In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Antiviral Molecule
2.3. Virus Titration
2.4. Maximum Non-Cytotoxic Dose
2.5. Cell Viability
2.6. Antiviral Assay
2.7. Plaque Reduction Assay
2.8. Time-of-Drug Addition Assays
2.8.1. Simultaneous Assay
2.8.2. Pre-Infection Assay
2.8.3. Virucidal Assay
2.9. Protein Extraction and Western Blot Analysis
2.10. Viral Genome Quantification
2.11. Statistical Analysis
3. Results
3.1. Cytotoxicity Assay to Identify the NDGA Maximum Non-Cytotoxic Dose
3.2. Antiviral Activity of NDGA
3.3. NDGA Mainly Acts at Early Stages of SARS-CoV-2 Infection
3.4. Western Blot Analysis
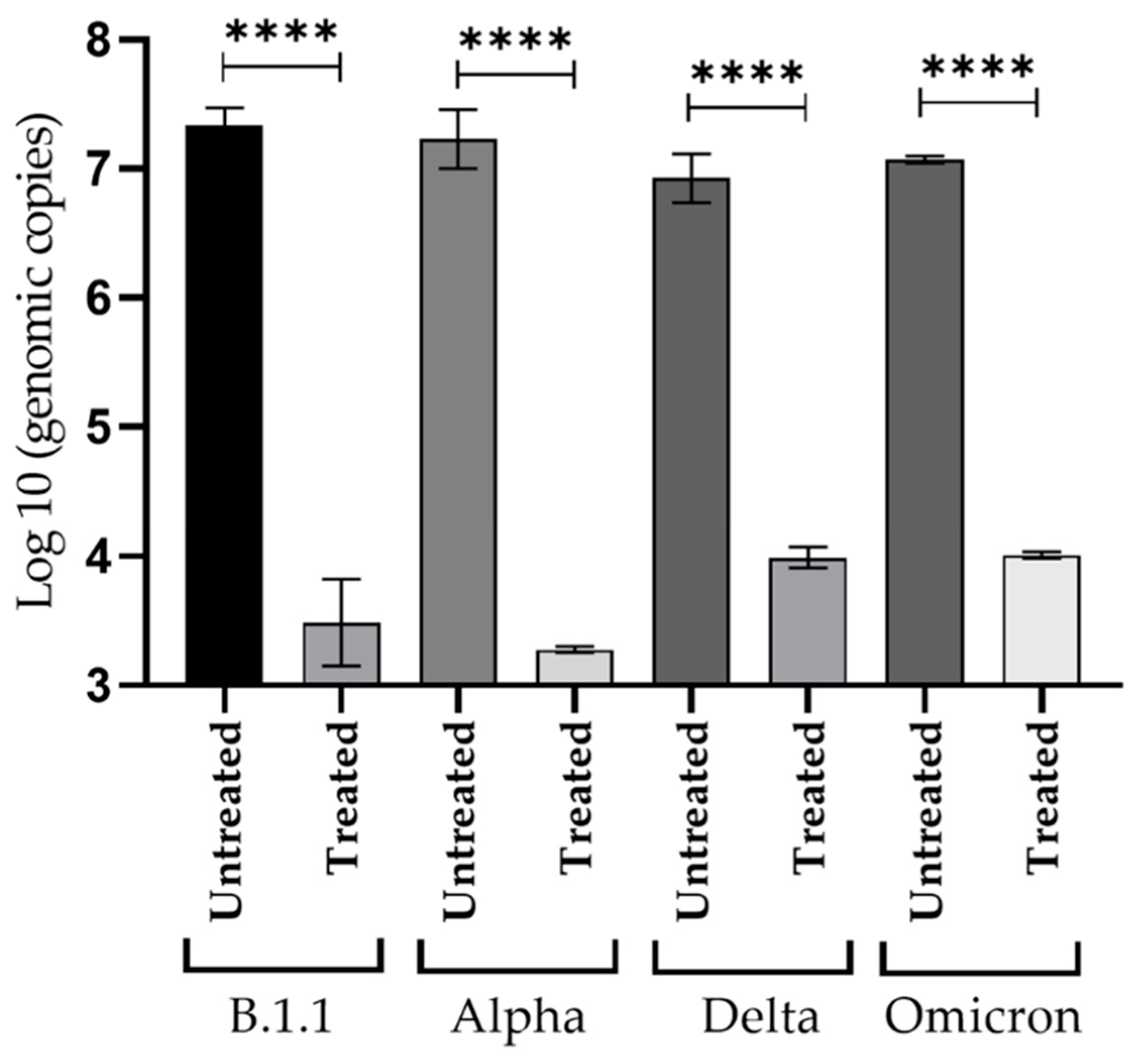
3.5. Inhibitory Effect of NDGA against Viral Replication of SARS-CoV-2 Variants
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-W.; Yuan, S.; Yuen, K.-S.; Fung, S.-Y.; Chan, C.-P.; Jin, D.-Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686. [Google Scholar] [CrossRef] [PubMed]
- Çalıca Utku, A.; Budak, G.; Karabay, O.; Güçlü, E.; Okan, H.D.; Vatan, A. Main symptoms in patients presenting in the COVID-19 period. Scott. Med. J. 2020, 65, 127–132. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Weng, L.-M.; Su, X.; Wang, X.-Q. Pain symptoms in patients with coronavirus disease (COVID-19): A literature review. J. Pain Res. 2021, 14, 147. [Google Scholar] [CrossRef]
- Ftiha, F.; Shalom, M.; Jradeh, H. Neurological symptoms due to Coronavirus disease 2019. Neurol. Int. 2020, 12, 15–18. [Google Scholar] [CrossRef]
- Ramachandran, P.; Onukogu, I.; Ghanta, S.; Gajendran, M.; Perisetti, A.; Goyal, H.; Aggarwal, A. Gastrointestinal symptoms and outcomes in hospitalized coronavirus disease 2019 patients. Dig. Dis. 2020, 38, 373–379. