.; Schols, D.; Herdewijn, P.; Desmyter, J.; De Clercq, E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 1988, 20, 309–321. [Google Scholar] [CrossRef]
- Wang, B.; Li, J.; Wu, S.; Wang, Y.; Chen, Y.; Zhai, Y.; Song, X.; Zhao, Z.; Zhang, Z.; Zhang, J.; et al. A seroepidemiological survey of adenovirus type 7 circulation among healthy adults in China and in Sierra Leone, West Africa. Front. Public Health 2023, 11, 1095343. [Google Scholar] [CrossRef]
- Mennechet, F.J.D.; Paris, O.; Ouoba, A.R.; Salazar Arenas, S.; Sirima, S.B.; Takoudjou Dzomo, G.R.; Diarra, A.; Traore, I.T.; Kania, D.; Eichholz, K.; et al. A review of 65 years of human adenovirus seroprevalence. Expert Rev. Vaccines 2019, 18, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Xiao, L.; Zheng, X.; Wang, J.; Shu, T.; Feng, Y.; Liu, X.; Su, W.; Wang, Q.; Li, C.; et al. Seroprevalence of neutralizing antibodies to human adenovirus type 4 and 7 in healthy populations from southern China. Front. Microbiol. 2018, 9, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, J.; Wu, S.; Chen, Y.; Zhang, Z.; Zhai, Y.; Guo, Q.; Zhang, J.; Song, X.; Zhao, Z.; et al. Seroepidemiological investigation of HAdV-4 infection among healthy adults in China and in Sierra Leone, West Africa. Emerg. Microbes Infect. 2018, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Rong, X.; Feng, Y.; Sun, X.; Li, L.; Wang, Q.; Wang, M.; Liu, W.; Li, C.; Yang, Y.; et al. Seroprevalence of neutralizing antibodies against adenovirus type 14 and 55 in healthy adults in Southern China. Emerg. Microbes Infect. 2017, 6, e43. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, X.; Liu, W.; Xian, Y.; Chen, W.; Zhou, R. A sensitive and high-throughput flow cytometry-based assay for measuring antibody neutralization of human adenovirus type 3. Virol. Sin. 2021, 36, 537–544. [Google Scholar] [CrossRef]
- Chemaly, R.F.; Hill, J.A.; Voigt, S.; Peggs, K.S. In vitro comparison of currently available and investigational antiviral agents against pathogenic human double-stranded DNA viruses: A systematic literature review. Antivir. Res 2019, 163, 50–58. [Google Scholar] [CrossRef]
- Kajon, A.E.; Dickson, L.M.; Metzgar, D.; Houng, H.S.; Lee, V.; Tan, B.H. Outbreak of febrile respiratory illness associated with adenovirus 11a infection in a Singapore military training cAMP. J. Clin. Microbiol. 2010, 48, 1438–1441. [Google Scholar] [CrossRef]
- Kajon, A.E.; Lamson, D.M.; St George, K. Emergence and re-emergence of respiratory adenoviruses in the United States. Curr. Opin. Virol. 2019, 34, 63–69. [Google Scholar] [CrossRef]
- Chmielewicz, B.; Benzler, J.; Pauli, G.; Krause, G.; Bergmann, F.; Schweiger, B. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 2005, 77, 232–237. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, J.; Dehghan, S.; Sridhar, S.; Lau, S.K.P.; Ou, J.; Woo, P.C.Y.; Zhang, Q.; Seto, D. A survey of recent adenoviral respiratory pathogens in Hong Kong reveals emergent and recombinant human adenovirus type 4 (HAdV-E4) circulating in civilian populations. Viruses 2019, 11, 129. [Google Scholar] [CrossRef]
- Syyam, A.; Nawaz, A.; Ijaz, A.; Sajjad, U.; Fazil, A.; Irfan, S.; Muzaffar, A.; Shahid, M.; Idrees, M.; Malik, K.; et al. Adenovirus vector system: Construction, history and therapeutic applications. BioTechniques 2022, 73, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mese, K.; Schellhorn, S.; Bahlmann, N.; Mach, N.; Bunz, O.; Dhingra, A.; Hage, E.; Lafon, M.E.; Wodrich, H.; et al. High-throughput cloning and characterization of emerging adenovirus yypes 70, 73, 74, and 75. Int. J. Mol. Sci. 2020, 21, 6370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fu, J.; Ehrhardt, A. Novel vector construction based on alternative adenovirus types via homologous recombination. Hum. Gene Ther. Methods 2018, 29, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Deng, Z.L.; Luo, X.; Tang, N.; Song, W.X.; Chen, J.; Sharff, K.A.; Luu, H.H.; Haydon, R.C.; Kinzler, K.W.; et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007, 2, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.; Resch, M.D.; Mazboudi, R.; Mulhall Maasz, H.; Galarza, J.M. Novel and efficient method for the reconstruction of adenoviruses through isothermal assembly and its potential applications. Front. Med. Technol. 2023, 5, 1095198. [Google Scholar] [CrossRef]
