Abstract
Gene-modification therapies are at the forefront of HIV-1 cure strategies. Chimeric antigen receptor (CAR)-T cells pose a potential approach to target infected cells during antiretroviral therapy or following analytical treatment interruption (ATI). However, there are technical challenges in the quantification of HIV-1-infected and CAR-T cells in the setting of lentiviral CAR gene delivery and also in the identification of cells expressing target antigens. First, there is a lack of validated techniques to identify and characterize cells expressing the hypervariable HIV gp120 in both ART-suppressed and viremic individuals. Second, close sequence homology between lentiviral-based CAR-T gene modification vectors and conserved regions of HIV-1 creates quantification challenges of HIV-1 and lentiviral vector levels. Consideration needs to be taken into standardizing HIV-1 DNA/RNA assays in the setting of CAR-T cell and other lentiviral vector-based therapies to avoid these confounding interactions. Lastly, with the introduction of HIV-1 resistance genes in CAR-T cells, there is a need for assays with single-cell resolution to determine the competence of the gene inserts to prevent CAR-T cells from becoming infected in vivo. As novel therapies continue to arise in the HIV-1 cure field, resolving these challenges in CAR-T-cell therapy will be crucial.
1. Introduction
Combination antiretroviral therapy (ART) has significantly reduced HIV-1 morbidity and mortality. However, latent viral reservoirs persist, composed largely of cells that do not express significant levels of viral antigens, thereby evading immune-mediated eradication [1]. These viral reservoirs persist indefinitely through a variety of homeostatic or other proliferative mechanisms in virally suppressed individuals, contributing to low level inflammation associated with numerous long-term comorbidities. In light of these ongoing comorbidities in the setting of ART and the massive global burden of HIV-1, developing curative therapeutic approaches remains a high research priority.
Gene modification therapies are at the forefront of HIV-1 cure strategies. While there has been success in CCR5-delta-32 mutation autologous stem cell transplants (SCT) in curing a handful of individuals [2,3,4,5], this strategy is practical only for those who require allogeneic stem cell transplantation for hematologic illnesses and have HIV-1 that exclusively uses CCR5 for cell entry [6]. To reduce residual HIV-1 burden and achieve long-term ART-free viral remission, various gene modification therapies have promise. For example, gene modification of stem cells during autologous stem cell transplant that disrupt one or more stages of the HIV-1 life cycle are currently being implemented. In addition, chimeric antigen receptor (CAR) T-cell therapy, which involves lentiviral vector delivery to autologous T cells, is a major HIV-1 cure strategy of interest due to the success of this approach for various hematologic malignancies [7]. This approach involves genetically engineering patients’ T-cells to express CARs on the cell surface which can recognize and bind to specific proteins expressed on HIV-infected cells leading to potential cell-mediated toxicity and immune-mediated clearance of infected cells [8,9,10].
CAR-T-cell strategies have potential advantages for boosting immune system response to HIV-1. This includes their ability to recognize cell surface proteins, given that they are based on heavy and light chain regions of the neutralizing antibodies or surface CD4 protein [11,12,13]. CAR-T cells may also be engineered to target other macromolecules apart from stably expressed cell surface proteins [14]. It is not entirely clear if classically designed CAR-Ts can recognize other antigens expressed with intracellular processing and major histocompatibility complex (MHC) presentation. As a result, novel strategies are being developed that target MHC-peptide complexes and do not compete with endogenous TCR for CD3 complex formation [15,16,17,18,19]. However, HIV-1 regulatory protein expression can lead to the downregulation of MHC 1, a process which occurs in more actively infected cells [20,21,22,23,24]. Nonetheless, prior research has demonstrated the potential for CAR-T cells to be able to both traffic to the diverse areas of tissue that compose the latent reservoir (e.g., lymph node, gut-associated lymphoid tissues) and play a vital role in long-term viral surveillance [25,26,27,28,29,30,31,32,33].
The potential for CAR-T-cell therapy to target HIV-1-infected cells through HIV-1 envelope gp120 regions in vivo comes with numerous technical challenges. HIV-1 envelope protein gp120 sequences are highly variable given antibody-mediated immune pressure [34,35,36], and thus, require CARs that recognize and engage a wide variety of envelope variants. The conserved region of gp120 is within this protein trimer and is not exposed until CD4 binding and identifying the target on latently infected cells that may differ across tissues [37,38,39]. While less of an issue for original CAR-T cells that used CD4 molecules to bind to HIV-1 envelope, CARs based on neutralization antibodies are likely to have variable binding to various viral strains. Another challenge is the identification and quantification of target cells when using lentiviral gene delivery to create CAR-T cells. This issue arises due to the sequence homology between HIV-1 and lentiviral vectors used to generate CAR-T cells [40]. This is an issue in lentiviral vector CAR-T cells as well as other non-CAR-T lentiviral gene modification strategies directed toward HIV-1 cure.
In this review, we aim to systematically describe the current challenges facing therapeutic development of CAR-T cells for potential use as a curative therapeutic [41] with an emphasis on the challenges of targeting gp120 and vector-HIV-1 sequence homology on the development and implementation of quantitative assays.
2. Materials and Methods
This systematic literature review was performed according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines. PubMed literature library was searched for articles pertaining to gene modification therapies in the context of HIV-1 infection.
The first search was conducted for background on the topic of gene modification and CAR-T cells. PubMed was searched for [“HIV-1”, “gene modification”, “therapies”, “CAR T-cell”]. The second search conducted was for the technical challenges associated with quantifying target cells. PubMed was searched for [“HIV-1”, “gp120”, “cell-surface”, “targeting”], [“HIV-1”, “sequence homology”, “lentivirus”], and [“HIV-1”, “lentivirus”, “CAR T-cell”, “resistance genes”]. The final search conducted was for gp120 targeting approaches and difficulties. Pub-Med was searched for [“gp120 conserved” “gp120 variable” “HIV bNAbs” “gp120 presentation” “HLA presentation” “CD4 inhibition” “gp120 conformation”.
3. Results
Due to the heterogeneous nature of the HIV-1 latent reservoir and scarcity of infected cells in people with HIV (PWH) on ART, there are many challenges with immune targeting of viral persistence. This review will address three different technical difficulties associated with quantifying and characterizing HIV-1-infected or lentiviral vector-transduced T cells.
3.1. Challenges of Targeting HIV-1 gp120 for Infected Cell Clearance
HIV-1 is a retrovirus with the capacity to integrate into the host chromosomes. Current treatments such as ART allow PWH to suppress a large majority of circulating plasma HIV-1 RNA and reduce replication in tissues [42,43,44]. Rebound viremia occurs rapidly, however, following ART discontinuation in most people [45,46]. Despite ART-mediated viral suppression, ongoing immune dysfunction and inflammation persist [47,48,49,50,51,52,53,54]. Identifying and targeting the persistent reservoir is tantamount to long-term ART-free HIV remission. While HIV-1-infected cells can express high levels of gp120 during active replication [55], these levels are far lower in the setting of ART suppression. Expression of HIV env proteins on a cell’s surface may occur during initial virus-cell binding and entry, viral assembly, and budding, or to a likely much lesser extent, intracellular antigen processing and HLA-mediated presentation [56,57]. However, immunoPET imaging has recently demonstrated that low levels of HIV-1 or SIV gp120 protein can be identified across a range of tissues in the setting of ART using gp120-specific antibodies [58,59,60,61]; whether or not this reflects cell surface expression of gp120 or free viral proteins or virions in close anatomical proximity to residual infected cells is not known. The potential paucity of gp120 expression on cell surfaces makes CAR-T-mediated immune targeting difficult in people on suppressive ART. Given the hypervariability of large portions of HIV-1 gp120 due to its surface location on the virion and humoral immune mediated pressure, this glycoprotein is difficult to target across PWH, even when using a HIV-specific broadly neutralizing antibodies (bNAbs).