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Lamb, Y.N. Remdesivir: First approval. Drugs 2020, 80, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Molnupiravir: First approval. Drugs 2022, 82, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, C.; Roviello, G.N. Potential Anti-SARS-CoV-2 Molecular Strategies. Molecules 2023, 28, 2118. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Roviello, G.N. Exploring the Protective Effect of Food Drugs against Viral Diseases: Interaction of Functional Food Ingredients and SARS-CoV-2, Influenza Virus, and HSV. Life 2023, 13, 402. [Google Scholar] [CrossRef]
- Behl, T.; Rocchetti, G.; Chadha, S.; Zengin, G.; Bungau, S.; Kumar, A.; Mehta, V.; Uddin, M.S.; Khullar, G.; Setia, D. Phytochemicals from plant foods as potential source of antiviral agents: An overview. Pharmaceuticals 2021, 14, 381. [Google Scholar] [CrossRef]
- Garcia-Ruiz, D.; Villalobos-Sánchez, E.; Alam-Escamilla, D.; Elizondo-Quiroga, D. In vitro inhibition of SARS-CoV-2 Infection by dry algae powders. Sci. Rep. 2022, 12, 17101. [Google Scholar] [CrossRef]
- Savoia, D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef]
- Mishra, K.; Sharma, N.; Diwaker, D.; Ganju, L.; Singh, S. Plant derived antivirals: A potential source of drug development. J. Virol. Antivir. Res. 2013, 2, 2. [Google Scholar]
- Arif, T.; Bhosale, J.; Kumar, N.; Mandal, T.; Bendre, R.; Lavekar, G.; Dabur, R. Natural products–antifungal agents derived from plants. J. Asian Nat. Prod. Res. 2009, 11, 621–638. [Google Scholar] [CrossRef]
- Bashyal, B.; Li, L.; Bains, T.; Debnath, A.; LaBarbera, D.V. Larrea tridentata: A novel source for anti-parasitic agents active against Entamoeba histolytica, Giardia lamblia and Naegleria fowleri. PLoS Negl. Trop. Dis. 2017, 11, e0005832. [Google Scholar] [CrossRef]
- Morales-Ubaldo, A.L.; Rivero-Perez, N.; Avila-Ramos, F.; Aquino-Torres, E.; Prieto-Méndez, J.; Hetta, H.F.; El-Saber Batiha, G.; Zaragoza-Bastida, A. Bactericidal activity of Larrea tridentata hydroalcoholic extract against phytopathogenic bacteria. Agronomy 2021, 11, 957. [Google Scholar] [CrossRef]