- Zou, X.; Zhu, Y.; Li, C.; Duan, Y.; Zhang, L.; Guo, X.; Hou, W.; Xie, Z.; Lu, Z. Construction of adenoviral vectors using DNA assembly technology. J. Vis. Exp. JoVE 2022, 184. [Google Scholar] [CrossRef]
- Guo, X.; Mei, L.; Yan, B.; Zou, X.; Hung, T.; Lu, Z. Site-directed modification of adenoviral vector with combined DNA assembly and restriction-ligation cloning. J. Biotechnol. 2020, 307, 193–201. [Google Scholar] [CrossRef]
- Ni, N.; Deng, F.; He, F.; Wang, H.; Shi, D.; Liao, J.; Zou, Y.; Wang, H.; Zhao, P.; Hu, X.; et al. A one-step construction of adenovirus (OSCA) system using the Gibson DNA Assembly technology. Mol. Ther. Oncolytics 2021, 23, 602–611. [Google Scholar] [CrossRef]
- Reddy, P.S.; Ganesh, S.; Hawkins, L.; Idamakanti, N. Generation of recombinant adenovirus using the Escherichia coli BJ5183 recombination system. Methods Mol. Med. 2007, 130, 61–68. [Google Scholar] [CrossRef]
- Haut, L.H.; Gill, A.L.; Kurupati, R.K.; Bian, A.; Li, Y.; Giles-Davis, W.; Xiang, Z.; Zhou, X.Y.; Ertl, H.C. A partial E3 deletion in replication-defective adenoviral vectors allows for stable expression of potentially toxic transgene products. Hum. Gene Ther. Methods 2016, 27, 187–196. [Google Scholar] [CrossRef]
- Robinson, M.; Ge, Y.; Ko, D.; Yendluri, S.; Laflamme, G.; Hawkins, L.; Jooss, K. Comparison of the E3 and L3 regions for arming oncolytic adenoviruses to achieve a high level of tumor-specific transgene expression. Cancer Gene Ther. 2008, 15, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kajon, A.E.; Xu, W.; Erdman, D.D. Sequence polymorphism in the E3 7.7K ORF of subspecies B1 human adenoviruses. Virus Res. 2005, 107, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Sprangers, M.C.; Lakhai, W.; Koudstaal, W.; Verhoeven, M.; Koel, B.F.; Vogels, R.; Goudsmit, J.; Havenga, M.J.; Kostense, S. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: Addressing preexisting immunity to vaccine and gene therapy vectors. J. Clin. Microbiol. 2003, 41, 5046–5052. [Google Scholar] [CrossRef] [PubMed]



| Plasmid | Primer | Sequence (5′-3′) # |
|---|---|---|
| pAd55 | P762-LARM-HindIII-AsiSI-F | gcgAAGCTTGCGATCGCcatcatcaataatataccttatag |
| P606-LARM-PmeI-R | ataGTTTAAACgtcttcatagcagtgcaaatcacag | |
| P607-RARM-PmeI-F | ataGTTTAAACcaccagtaatgtcatcaaagttgctg | |
| P763-RARM-EcoRI-AsiSI-R | ataGAATTCGCGATCGCcatcatcaataatataccttatag | |
| pEGFP-E3LR | E3L1000-F: | CGTATTACCGCCATGCATTAGTTATTAATACGCGTctggttagctgcgcagccggcatc |
| E3L1000-R: | CTAATGACCCCGTAATTGATTACTATTAATgcagtggtctaaatgtcgcagccgag | |
| E3R1000-F: | GTTTGTCCAAACTCATCAATGTATCTTAAGatgcggactaagagacctgctac | |
| E3R1000-R: | AATATTAACGCTTACAATTTACGCCTTAAGACGCGTgatgagacacaatcgcccgatcc | |
| PCR | E3D-Det-F2 | cacccctcgtcagactgttttgac |
| AVRII-R | cactggagtccatcatttgacag | |
| qPCR | qHAdV-UniF | ATGGCCACCCCATCGAT |
| qHAdV-UniR | ACTCAGGTACTCCGAAGCATCCT | |
| qHAdV-UniProbe | FAM-TGGGCATACATGCACATCGCCG-BHQ1 | |
| pAdBone0 | P617-AdBackbone-F-EcoRI | ataGAATTCTCGACCGATGCCCTTGAGAGCCTTC |
| P618-AdBackbone-R-HindIII: | gcgAAGCTTCAGGTGGCACTTTTCGGGGAAATG | |
| EGFP | EGFP-F | cgttacataacttacggtaaatgg |
| EGFP-R | taagatacattgatgagtttggacaaac |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Chen, Y.; Feng, Y.; Li, J.; Kang, X.; Zhang, S.; Li, Y.; Zhao, Z.; Yang, W.; Zhao, L.; et al. Generation and Characterization of a Replication-Competent Human Adenovirus Type 55 Encoding EGFP. Viruses 2023, 15, 1192. https://doi.org/10.3390/v15051192
Li W, Chen Y, Feng Y, Li J, Kang X, Zhang S, Li Y, Zhao Z, Yang W, Zhao L, et al. Generation and Characterization of a Replication-Competent Human Adenovirus Type 55 Encoding EGFP. Viruses. 2023; 15(5):1192. https://doi.org/10.3390/v15051192
Chicago/Turabian StyleLi, Wei, Yuehong Chen, Ye Feng, Jing Li, Xiaoping Kang, Sen Zhang, Yuchang Li, Zhiyan Zhao, Wenguang Yang, Lu Zhao, and et al. 2023. "Generation and Characterization of a Replication-Competent Human Adenovirus Type 55 Encoding EGFP" Viruses 15, no. 5: 1192. https://doi.org/10.3390/v15051192
APA StyleLi, W., Chen, Y., Feng, Y., Li, J., Kang, X., Zhang, S., Li, Y., Zhao, Z., Yang, W., Zhao, L., Wang, H., & Jiang, T. (2023). Generation and Characterization of a Replication-Competent Human Adenovirus Type 55 Encoding EGFP. Viruses, 15(5), 1192. https://doi.org/10.3390/v15051192