Challenges in Identifying and Quantifying HIV-1 gp120 in the Setting of ART
Most PWH have increased gp120 expression after ART cessation as virus begins to emerge and replicate in blood and tissues [58,61]. Ultimately, HIV-1 expression that is recognizable to the human immune system remains vital in the immune targeting HIV-1 cure strategies such as CAR-T approaches.
Gp120 is a large trimeric envelope protein expressed on the surface of the HIV-1 virion and infected cells, with three glycoprotein subunits including V1, V2, and V3. These variable loop regions protect HIV from immune recognition and assist in HIV virion binding for invasion into CD4 cells [37,62]. During virion binding to CD4, portions of this trimeric protein are released, and expose a conserved region of the envelope. This region is protected from recognition due to its sequence and functional consistency. Due to the exposure of these epitopes only in the late stages of cell infection, it is difficult to target this region for viral integration prevention and early neutralization. While immunotherapies have been brought into the HIV-1 cure field, the high level of immune escape has hindered the success of targeting these infected cells. Original CAR-T cells designed to recognize HIV envelope utilized CD4 proteins, but more recent CAR-T constructs have evolved to have broader and longer lasting antiviral activity by presenting costimulatory molecules (i.e., 4-1BB and CD28) combined with bNAbs as seen in Table 1 [41,63,64]. Another application of bNAbs is through leveraging single chain variable fragments (scFv) in CAR-T constructs. Increased success has been seen in persistence and protection in vivo of combination scFv CAR-T cells versus monotherapy. These scFv CAR-T cells allow for the recognition of conformationally available HIV-1 gp120 activated in an MHC-independent manner [65]. The affinity and specificity of the groups are also dependent on the positioning of the various domains as determined by the linker. This positioning is crucial in developing effective CAR-T therapies while preventing off-target effects or toxicities [66]. In addition, these variable domains fall victim to the same shortcomings that their parent monoclonal and bNAbs faced, an inability to accurately target the gp120 protein. A current study is using lentiviral vectors to mediate CAR-modification of T cells to present these anti-HIV CARs (i.e., duoCARs) and make cells resistant to HIV-1 [41,67]. Other novel CAR-T-cell technologies are being developed to recognize, bind, and kill MHC–antigen complexes, but these methods are predominantly in the pre-clinical development stage for various malignancies [15,16,17,18,19].

Table 1.
Generation of CAR-T cells [10]. Each panel describes the basic construction of the four generations of CAR-T cells and demonstrates their advancements.
There is a paucity of data regarding the frequency and density of gp120 expression on latently infected cells. In addition, quantifying the density of gp120 expression presents a distinct challenge as there is a major dearth of literature demonstrating surface detection of HIV-1 gp120 in the setting of ART outside of whole body immunoPET studies [58,68]. Without understanding its expression or density on persistently infected cells or the dynamics of viral envelope expression following treatment cessation, therapeutic development of CAR-T cells will be hampered. This also holds true outside of PET imaging. The inability to target gp120 consistently and efficiently, especially in latent tissue reservoirs, creates a barrier for virus and infection clearance [69]. As previously mentioned, the monoclonal antibodies and bNAbs developed thus far have difficulty targeting the full range of envelope subunit diversity [1,70,71]. The conserved regions of the HIV envelope are hidden within the tripod-like structure of gp120 and is only visible when actively bound and infecting a CD4 cell, as depicted in Figure 1. The lack of ability to use standardized antibodies to target the major region for HIV identification greatly reduces the ability to both identify and quantify expression infected human cells as well as for in vivo immune targeting. Similarly, bNAbs sometimes fail to target a conserved region across all infected cells in the human population [72,73,74], even if expressed in the setting of robust latency reversal or analytical treatment interruption. As CAR-T-cell therapeutics are commonly composed of variable antibody heavy and light chain regions, they are likely to have similar targeting issues. However, as a “living drug”, they have the capacity to expand in the presence of antigen recognition and may have different tissue penetration patterns to bnAbs or native T cells (e.g., central nervous system, lymph nodes, bone [58]). As a result, CAR-T-cell therapies (and bnAbs) are commonly designed to be used in combination with analytical treatment interruptions (ATI; i.e. highly monitored antiviral pauses) in which gp120 expression may increase dramatically [75].
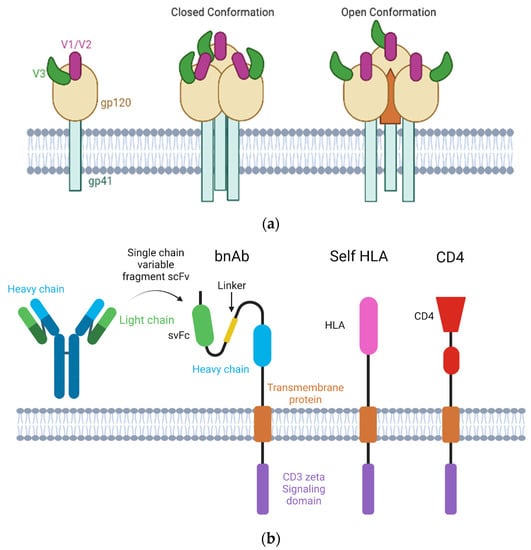
Figure 1.
Structure of HIV envelope protein and TCR structures for CAR-T cells. (a) (Left) structure of HIV-1 envelope gp120 and gp140 proteins alone, (middle) full trimeric structure of the closed conformation before CD4 binding, and (right) trimeric protein in the open conformation exposing the internal epitope after CD4 binding (for simplicity, the release of gp120 and gp140 is excluded in this diagram). (b) Depiction of the composition of the CAR-T-cell receptor using the light and heavy chain of the antibody, as well as the transmembrane and intracellular proteins. The proceeding receptors show the hypothesized self HLA-presenting receptor model, then, the CD4-presenting model. Figure was created with BioRender.com (accessed on 28 March 2023).
3.2. Sequence Homology Challenges
Another technical challenge associated with CAR-T-cell development is using vector backbones for the integration into the host genome. While there are other ways to genetically engineer CAR-T cells, lentiviral vectors are used for the transduction efficiency and accuracy of multi-gene integration. In cancer therapy, the genus and family of lentiviruses do not create difficulty in quantifying and assessing therapeutic results; however, this is not the case for HIV-1 cure therapies. Here, we detail an overview of lentiviruses and the challenges their sequence similarities bring to therapeutic action.
3.2.1. Retroviruses and Their Conserved Regions
The family Retroviridae is composed mainly of simple (gamma-retroviruses) and complex (lentiviruses) retroviruses [76]. The main difference between simple and complex retroviruses is the amount of polyproteins encoded that affect viral synthesis (e.g., viral RNA replication) [76,77,78]. Ubiquitous polyproteins encoded for in all retroviruses are gag, pol, env genes; while lentiviruses also are comprised of various other proteins, depending on the specific virus and strain, and include tat, tax, rev, rex, nef, etc., [76,77,78]. Both subfamilies contain gag, which maintains viral structure, pol, which encodes enzymatic ssRNA, and env, which encodes viral envelope proteins (gp120) [79]. These three regions are quite homologous across retroviral genomes.
Common retroviruses used as vector backbones today are HIV-1 and murine leukemia virus (MLV) [80,81,82,83,84]. In this review, we will focus on the lentivirus subgroup, which includes HIV, due to the multiple advantages of lentiviral vectors that tend to be lacking in gamma-retroviral vectors.
In addition to proximal HIV-1 Gag [85], a conserved region that is shared by many lentiviruses is that of the long-terminal repeat (LTR) region that acts as a promoter and modulator of viral transcription [86]. These conserved regions of lentiviruses are shown in Figure 2. As depicted, the conserved regions tend to be consistent across the genus with little variation.
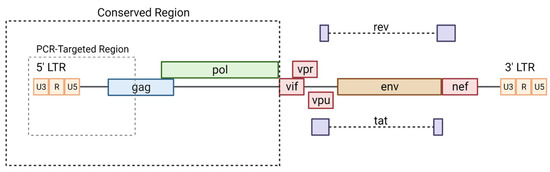
Figure 2.