- Skouta, R.; Morán-Santibañez, K.; Valenzuela, C.A.; Vasquez, A.H.; Fenelon, K. Assessing the antioxidant properties of Larrea tridentata extract as a potential molecular therapy against oxidative stress. Molecules 2018, 23, 1826. [Google Scholar] [CrossRef] [PubMed]
- Morán-Santibañez, K.; Vasquez, A.H.; Varela-Ramirez, A.; Henderson, V.; Sweeney, J.; Odero-Marah, V.; Fenelon, K.; Skouta, R. Larrea tridentata extract mitigates oxidative stress-induced cytotoxicity in human neuroblastoma SH-SY5Y cells. Antioxidants 2019, 8, 427. [Google Scholar] [CrossRef]
- Manda, G.; Rojo, A.I.; Martínez-Klimova, E.; Pedraza-Chaverri, J.; Cuadrado, A. Nordihydroguaiaretic acid: From herbal medicine to clinical development for cancer and chronic diseases. Front. Pharmacol. 2020, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Saldívar, R.H.L. Estado actual del conocimiento sobre las propiedades biocidas de la gobernadora [Larrea tridentata (DC) Coville]. Rev. Mex. Fitopatol. 2003, 21, 214–222. [Google Scholar]
- Hyder, P.W.; Fredrickson, E.; Estell, R.E.; Tellez, M.; Gibbens, R.P. Distribution and concentration of total phenolics, condensed tannins, and nordihydroguaiaretic acid (NDGA) in creosotebush (Larrea tridentata). Biochem. Syst. Ecol. 2002, 30, 905–912. [Google Scholar] [CrossRef]
- Merino-Ramos, T.; Jiménez de Oya, N.; Saiz, J.-C.; Martín-Acebes, M.A. Antiviral activity of nordihydroguaiaretic acid and its derivative tetra-O-methyl nordihydroguaiaretic acid against West Nile virus and Zika virus. Antimicrob. Agents Chemother. 2017, 61, e00376-17. [Google Scholar] [CrossRef]