Conserved regions of HIV-1. The conserved regions of HIV-1 are targeted in PCR assays as well as commonly used in lentiviral vector production. Modified from [87] and created with BioRender.com (accessed on 1 May 2023).
These conserved sequences are the core of the lentiviral backbone. This framework proves to be useful for a few reasons. One of them being the ability to carry and integrate large amounts of transgenes into the human genome. The size of the lentiviral vector backbone tends to be around 3.8 kb without any gene inserts. And while with increasing size of genes added tend to decrease transduction efficiency, each vector can hold around 9 kb in gene inserts, totaling approximately 14 kb [88,89]. This is more than the packaging capacity of typical adeno-associated viral vectors (4.8 kb of added DNA) [90]. More gene combination possibilities may allow a broader range of potential therapeutic effects.
Another useful ability of lentiviral vectors is that they have long-term transgene expression and consistent transduction in replicating and non-replicating cells which is a core reason they are widely used in the field of gene-engineering [91,92,93]. In the past, viral vectors used on target cells that have had inconsistent expression of genes caused off-target effects on non-targeted tissues [94,95,96,97]. However, there are more updated lentiviral vector backbones that allow for the regulation of specific gene expression, such as the tet system [98]. As lentiviral vectors continue to be researched, integrated genes have sustained expression allowing for greater therapeutic effect in gene-modification therapies [99,100].
Due to the advances and advantages of lentiviral vector backbone usage, their use is widespread in developing CAR-T-cell therapies, including the HIV-1 cure field. These vectors are currently being used in human studies (ClinicalTrials.gov #NCT02797470, #NCT04648046, #NCT02343666, #NCT00569985, #NCT05529342). For instance, AIDS Malignancy Consortium 097 study (AMC097) modifies autologous stem cells of HIV-1 infected individuals with a lentiviral vector backbone that carries a CCR5 shRNA, chimeric macaque-human Trim5α, and a HIV-1 TAR decoy [101,102,103]. However, with the many lentiviral vector backbones being created from HIV-1, this creates downstream sequence homology issues with HIV-1.
3.2.2. Co-Quantification of HIV and Lentiviral Vector in HIV-1 Infected Individuals
In the setting of lentiviral vector or HIV-1 quantification in samples from participants by various sequence specific PCR methods in gene therapy studies, the conserved regions of both viral sequences are often used, as hypervariable regions are not reliable enough to ensure accurate counts across participants [104]. For the quantification of HIV-1, the LTR-proximal Gag region is typically used for consistency and to reliably quantitate target copy numbers across the diverse viral strains (of note, lentiviral vector DNA and RNA is conserved, although infecting HIV-1 strains are highly diverse). Since primers and probes in PCR techniques are usually designed for conserved sequences, the vast majority of highly tested assays to define the HIV-1 reservoir correspond to these conserved regions common to lentiviral vectors [85].
To be able to quantify lentiviral vector and HIV-1 infection separately, there is a challenge to design assays that will not have cross-reactivity between conserved regions of HIV-1 and lentiviral vector sequences. If there is overlap in these regions, current assays are not able to differentiate between cell-associated lentiviral vector DNA and integrated HIV-1. Possibilities such as adding viral genes in the backbone or targeting smaller regions within the vector could lead to less cross-reactivity. It is also possible for viral load assays that detect lentiviral RNAs from the proximal Gag region to detect certain lentiviral vector transcripts. Not knowing the difference between the HIV reservoir and the presence of a lentiviral vector can cause a multitude of issues that affect the overall knowledge of therapeutic efficacy.
There are both clinical and research assays for quantification of HIV-1 that depend on conserved regions of HIV-1 sequence. While there are antibody, antigen, and nucleic acid tests in clinical use to detect and quantify virus in plasma, the HIV-1 reservoir is typically detected via RNA and DNA PCR of peripheral mononuclear blood cells (PBMCs) [105,106,107,108,109,110]. In PWH on suppressive ART, blood plasma virus is typically undetectable by routine viral load testing [111,112,113]. Cell-associated HIV infection can nearly always be detected, however. With the sequence homology of HIV-1 and various lentiviral constructs there is no current standard assay to distinguish between the two, prohibiting accurate monitoring of HIV-1 reservoirs following lentiviral-based CAR-T-cell therapy. This applies to the recently described HIV-1 Intact Proviral DNA Assay (IDPA) [114]. Furthermore, the inability to distinguish certain regions of HIV-1 from various lentiviral vectors presents challenges in characterizing the expansion of the dynamics of CAR-T-cell populations within the body following infusion with or without ATI.
When conducting studies or clinical trials for gene-modification therapies, it is important to have a method to understand how these cells are expanding and exerting therapeutic and off-target effects on the body. There is an urgent need for standardization of quantitation assays that will be used for CAR-T-cell therapies for the current and future HIV-1 cure investigations.
3.2.3. Lentiviral Vector Uses in HIV Cure
The field of gene modification therapies is a rapidly expanding field across diseases, and the HIV-1 cure space is currently adapting many of these techniques [115,116,117,118]. While there are a multitude of ways to genetically modify cells, the exploration of lentiviral vectors is being employed toward HIV cure. As previously discussed, use of lentiviral vector backbones in these therapies has several advantages. These uses include ease in which they can integrate large transgenes and facilitate expression within dividing target cell types [79,119]. However, in the clinical space, they are also ideal due to the thoroughly conducted safety and efficacy studies in humans [120].
Lentiviral-based gene modification is used to generate CAR-T cells for HIV-1 cure in patients who are otherwise healthy and stably suppressed on ART (ClinicalTrials.gov #NCT04648046) [41,67]. While the cells transduced with lentiviral vectors make RNA transcripts that code for CARs or other products of interest in gene therapy studies, they are engineered to avoid lentiviral replication [79,121]. Nonetheless, there is a possibility that gene modified CD4+ T cells become infected with HIV-1 in vivo. Although the potential for recombination between HIV-1 and lentiviral vector DNA is highly improbable due to advances in lentiviral vector production, the potential is there, which could theoretically pose safety and efficacy issues in clinical spaces [92,122,123,124].
3.3. Challenges of Single-Cell Resolution of CAR-T Cell Resistance Genes
Quantifying CAR-T-cell expansion and the correlative HIV-1 burden is critical to determining therapeutic success. Several major questions remain unanswered, however. For example, despite strategies to engineer CAR-T cells to be resistant to HIV infection (CAR-T cells are often CD4-T cells of origin) by using C46 or non-CD4-based single chain variable fragments (scFv) [41,67,125], it is not known if CAR-T cells or other gene modified cells (e.g., the multiple gene-modified stem cell transplant strategy as above) become infected in vivo, especially during potentially high levels of HIV-1 plasma viremia and replication in studies involving ATIs.
Limitations of Current Single-Cell Analysis Methods
Single-cell characterization of HIV-1-infected and CAR-T cells will be required in order to determine the dynamics of CAR-T cells (or other gene modified cell strategies that involve engineered CD4+ T cells or hematopoietic stem cells [101,102,103,126]) over time in vivo with precision and accuracy as well as to determine if these modified cells become infected in vivo. Whereas there are numerous methods to investigate host and viral transcriptional activity on a single cell level, there are limitations associated with them when looking specifically at lentiviral vector-mediated CAR-T cell and other cellular therapies in PWH. The main techniques that can distinguish attributes of individual cells are single-cell RNA sequencing, fluorescence microscopy, and flow cytometry [127,128,129,130].
Single-cell RNA sequencing surveys the sequence of individual cellular RNA transcript expression. scRNA seq has advantages in that it can identify distinct genetic and phenotypic cell populations. However, it is limited by cost because the relatively fewer number of cells that can be analyzed at once adds to the difficulty in detecting non-polyadenylated HIV-1 or lentiviral transcripts [131,132,133]. Fluorescence microscopy can detect the genetic profile and protein expression in culture and tissues samples, while flow cytometry utilizes antibody kinetics to analyze surface proteins [98]. An additional technique utilizing both scRNA seq and flow cytometry is cellular indexing of transcriptomes and epitopes (CITE) sequencing [99]. The application of these methodologies enables single-cell resolution for different research targets. CAR-T cells used in oncology research were previously used in methods to identify and track cells by using flow cytometry [67,100,134]. When there is optimal engineering of the CAR to have a uniquely identifiable biomarker (i.e., CD19 CAR-T cells), flow cytometry can quantify the changes in CAR-T-cell reservoir. An activated and discrete biomarker is also needed in other lentiviral gene modification strategies when there is no distinct surface protein that delineates modified cells. Being able to identify modified cells infected with HIV-1 is critically important as this indicates that modifications that are engineered to protect cells from HIV infection (e.g., C46 with CAR-T, multiple gene modification in AMC097) are not effectively preventing de novo infection. So while these assays provide useful information on single-cell dynamics and biology, there are limitations to each in the context of developing lentiviral-based CAR-T-cell therapeutics.
Assays that allow for high throughput quantification of HIV-infected cells (which can be as rare as one in a million CD4 T cells in peripheral blood) and gene-modified lentiviral vector at a single-cell level are highly desirable to advance the development of lentiviral CAR-T-cell therapies in the HIV field. These assays need not be sophisticated in terms of the multiomic capabilities, but rather allow the survey of a large number of cells for simultaneous characterization of HIV-1 and lentiviral vector insert DNA or RNA transcripts. Single-cell encapsulation in microfluidic droplets with subsequent multiplexed PCR detection of distinct target sequences that do not overlap is one potential, cost-effective solution.
4. Discussion
CAR-T-cell therapies are gaining traction in the HIV-1 cure field and hold some promise to target and remove infected cells expressing HIV-1 envelope proteins. While the safety, efficacy, and efficiency of lentiviral vector-generated CAR-T cells have been proven in other therapeutic areas, such as oncology, there are many practical assay issues that arise with the introduction of these modalities for HIV cure. HIV-1 envelope surface expression is likely low and metastable in the setting of suppressive ART and it is not clear if CAR-T cells can effectively target infected cells in vivo across a range of tissues. As a result, many studies involve ATI following CAR-T infusion to allow for easier recognition of infected cells and to maintain or expand the CAR-T cell pool. There are also many technical difficulties in measuring target protein expression (e.g., Env gp120) and to quantify lentiviral vector and HIV-1 DNA or RNAs both in bulk samples with single-cell resolution.
Author Contributions
Conceptualization, A.M.B., T.-M.D. and T.J.H.; methodology, A.M.B. and T.-M.D.; validation, A.M.B., T.-M.D., A.N.D. and T.J.H.; writing—original draft preparation, A.M.B. and T.-M.D.; writing—review and editing, A.M.B., T.-M.D., A.N.D. and T.J.H.; supervision, A.N.D. and T.J.H.; project administration, A.M.B.; funding acquisition, A.N.D. and T.J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the NIH/National Institute of Allergy and Infectious Diseases (R01AI176951 to TJH; K23AI162249 to AND), as well as through the California HIV Research Program (H22BD4484).
Data Availability Statement
No new data were created.
Conflicts of Interest
TJH receives grant support from Merck and Co. and has consulted for Roche and Regeneron.
References
- Halper-Stromberg, A.; Lu, C.-L.; Klein, F.; Horwitz, J.A.; Bournazos, S.; Nogueira, L.; Eisenreich, T.R.; Liu, C.; Gazumyan, A.; Schaefer, U.; et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 2014, 158, 989–999. [Google Scholar] [CrossRef]
- Gupta, R.K.; Peppa, D.; Hill, A.L.; Gálvez, C.; Salgado, M.; Pace, M.; McCoy, L.E.; Griffith, S.A.; Thornhill, J.; Alrubayyi, A.; et al. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: A case report. Lancet HIV 2020, 7, e340–e347. [Google Scholar] [CrossRef] [PubMed]
- Hütter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müßig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kücherer, C.; Blau, O.; et al. Long-Term Control of HIV by CCR5 Delta32/Delta32 Stem-Cell Transplantation. N. Engl. J. Med. 2009, 360, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.-E.O.; Knops, E.; Cords, L.; Lübke, N.; Salgado, M.; Busman-Sahay, K.; Estes, J.D.; Huyveneers, L.E.P.; Perdomo-Celis, F.; Wittner, M.; et al. In-depth virological and immunological characterization of HIV-1 cure after CCR5Δ32/Δ32 allogeneic hematopoietic stem cell transplantation. Nat. Med. 2023, 29, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.; Glesby, M.; Shore, T.B.; Small, C.; Gergis, U. HIV-1 remission with CCR5∆ 32∆ 32 haplo-cord transplant in a US woman: IMPAACT P1107. In Proceedings of the Conference on Retroviruses and Opportunistic Infections (CROI), Denver, CO, USA, 12–16 February 2022. [Google Scholar]
- Prator, C.A.; Donatelli, J.; Henrich, T.J. From Berlin to London: HIV-1 reservoir reduction following stem cell transplantation. Curr. HIV/AIDS Rep. 2020, 17, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Seif, M.; Einsele, H.; Löffler, J. CAR T Cells Beyond Cancer: Hope for Immunomodulatory Therapy of Infectious Diseases. Front. Immunol. 2019, 10, 2711. [Google Scholar] [CrossRef]
- Ali, A.; Kitchen, S.G.; Chen, I.S.Y.; Ng, H.L.; Zack, J.A.; Yang, O.O. HIV-1-Specific Chimeric Antigen Receptors Based on Broadly Neutralizing Antibodies. J. Virol. 2016, 90, 6999–7006. [Google Scholar] [CrossRef]
- Hale, M.; Mesojednik, T.; Romano Ibarra, G.S.; Sahni, J.; Bernard, A.; Sommer, K.; Scharenberg, A.M.; Rawlings, D.J.; Wagner, T.A. Engineering HIV-Resistant, Anti-HIV Chimeric Antigen Receptor T Cells. Mol. Ther. 2017, 25, 570–579. [Google Scholar] [CrossRef]
- Hiltensperger, M.; Krackhardt, A.M. Current and future concepts for the generation and application of genetically engineered CAR-T and TCR-T cells. Front. Immunol. 2023, 14, 1121030. [Google Scholar] [CrossRef]
- Jin, K.-T.; Chen, B.; Liu, Y.-Y.; Lan, H.U.-R.; Yan, J.-P. Monoclonal antibodies and chimeric antigen receptor (CAR) T cells in the treatment of colorectal cancer. Cancer Cell Int. 2021, 21, 83. [Google Scholar] [CrossRef]