- Martinez, F.; Mugas, M.L.; Aguilar, J.J.; Marioni, J.; Contigiani, M.S.; Montoya, S.C.N.; Konigheim, B.S. First report of antiviral activity of nordihydroguaiaretic acid against Fort Sherman virus (Orthobunyavirus). Antivir. Res. 2021, 187, 104976. [Google Scholar] [CrossRef]
- Soto-Acosta, R.; Bautista-Carbajal, P.; Syed, G.H.; Siddiqui, A.; Del Angel, R.M. Nordihydroguaiaretic acid (NDGA) inhibits replication and viral morphogenesis of dengue virus. Antivir. Res. 2014, 109, 132–140. [Google Scholar] [CrossRef]
- Uchide, N.; Ohyama, K.; Bessho, T.; Toyoda, H. Inhibition of influenza-virus-induced apoptosis in chorion cells of human fetal membranes by nordihydroguaiaretic Acid. Intervirology 2005, 48, 336–340. [Google Scholar] [CrossRef]
- Hwu, J.R.; Tseng, W.N.; Gnabre, J.; Giza, P.; Huang, R.C.C. Antiviral activities of methylated nordihydroguaiaretic acids. 1. Synthesis, structure identification, and inhibition of tat-regulated HIV transactivation. J. Med. Chem. 1998, 41, 2994–3000. [Google Scholar] [CrossRef] [PubMed]
- Park, R. Inhibition of the Herpes Simplex Virus Type I by Three NDGA Derivatives: Mal.4, M4N, and G4N. Ph.D. Thesis, The Johns Hopkins University, Baltimore, MD, USA, 2003. [Google Scholar]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85. [Google Scholar] [CrossRef] [PubMed]
- Janik, E.; Niemcewicz, M.; Podogrocki, M.; Majsterek, I.; Bijak, M. The emerging concern and interest SARS-CoV-2 variants. Pathogens 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Melo-Filho, C.C.; Bobrowski, T.; Martin, H.-J.; Sessions, Z.; Popov, K.I.; Moorman, N.J.; Baric, R.S.; Muratov, E.N.; Tropsha, A. Conserved coronavirus proteins as targets of broad-spectrum antivirals. Antivir. Res. 2022, 204, 105360. [Google Scholar] [CrossRef]
- Zhong, W.; Jiang, X.; Yang, X.; Feng, T.; Duan, Z.; Wang, W.; Sun, Z.; Chen, L.; Nie, X.; Zhu, C. The efficacy of paxlovid in elderly patients infected with SARS-CoV-2 omicron variants: Results of a non-randomized clinical trial. Front. Med. 2022, 9, 980002. [Google Scholar] [CrossRef]