- Ramos, C.A.; Dotti, G. Chimeric antigen receptor (CAR)-engineered lymphocytes for cancer therapy. Expert Opin. Biol. Ther. 2011, 11, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.J.; Dougan, M.; Jailkhani, N.; Ingram, J.; Fang, T.; Kummer, L.; Momin, N.; Pishesha, N.; Rickelt, S.; Hynes, R.O.; et al. Nanobody-based CAR T cells that target the tumor microenvironment inhibit the growth of solid tumors in immunocompetent mice. Proc. Natl. Acad. Sci. USA 2019, 116, 7624–7631. [Google Scholar] [CrossRef]
- Keane, J.T.; Posey, A.D., Jr. Chimeric Antigen Receptors Expand the Repertoire of Antigenic Macromolecules for Cellular Immunity. Cells 2021, 10, 3356. [Google Scholar] [CrossRef] [PubMed]
- Akatsuka, Y. TCR-Like CAR-T Cells Targeting MHC-Bound Minor Histocompatibility Antigens. Front. Immunol. 2020, 11, 257. [Google Scholar] [CrossRef]
- Raskin, S.; Van Pelt, S.; Toner, K.; Balakrishnan, P.B.; Dave, H.; Bollard, C.M.; Yvon, E. Novel TCR-like CAR-T cells targeting an HLA∗0201-restricted SSX2 epitope display strong activity against acute myeloid leukemia. Mol. Ther. Methods Clin. Dev. 2021, 23, 296–306. [Google Scholar] [CrossRef]
- Ma, Q.; Garber, H.R.; Lu, S.; He, H.; Tallis, E.; Ding, X.; Sergeeva, A.; Wood, M.S.; Dotti, G.; Salvado, B.; et al. A novel TCR-like CAR with specificity for PR1/HLA-A2 effectively targets myeloid leukemia in vitro when expressed in human adult peripheral blood and cord blood T cells. Cytotherapy 2016, 18, 985–994. [Google Scholar] [CrossRef]
- Smith, H.A.; McNeel, D.G. Vaccines targeting the cancer-testis antigen SSX-2 elicit HLA-A2 epitope-specific cytolytic T cells. J. Immunother. 2011, 34, 569–580. [Google Scholar] [CrossRef]
- Chabannon, C.; Bonini, C. Structure of and Signalling Through Chimeric Antigen Receptor. In The EBMT/EHA CAR-T Cell Handbook; Kröger, N., Gribben, J., Chabannon, C., Yakoub-Agha, I., Einsele, H., Eds.; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Blagoveshchenskaya, A.D.; Thomas, L.; Feliciangeli, S.F.; Hung, C.-H.; Thomas, G. HIV-1 Nef Downregulates MHC-I by a PACS-1- and PI3K-Regulated ARF6 Endocytic Pathway. Cell 2002, 111, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef]
- Usami, Y.; Wu, Y.; Göttlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef]
- Michel, N.; Allespach, I.; Venzke, S.; Fackler, O.T.; Keppler, O.T. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 2005, 15, 714–723. [Google Scholar] [CrossRef]
- Fackler, O.T.; Baur, A.S. Live and let die: Nef functions beyond HIV replication. Immunity 2002, 16, 493–497. [Google Scholar] [CrossRef]
- Marban, C.; Forouzanfar, F.; Ait-Ammar, A.; Fahmi, F.; El Mekdad, H.; Daouad, F.; Rohr, O.; Schwartz, C. Targeting the Brain Reservoirs: Toward an HIV Cure. Front. Immunol. 2016, 7, 397. [Google Scholar] [CrossRef] [PubMed]
- Grupp, S.A.; Kalos, M.; Barrett, D.; Aplenc, R.; Porter, D.L.; Rheingold, S.R.; Teachey, D.T.; Chew, A.; Hauck, B.; Wright, J.F.; et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. N. Engl. J. Med. 2013, 368, 1509–1518. [Google Scholar] [CrossRef]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Salmon, H.; Franciszkiewicz, K.; Damotte, D.; Dieu-Nosjean, M.-C.; Validire, P.; Trautmann, A.; Mami-Chouaib, F.; Donnadieu, E. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J. Clin. Investig. 2012, 122, 899–910. [Google Scholar] [CrossRef]
- Newick, K.; O’Brien, S.; Sun, J.; Kapoor, V.; Maceyko, S.; Lo, A.; Puré, E.; Moon, E.; Albelda, S.M. Augmentation of CAR T-cell Trafficking and Antitumor Efficacy by Blocking Protein Kinase A LocalizationPKA Blockade Improves CAR Therapy. Cancer Immunol. Res. 2016, 4, 541–551. [Google Scholar] [CrossRef]
- Caruana, I.; Savoldo, B.; Hoyos, V.; Weber, G.; Liu, H.; Kim, E.S.; Ittmann, M.M.; Marchetti, D.; Dotti, G. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat. Med. 2015, 21, 524–529. [Google Scholar] [CrossRef]
- Mu, W.; Carrillo, M.A.; Kitchen, S.G. Engineering CAR T Cells to Target the HIV Reservoir. Front. Cell. Infect. Microbiol. 2020, 10, 410. [Google Scholar] [CrossRef]
- Kochenderfer, J.N.; Somerville, R.P.T.; Lu, T.; Shi, V.; Bot, A.; Rossi, J.; Xue, A.; Goff, S.L.; Yang, J.C.; Sherry, R.M.; et al. Lymphoma Remissions Caused by Anti-CD19 Chimeric Antigen Receptor T Cells Are Associated With High Serum Interleukin-15 Levels. J. Clin. Oncol. 2017, 35, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Kalos, M.; Levine, B.L.; Porter, D.L.; Katz, S.; Grupp, S.A.; Bagg, A.; June, C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011, 3, 95ra73. [Google Scholar] [CrossRef]
- Moscoso, C.G.; Xing, L.; Hui, J.; Hu, J.; Kalkhoran, M.B.; Yenigun, O.M.; Sun, Y.; Paavolainen, L.; Martin, L.; Vahlne, A.; et al. Trimeric HIV Env provides epitope occlusion mediated by hypervariable loops. Sci. Rep. 2014, 4, 7025. [Google Scholar] [CrossRef]
- Wang, W.-K.; Dudek, T.; Essex, M.; Lee, T.-H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: Therapeutic and prophylactic implications. Proc. Natl. Acad. Sci. USA 1999, 96, 4558–4562. [Google Scholar] [CrossRef] [PubMed]
- Haigwood, N.L.; Shuster, J.R.; Moore, G.K.; Lee, H.; Skiles, P.V.; Higgins, K.W.; Barr, P.J.; George-Nascimento, C.; Steimer, K.S. Importance of hypervariable regions of HIV-1 gp120 in the generation of virus neutralizing antibodies. AIDS Res. Hum. Retrovir. 1990, 6, 855–869. [Google Scholar] [CrossRef]
- Acharya, P.; Lusvarghi, S.; Bewley, C.A.; Kwong, P.D. HIV-1 gp120 as a therapeutic target: Navigating a moving labyrinth. Expert Opin. Ther. Targets 2015, 19, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Clapham, P.R.; McKnight, Á. HIV-1 receptors and cell tropism. Br. Med Bull. 2001, 58, 43–59. [Google Scholar] [CrossRef]
- Wang, X.; Cao, M.; Wu, Y.; Xu, W.; Wang, Q.; Ying, T.; Lu, L.; Jiang, S. Synergistic Effect by Combining a gp120-Binding Protein and a gp41-Binding Antibody to Inactivate HIV-1 Virions and Inhibit HIV-1 Infection. Molecules 2021, 26, 1964. [Google Scholar] [CrossRef]
- Johnson, N.M.; Alvarado, A.F.; Moffatt, T.N.; Edavettal, J.M.; Swaminathan, T.A.; Braun, S.E. HIV-based lentiviral vectors: Origin and sequence differences. Mol. Ther. Methods Clin. Dev. 2021, 21, 451–465. [Google Scholar] [CrossRef]
- Anthony-Gonda, K.; Bardhi, A.; Ray, A.; Flerin, N.; Li, M.; Chen, W.; Ochsenbauer, C.; Kappes, J.C.; Krueger, W.; Worden, A.; et al. Multispecific anti-HIV duoCAR-T cells display broad in vitro antiviral activity and potent in vivo elimination of HIV-infected cells in a humanized mouse model. Sci. Transl. Med. 2019, 11, 504. [Google Scholar] [CrossRef]
- Zhang, L.; Ramratnam, B.; Tenner-Racz, K.; He, Y.; Vesanen, M.; Lewin, S.; Talal, A.; Racz, P.; Perelson, A.S.; Korber, B.T.; et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 1999, 340, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Mocroft, A.; Phillips, A.N.; Gatell, J.; Ledergerber, B.; Fisher, M.; Clumeck, N.; Losso, M.; Lazzarin, A.; Fatkenheuer, G.; Lundgren, J. Normalisation of CD4 counts in patients with HIV-1 infection and maximum virological suppression who are taking combination antiretroviral therapy: An observational cohort study. Lancet 2007, 370, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.D.; Chaisson, R.E. Natural history of HIV infection in the_era of combination antiretroviral therapy. AIDS 1999, 13, 1933. [Google Scholar] [CrossRef]
- Fromentin, R.; DaFonseca, S.; Costiniuk, C.T.; El-Far, M.; Procopio, F.A.; Hecht, F.M.; Hoh, R.; Deeks, S.G.; Hazuda, D.J.; Lewin, S.R.; et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4+ T cells from ART-suppressed individuals. Nat. Commun. 2019, 10, 814. [Google Scholar] [CrossRef]
- Hill, A.L.; Rosenbloom, D.I.S.; Goldstein, E.; Hanhauser, E.; Kuritzkes, D.R.; Siliciano, R.F.; Henrich, T.J. Real-time predictions of reservoir size and rebound time during antiretroviral therapy interruption trials for HIV. PLoS Pathog. 2016, 12, e1005535. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.W.; Sereti, I. Residual immune dysfunction under antiretroviral therapy. Semin. Immunol. 2021, 51, 101471. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.W. Very Early ART and Persistent Inflammation in Treated HIV. Clin. Infect. Dis. 2017, 64, 132–133. [Google Scholar] [CrossRef]
- Klatt, N.R.; Chomont, N.; Douek, D.C.; Deeks, S.G. Immune activation and HIV persistence: Implications for curative approaches to HIV infection. Immunol. Rev. 2013, 254, 326–342. [Google Scholar] [CrossRef]
- Neuhaus, J.; Jacobs, D.R., Jr.; Baker, J.V.; Calmy, A.; Duprez, D.; La Rosa, A.; Kuller, L.H.; Pett, S.L.; Ristola, M.; Ross, M.J.; et al. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J. Infect. Dis. 2010, 201, 1788–1795. [Google Scholar] [CrossRef]
- Marchetti, G.; Bellistrì, G.M.; Borghi, E.; Tincati, C.; Ferramosca, S.; La Francesca, M.; Morace, G.; Gori, A.; Monforte, A.D. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS 2008, 22, 2035–2038. [Google Scholar] [CrossRef]
- Lederman, M.M.; Calabrese, L.; Funderburg, N.T.; Clagett, B.; Medvik, K.; Bonilla, H.; Gripshover, B.; Salata, R.A.; Taege, A.; Lisgaris, M.; et al. Immunologic Failure Despite Suppressive Antiretroviral Therapy Is Related to Activation and Turnover of Memory CD4 Cells. J. Infect. Dis. 2011, 204, 1217–1226. [Google Scholar] [CrossRef]
- Hayes, T.L.; Asmuth, D.M.; Critchfield, J.W.; Knight, T.H.; McLaughlin, B.E.; Yotter, T.; McConnell, D.H.; Garcia, J.C.; Pollard, R.B.; Shacklett, B.L. Impact of highly active antiretroviral therapy initiation on CD4+ T-cell repopulation in duodenal and rectal mucosa. AIDS 2013, 27, 867–877. [Google Scholar] [CrossRef]
- Tincati, C.; Biasin, M.; Bandera, A.; Violin, M.; Marchetti, G.; Piacentini, L.; Vago, G.L.; Balotta, C.; Moroni, M.; Franzetti, F.; et al. Early initiation of highly active antiretroviral therapy fails to reverse immunovirological abnormalities in gut-associated lymphoid tissue induced by acute HIV infection. Antivir. Ther. 2009, 14, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Missé, D.; Gajardo, J.; Oblet, C.; Religa, A.; Riquet, N.; Mathieu, D.; Yssel, H.; Veas, F. Soluble HIV-1 gp120 enhances HIV-1 replication in non-dividing CD4+ T cells, mediated via cell signaling and Tat cofactor overexpression. AIDS 2005, 19, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. HIV: Cell binding and entry. Cold Spring Harb. Perspect. Med. 2012, 2, a006866. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Kräusslich, H.-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef]
- Beckford-Vera, D.R.; Flavell, R.R.; Seo, Y.; Martinez-Ortiz, E.; Aslam, M.; Thanh, C.; Fehrman, E.; Pardons, M.; Kumar, S.; Deitchman, A.N.; et al. First-in-human immunoPET imaging of HIV-1 infection using 89Zr-labeled VRC01 broadly neutralizing antibody. Nat. Commun. 2022, 13, 1219. [Google Scholar] [CrossRef]
- Samer, S.; Thomas, Y.; Araínga, M.; Carter, C.; Shirreff, L.M.; Arif, M.S.; Avita, J.M.; Frank, I.; McRaven, M.D.; Thuruthiyil, C.T.; et al. Blockade of TGF-β signaling reactivates HIV-1/SIV reservoirs and immune responses in vivo. JCI Insight 2022, 7, e162290. [Google Scholar] [CrossRef]
- Song, J.; Cai, Z.; White, A.G.; Jin, T.; Wang, X.; Kadayakkara, D.; Anderson, C.J.; Ambrose, Z.; Young, W.-B. Visualization and quantification of simian immunodeficiency virus-infected cells using non-invasive molecular imaging. J. Gen. Virol. 2015, 96, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, P.J.; Rogers, K.A.; Zurla, C.; Blanchard, E.L.; Gumber, S.; Strait, K.; Connor-Stroud, F.; Schuster, D.M.; Amancha, P.K.; Hong, J.J.; et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat. Methods 2015, 12, 427–432. [Google Scholar] [CrossRef]
- Klein, F.; Halper-Stromberg, A.; Horwitz, J.A.; Gruell, H.; Scheid, J.F.; Bournazos, S.; Mouquet, H.; Spatz, L.A.; Diskin, R.; Abadir, A.; et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 2012, 492, 118–122. [Google Scholar] [CrossRef]
- Leibman, R.S.; Richardson, M.W.; Ellebrecht, C.T.; Maldini, C.R.; Glover, J.A.; Secreto, A.J.; Kulikovskaya, I.; Lacey, S.F.; Akkina, S.R.; Yi, Y.; et al. Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor. PLoS Pathog. 2017, 13, e1006613. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Lederle, A.; Laumond, G.; Ducloy, C.; Schmidt, S.; Decoville, T.; Moog, C. Broadly neutralizing antibody VRC01 prevents HIV-1 transmission from plasmacytoid dendritic cells to CD4 T lymphocytes. J. Virol. 2014, 88, 10975–10981. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, A.; Karulkar, A.; Ghosh, S.; Rafiq, A.; Purwar, R. Lymphocytes in Cellular Therapy: Functional Regulation of CAR T Cells. Front. Immunol. 2018, 9, 3180. [Google Scholar] [CrossRef] [PubMed]
- Anthony-Gonda, K.; Ray, A.; Su, H.; Wang, Y.; Xiong, Y.; Lee, D.; Block, A.; Chilunda, V.; Weiselberg, J.; Zemelko, L.; et al. In vivo killing of primary HIV-infected cells by peripheral-injected early memory–enriched anti-HIV duoCAR T cells. JCI Insight 2022, 7, e161698. [Google Scholar] [CrossRef]
- Cohn, L.B.; Chomont, N.; Deeks, S.G. The Biology of the HIV-1 Latent Reservoir and Implications for Cure Strategies. Cell Host Microbe 2020, 27, 519–530. [Google Scholar] [CrossRef]
- Brozy, J.; Schlaepfer, E.; Mueller, C.K.S.; Rochat, M.-A.; Rampini, S.K.; Myburgh, R.; Raum, T.; Kufer, P.; Baeuerle, P.A.; Muenz, M.; et al. Antiviral Activity of HIV gp120-Targeting Bispecific T Cell Engager Antibody Constructs. J. Virol. 2018, 92, 14. [Google Scholar] [CrossRef]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suárez, I.; Oliveira, T.Y.; Lorenzi, J.C.C.; et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef]
- Bar-On, Y.; Gruell, H.; Schoofs, T.; Pai, J.A.; Nogueira, L.; Butler, A.L.; Millard, K.; Lehmann, C.; Suárez, I.; Oliveira, T.Y.; et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 2018, 24, 1701–1707. [Google Scholar] [CrossRef]
- Mascola, J.R.; Haynes, B.F. HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol. Rev. 2013, 254, 225–244. [Google Scholar] [CrossRef]
- Haynes, B.F.; Wiehe, K.; Borrow, P.; Saunders, K.O.; Korber, B.; Wagh, K.; McMichael, A.J.; Kelsoe, G.; Hahn, B.H.; Alt, F.; et al. Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies. Nat. Rev. Immunol. 2022, 23, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R.; Desrosiers, R.C.; Doms, R.W.; Koff, W.C.; Kwong, P.D.; Moore, J.P.; Nabel, G.J.; Sodroski, J.; Wilson, I.A.; Wyatt, R.T. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 2004, 5, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Prator, C.A.; Thanh, C.; Kumar, S.; Pan, T.; Peluso, M.J.; Bosch, R.; Jones, N.; Milush, J.M.; Bakkour, S.; Stone, M.; et al. Circulating CD30+CD4+ T Cells Increase Before Human Immunodeficiency Virus Rebound After Analytical Antiretroviral Treatment Interruption. J. Infect. Dis. 2020, 221, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, C. Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. Retroviral “Lifestyles”: Simple versus Complex; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Dufait, I.; Liechtenstein, T.; Lanna, A.; Bricogne, C.; Laranga, R.; Padella, A.; Breckpot, K.; Escors, D. Retroviral and lentiviral vectors for the induction of immunological tolerance. Scientifica 2012, 2012, 694137. [Google Scholar] [CrossRef]