- Hosseini, E.S.; Kashani, N.R.; Nikzad, H.; Azadbakht, J.; Bafrani, H.H.; Kashani, H.H. The novel coronavirus Disease-2019 (COVID-19): Mechanism of action, detection and recent therapeutic strategies. Virology 2020, 551, 1–9. [Google Scholar] [CrossRef]
- Koob, T.J.; Willis, T.A.; Hernandez, D.J. Biocompatibility of NDGA‐polymerized collagen fibers. I. Evaluation of cytotoxicity with tendon fibroblasts in vitro. J. Biomed. Mater. Res. 2001, 56, 31–39. [Google Scholar] [CrossRef]
- Unal, M.A.; Bitirim, C.V.; Summak, G.Y.; Bereketoglu, S.; Cevher Zeytin, I.; Besbinar, O.; Gurcan, C.; Aydos, D.; Goksoy, E.; Kocakaya, E. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can. J. Physiol. Pharmacol. 2021, 99, 449–460. [Google Scholar] [CrossRef]
- Signer, J.; Jonsdottir, H.R.; Albrich, W.C.; Strasser, M.; Züst, R.; Ryter, S.; Ackermann-Gäumann, R.; Lenz, N.; Siegrist, D.; Suter, A. In vitro antiviral activity of Echinaforce®, an Echinacea purpurea preparation, against common cold coronavirus 229E and highly pathogenic MERS-CoV and SARS-CoV. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Li, R.; Hou, Y.; Huang, J.; Pan, W.; Ma, Q.; Shi, Y.; Li, C.; Zhao, J.; Jia, Z.; Jiang, H.; et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. [Google Scholar]
- Aoki-Utsubo, C.; Chen, M.; Hotta, H. Time-of-addition and temperature-shift assays to determine particular step(s) in the viral life cycle that is blocked by antiviral substance(s). Bio-Protocol 2018, 8, e2830. [Google Scholar] [CrossRef] [PubMed]
- Syed, G.H.; Siddiqui, A. Effects of hypolipidemic agent nordihydroguaiaretic acid on lipid droplets and hepatitis C virus. Hepatology 2011, 54, 1936–1946. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Moghadasi, S.A.; Heilmann, E.; Khalil, A.M.; Nnabuife, C.; Kearns, F.L.; Ye, C.; Moraes, S.N.; Costacurta, F.; Esler, M.A.; Aihara, H. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. Sci. Adv. 2023, 9, eade8778. [Google Scholar] [CrossRef]