- Escors, D.; Breckpot, K. Lentiviral vectors in gene therapy: Their current status and future potential. Arch. Immunol. Ther. Exp. 2010, 58, 107–119. [Google Scholar] [CrossRef]
- Milone, M.C.; O’Doherty, U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef]
- Chen, B.K.; Rousso, I.; Shim, S.; Kim, P.S. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 2001, 98, 15239–15244. [Google Scholar] [CrossRef]
- Shunaeva, A.; Potashnikova, D.; Pichugin, A.; Mishina, A.; Filatov, A.; Nikolaitchik, O.; Hu, W.-S.; Mazurov, D. Improvement of HIV-1 and Human T Cell Lymphotropic Virus Type 1 Replication-Dependent Vectors via Optimization of Reporter Gene Reconstitution and Modification with Intronic Short Hairpin RNA. J. Virol. 2015, 89, 10591–10601. [Google Scholar] [CrossRef]
- Nowrouzi, A.; Glimm, H.; von Kalle, C.; Schmidt, M. Retroviral vectors: Post entry events and genomic alterations. Viruses 2011, 3, 429–455. [Google Scholar] [CrossRef]
- Baum, C.; Schambach, A.; Bohne, J.; Galla, M. Retrovirus Vectors: Toward the Plentivirus? Mol. Ther. 2006, 13, 1050–1063. [Google Scholar] [CrossRef]
- Lambrechts, L.; Cole, B.; Rutsaert, S.; Trypsteen, W.; Vandekerckhove, L. Emerging PCR-Based Techniques to Study HIV-1 Reservoir Persistence. Viruses 2020, 12, 149. [Google Scholar] [CrossRef]
- Ramírez de Arellano, E.; Martín, C.; Soriano, V.; Alcamí, J.; Holguín, A. Genetic analysis of the long terminal repeat (LTR) promoter region in HIV-1-infected individuals with different rates of disease progression. Virus Genes 2007, 34, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Kalinichenko, S.; Komkov, D.; Mazurov, D. HIV-1 and HTLV-1 Transmission Modes: Mechanisms and Importance for Virus Spread. Viruses 2022, 14, 152. [Google Scholar] [CrossRef]
- Ramezani, A.; Hawley, R.G. Strategies to insulate lentiviral vector-expressed transgenes. Methods Mol. Biol. 2010, 614, 77–100. [Google Scholar] [PubMed]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Mali, S. Delivery systems for gene therapy. Indian J. Hum. Genet. 2013, 19, 3–8. [Google Scholar] [CrossRef]
- Naldini, L.; Blömer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Zufferey, R.; Dull, T.; Mandel, R.J.; Bukovsky, A.; Quiroz, D.; Naldini, L.; Trono, D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998, 72, 9873–9880. [Google Scholar] [CrossRef] [PubMed]
- Blömer, U.; Naldini, L.; Kafri, T.; Trono, D.; Verma, I.M.; Gage, F.H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 1997, 71, 6641–6649. [Google Scholar] [CrossRef]
- Challita, P.M.; Kohn, D.B. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc. Natl. Acad. Sci. USA 1994, 91, 2567–2571. [Google Scholar] [CrossRef]
- Münch, R.C.; Muth, A.; Muik, A.; Friedel, T.; Schmatz, J.; Dreier, B.; Trkola, A.; Plückthun, A.; Büning, H.; Buchholz, C.J. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 2015, 6, 6246. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Yuan, Z.; Li, J.; He, B.; Zheng, H.; Mayer, C.; Li, J.; Xiao, X. Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene Ther. 2011, 18, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Manno, C.S.; Pierce, G.F.; Arruda, V.R.; Glader, B.; Ragni, M.; Rasko, J.J.; Ozelo, M.C.; Hoots, K.; Blatt, P.; Konkle, B.; et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006, 12, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Kafri, T.; van Praag, H.; Gage, F.H.; Verma, I.M. Lentiviral vectors: Regulated gene expression. Mol. Ther. 2000, 1, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; La Russa, V.F.; Bao, L.; Kolls, J.; Schwarzenberger, P.; Reiser, J. Lentiviral vectors for sustained transgene expression in human bone marrow-derived stromal cells. Mol. Ther. 2002, 5, 555–565. [Google Scholar] [CrossRef]
- Kafri, T.; Blömer, U.; Peterson, D.A.; Gage, F.H.; Verma, I.M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat. Genet. 1997, 17, 314–317. [Google Scholar] [CrossRef]
- Anderson, J.S.; Javien, J.; Nolta, J.A.; Bauer, G. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5 alpha protein, a CCR5 shRNA, and a TAR decoy. Mol. Ther. 2009, 17, 2103–2114. [Google Scholar] [CrossRef]
- Barclay, S.L.; Yang, Y.; Zhang, S.; Fong, R.; Barraza, A.; Nolta, J.A.; Abedi, M.; Anderson, J.S. 296. Pre-Selection of Anti-HIV Lentiviral Vector Gene Modified Hematopoietic Stem Cells Significantly Improves Protection from HIV Infection: The Basis for a Future Clinical Trial. Mol. Ther. 2015, 23, S119. [Google Scholar] [CrossRef]
- Anderson, J.; Li, M.-J.; Palmer, B.; Remling, L.; Li, S.; Yam, P.; Yee, J.-K.; Rossi, J.; Zaia, J.; Akkina, R. Safety and Efficacy of a Lentiviral Vector Containing Three Anti-HIV Genes—CCR5 Ribozyme, Tat-rev siRNA, and TAR Decoy—In SCID-hu Mouse–Derived T Cells. Mol. Ther. 2007, 15, 1182–1188. [Google Scholar] [CrossRef]
- Braun, M.J.; Clements, J.E.; Gonda, M.A. The visna virus genome: Evidence for a hypervariable site in the env gene and sequence homology among lentivirus envelope proteins. J. Virol. 1987, 61, 4046–4054. [Google Scholar] [CrossRef]
- Hong, F.; Jacobs, J.L.; Aga, E.; Cillo, A.R.; Fyne, E.; Koontz, D.L.; Zheng, L.; Mellors, J.W. Associations between HIV-1 DNA copy number, proviral transcriptional activity, and plasma viremia in individuals off or on suppressive antiretroviral therapy. Virology 2018, 521, 51–57. [Google Scholar] [CrossRef]
- Avettand-Fènoël, V.; Hocqueloux, L.; Ghosn, J.; Cheret, A.; Frange, P.; Melard, A.; Viard, J.-P.; Rouzioux, C. Total HIV-1 DNA, a Marker of Viral Reservoir Dynamics with Clinical Implications. Clin. Microbiol. Rev. 2016, 29, 859–880. [Google Scholar] [CrossRef]
- Schnittman, S.M.; Psallidopoulos, M.C.; Lane, H.C.; Thompson, L.; Baseler, M.; Massari, F.; Fox, C.H.; Salzman, N.P.; Fauci, A.S. The reservoir for HIV-1 in human peripheral blood is a T cell that maintains expression of CD4. Science 1989, 245, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Henrich, T.J.; Gallien, S.; Li, J.Z.; Pereyra, F.; Kuritzkes, D.R. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods 2012, 186, 68–72. [Google Scholar] [CrossRef]
- Hong, F.; Aga, E.; Cillo, A.R.; Yates, A.L.; Besson, G.; Fyne, E.; Koontz, D.L.; Jennings, C.; Zheng, L.; Mellors, J.W. Novel Assays for Measurement of Total Cell-Associated HIV-1 DNA and RNA. J. Clin. Microbiol. 2016, 54, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Gibellini, D.; Vitone, F.; Schiavone, P.; Ponti, C.; La Placa, M.; Re, M.C. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. J. Clin. Virol. 2004, 29, 282–289. [Google Scholar] [CrossRef]
- Benveniste, O.; Flahault, A.; Rollot, F. Level Regeneration of CD4+ Cells in HIV-1—Infected Patients Receiving Highly Active Antiretroviral Therapy Who Have Prolonged Undetectable Plasma Viral Loads. J. Infect. 2005, 191, 1670–1679. Available online: https://academic.oup.com/jid/article-abstract/191/10/1670/789138 (accessed on 28 March 2023).
- Poizot-Martin, I.; Faucher, O.; Obry-Roguet, V.; Nicolino-Brunet, C.; Ronot-Bregigeon, S.; Dignat-George, F.; Tamalet, C. Lack of correlation between the size of HIV proviral DNA reservoir and the level of immune activation in HIV-infected patients with a sustained undetectable HIV viral load for 10 years. J. Clin. Virol. 2013, 57, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, P.R.; Whaley, M.; Montaner, J.S. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS 1999, 13, F59–F62. [Google Scholar] [CrossRef]
- Bruner, K.M.; Wang, Z.; Simonetti, F.R.; Bender, A.M.; Kwon, K.J.; Sengupta, S.; Fray, E.J.; Beg, S.A.; Antar, A.A.R.; Jenike, K.M.; et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019, 566, 120–125. [Google Scholar] [CrossRef]
- Hoxie, J.A.; June, C.H. Novel cell and gene therapies for HIV. Cold Spring Harb. Perspect. Med. 2012, 2, a007179. [Google Scholar] [CrossRef]
- Haworth, K.G.; Peterson, C.W.; Kiem, H.-P. CCR5-edited gene therapies for HIV cure: Closing the door to viral entry. Cytotherapy 2017, 19, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Strayer, D.S.; Akkina, R.; Bunnell, B.A.; Dropulic, B.; Planelles, V.; Pomerantz, R.J.; Rossi, J.J.; Zaia, J.A. Current status of gene therapy strategies to treat HIV/AIDS. Mol. Ther. 2005, 11, 823–842. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Guo, D.; Chen, S. Application of CRISPR/Cas9-Based Gene Editing in HIV-1/AIDS Therapy. Front. Cell Infect. Microbiol. 2019, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Mautino, M.R. Lentiviral vectors for gene therapy of HIV-1 infection. Curr. Gene Ther. 2002, 2, 23–43. [Google Scholar]
- Scholler, J.; Brady, T.L.; Binder-Scholl, G.; Hwang, W.-T.; Plesa, G.; Hege, K.M.; Vogel, A.N.; Kalos, M.; Riley, J.L.; Deeks, S.G.; et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med. 2012, 4, 132ra53. [Google Scholar] [CrossRef] [PubMed]
- Labbé, R.P.; Vessillier, S.; Rafiq, Q.A. Lentiviral Vectors for T Cell Engineering: Clinical Applications, Bioprocessing and Future Perspectives. Viruses 2021, 13, 1528. [Google Scholar] [CrossRef]
- Hongeng, S.; Anurathapan, U.; Songdej, D.; Phuphuakrat, A.; Jongrak, K.; Parsons, G.; Deary, B.; Bonner, M.; Veres, G.; Asmal, M. Wild-type HIV infection after treatment with lentiviral gene therapy for β-thalassemia. Blood Adv. 2021, 5, 2701–2706. [Google Scholar] [CrossRef]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef]
- Wu, X.; Wakefield, J.K.; Liu, H.; Xiao, H.; Kralovics, R.; Prchal, J.T.; Kappes, J.C. Development of a novel trans-lentiviral vector that affords predictable safety. Mol. Ther. 2000, 2, 47–55. [Google Scholar] [CrossRef]
- Zhen, A.; Peterson, C.W.; Carrillo, M.A.; Reddy, S.S.; Youn, C.S.; Lam, B.B.; Chang, N.Y.; Martin, H.A.; Rick, J.W.; Kim, J.; et al. Long-term persistence and function of hematopoietic stem cell-derived chimeric antigen receptor T cells in a nonhuman primate model of HIV/AIDS. PLoS Pathog. 2017, 13, e1006753. [Google Scholar] [CrossRef]
- Walker, J.E.; Chen, R.X.; McGee, J.; Nacey, C.; Pollard, R.B.; Abedi, M.; Bauer, G.; Nolta, J.A.; Anderson, J.S. Generation of an HIV-1-Resistant Immune System with CD34+ Hematopoietic Stem Cells Transduced with a Triple-Combination Anti-HIV Lentiviral Vector. J. Virol. 2012, 86, 5719–5729. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Carter, N.P. The new cytogenetics: Blurring the boundaries with molecular biology. Nat. Rev. Genet. 2005, 6, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Hanley, M.B.; Lomas, W.; Mittar, D.; Maino, V.; Park, E. Detection of low abundance RNA molecules in individual cells by flow cytometry. PLoS ONE 2013, 8, e57002. [Google Scholar] [CrossRef] [PubMed]
- Bauman, J.G.; Bayer, J.A.; van Dekken, H. Fluorescent in-situ hybridization to detect cellular RNA by flow cytometry and confocal microscopy. J. Microsc. 1990, 157, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Lassen, K.G.; Bailey, J.R.; Siliciano, R.F. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 2004, 78, 9105–9114. [Google Scholar] [CrossRef]
- Li, H.; Tang, Y.; Wang, Y.; Li, Y.; Yang, Y.; Liao, K.; Song, F.; Deng, S.; Chen, Y. Single-cell sequencing resolves the landscape of immune cells and regulatory mechanisms in HIV-infected immune non-responders. Cell Death Dis. 2022, 13, 1–12. [Google Scholar] [CrossRef]
- Svoboda, M.; Frost, H.R.; Bosco, G. Internal oligo (dT) priming introduces systematic bias in bulk and single-cell RNA sequencing count data. NAR Genom. Bioinform. 2022, 4, lqac035. [Google Scholar] [CrossRef]
- Blache, U.; Weiss, R.; Boldt, A.; Kapinsky, M.; Blaudszun, A.-R.; Quaiser, A.; Pohl, A.; Miloud, T.; Burgaud, M.; Vucinic, V.; et al. Advanced Flow Cytometry Assays for Immune Monitoring of CAR-T Cell Applications. Front. Immunol. 2021, 12, 658314. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).