- Zhou, Y.; Gammeltoft, K.A.; Ryberg, L.A.; Pham, L.V.; Fahnoe, U.; Binderup, A.; Hernandez, C.R.D.; Offersgaard, A.; Fernandez-Antunez, C.; Peters, G.H.J. Nirmatrelvir resistant SARS-CoV-2 variants with high fitness in vitro. bioRxiv 2022. [Google Scholar] [CrossRef]
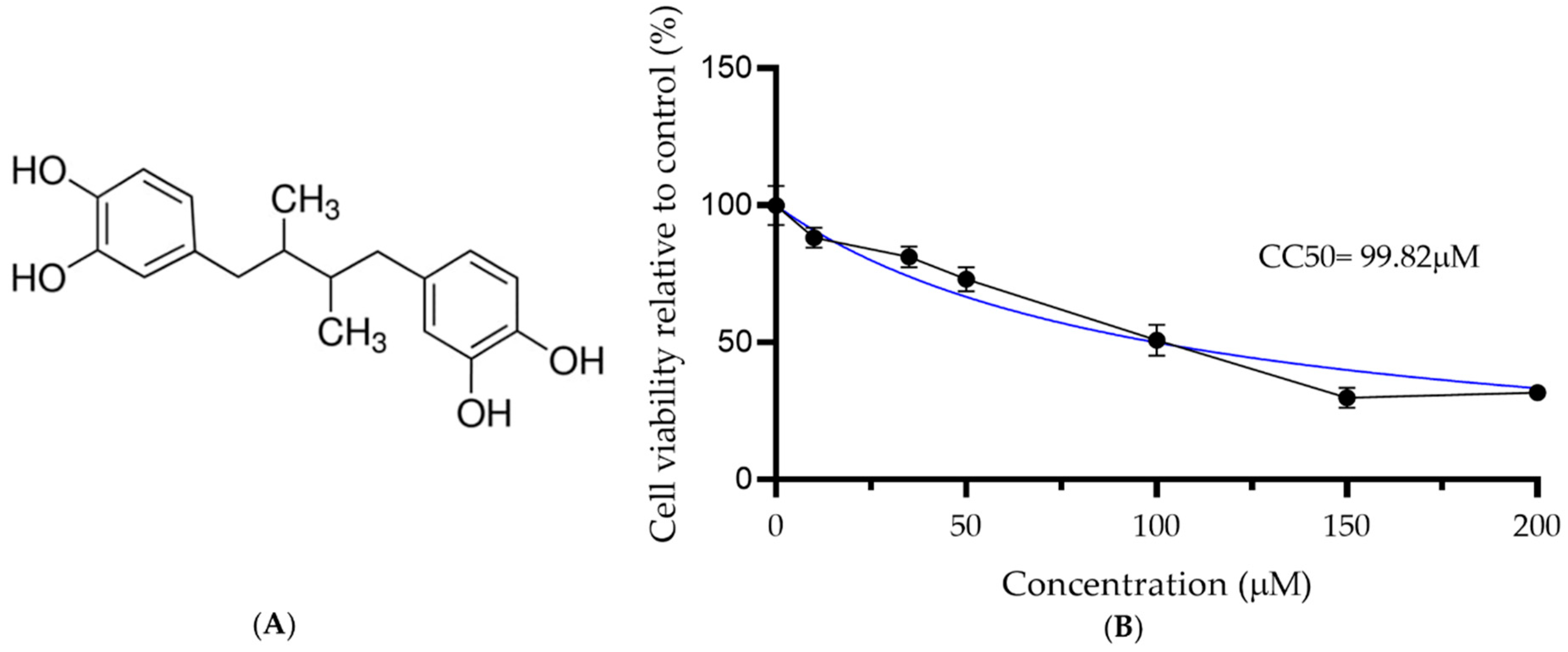
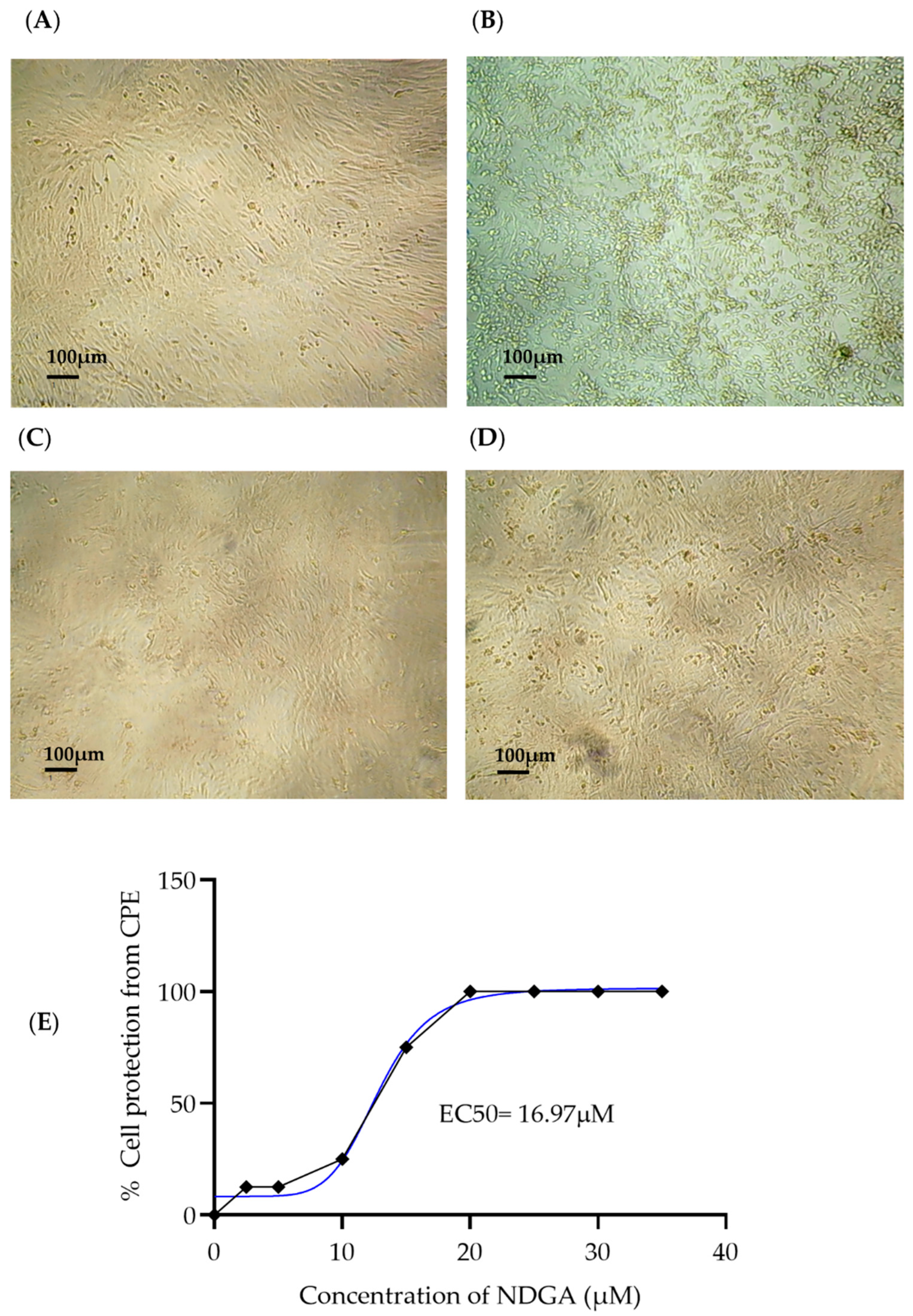

| Compound | CC50 | EC50 | EC90 | SI |
|---|---|---|---|---|
| NDGA | 99.82 | 16.97 | 25.64 | 5.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalobos-Sánchez, E.; García-Ruiz, D.; Camacho-Villegas, T.A.; Canales-Aguirre, A.A.; Gutiérrez-Ortega, A.; Muñoz-Medina, J.E.; Elizondo-Quiroga, D.E. In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2. Viruses 2023, 15, 1155. https://doi.org/10.3390/v15051155
Villalobos-Sánchez E, García-Ruiz D, Camacho-Villegas TA, Canales-Aguirre AA, Gutiérrez-Ortega A, Muñoz-Medina JE, Elizondo-Quiroga DE. In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2. Viruses. 2023; 15(5):1155. https://doi.org/10.3390/v15051155
Chicago/Turabian StyleVillalobos-Sánchez, Erendira, Daniel García-Ruiz, Tanya A. Camacho-Villegas, Alejandro A. Canales-Aguirre, Abel Gutiérrez-Ortega, José E. Muñoz-Medina, and Darwin E. Elizondo-Quiroga. 2023. "In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2" Viruses 15, no. 5: 1155. https://doi.org/10.3390/v15051155
APA StyleVillalobos-Sánchez, E., García-Ruiz, D., Camacho-Villegas, T. A., Canales-Aguirre, A. A., Gutiérrez-Ortega, A., Muñoz-Medina, J. E., & Elizondo-Quiroga, D. E. (2023). In Vitro Antiviral Activity of Nordihydroguaiaretic Acid against SARS-CoV-2. Viruses, 15(5), 1155. https://doi.org/10.3390/v15051155









