Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Plasmids That Express ASFV Proteins Fused to EGFP
2.2. Cell Culture and Transfections
2.3. EGFP Coimmunoprecipitations
2.4. Sample Digestion
2.5. Mass Spectrometry
2.6. Mass Spectrometry Data Analysis
2.7. Identification of the Interacting Partners of ASFV Proteins
2.8. Identification of Protein Networks and Common and Unique Pathways in the Interactomes Using Software Analysis
2.9. Validation of the MS Results
2.10. Viruses and Infection
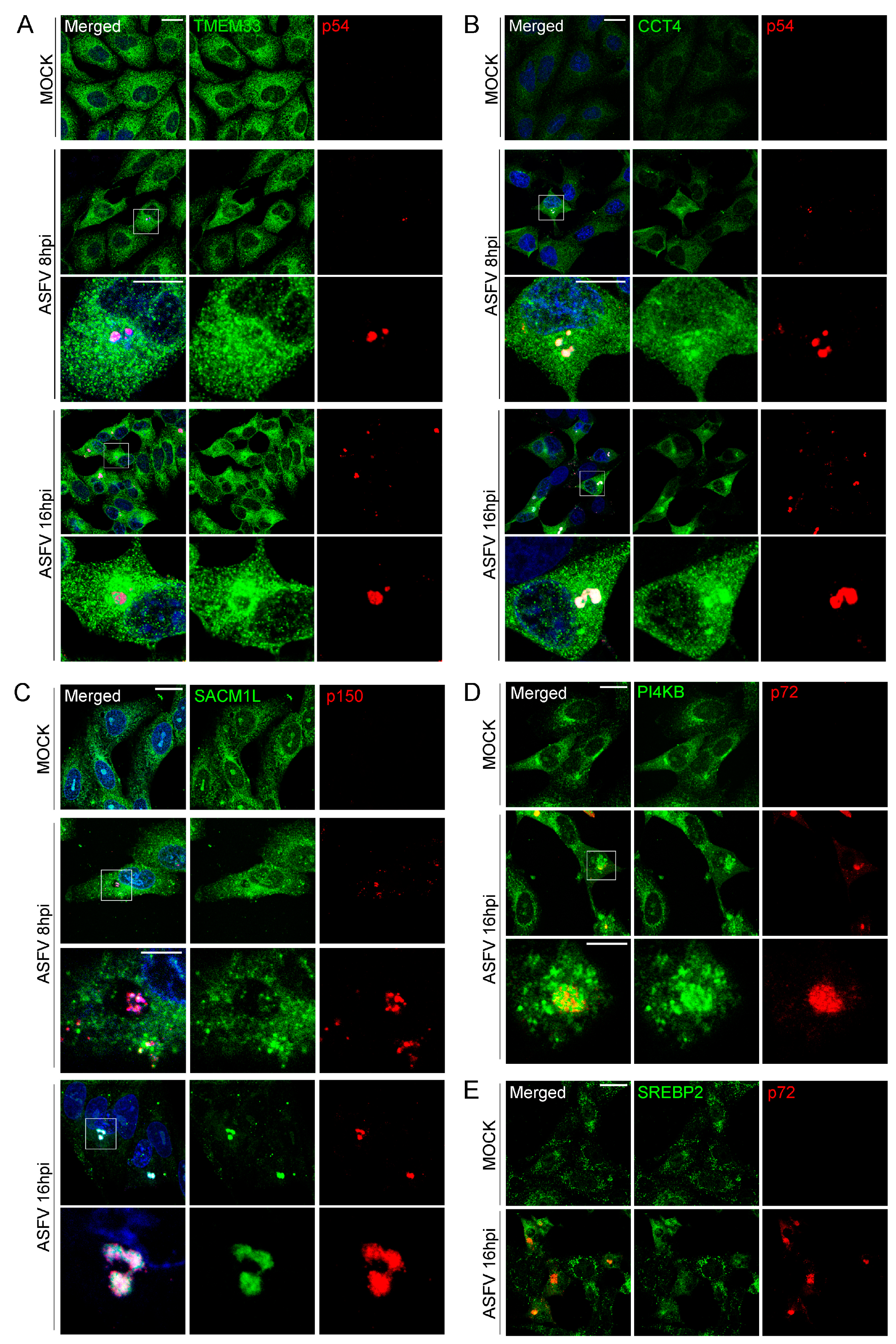
2.11. Indirect Immunofluorescence and Antibodies, Conventional and Confocal Microscopy
2.12. Experiments with Inhibitors
3. Results
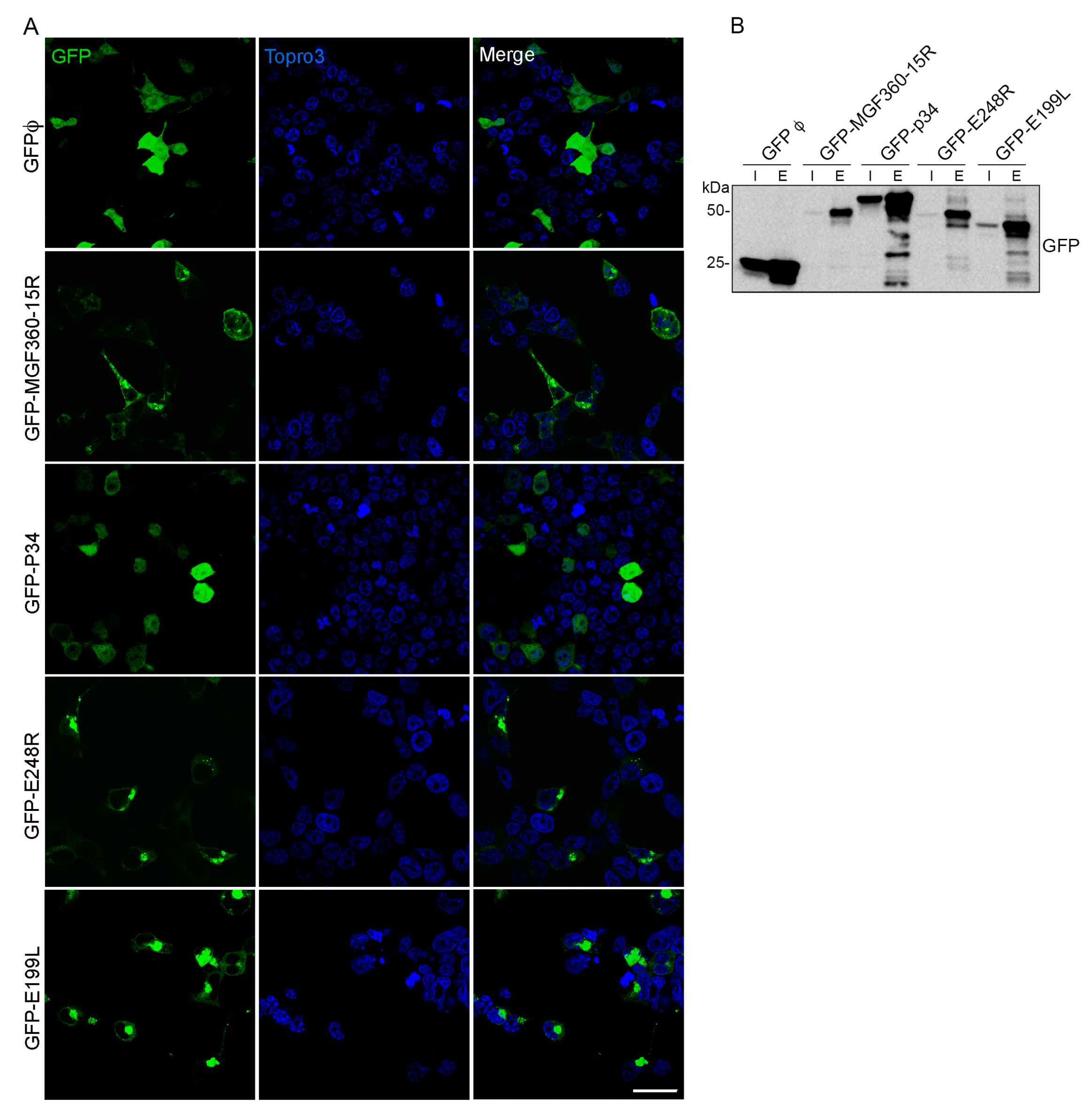
3.1. Expression of ASFV VP in Cells
3.2. Identification of the Potential Cellular Interacting Partners of MGF360-15R, P34, E248R and E199L ASFV Proteins
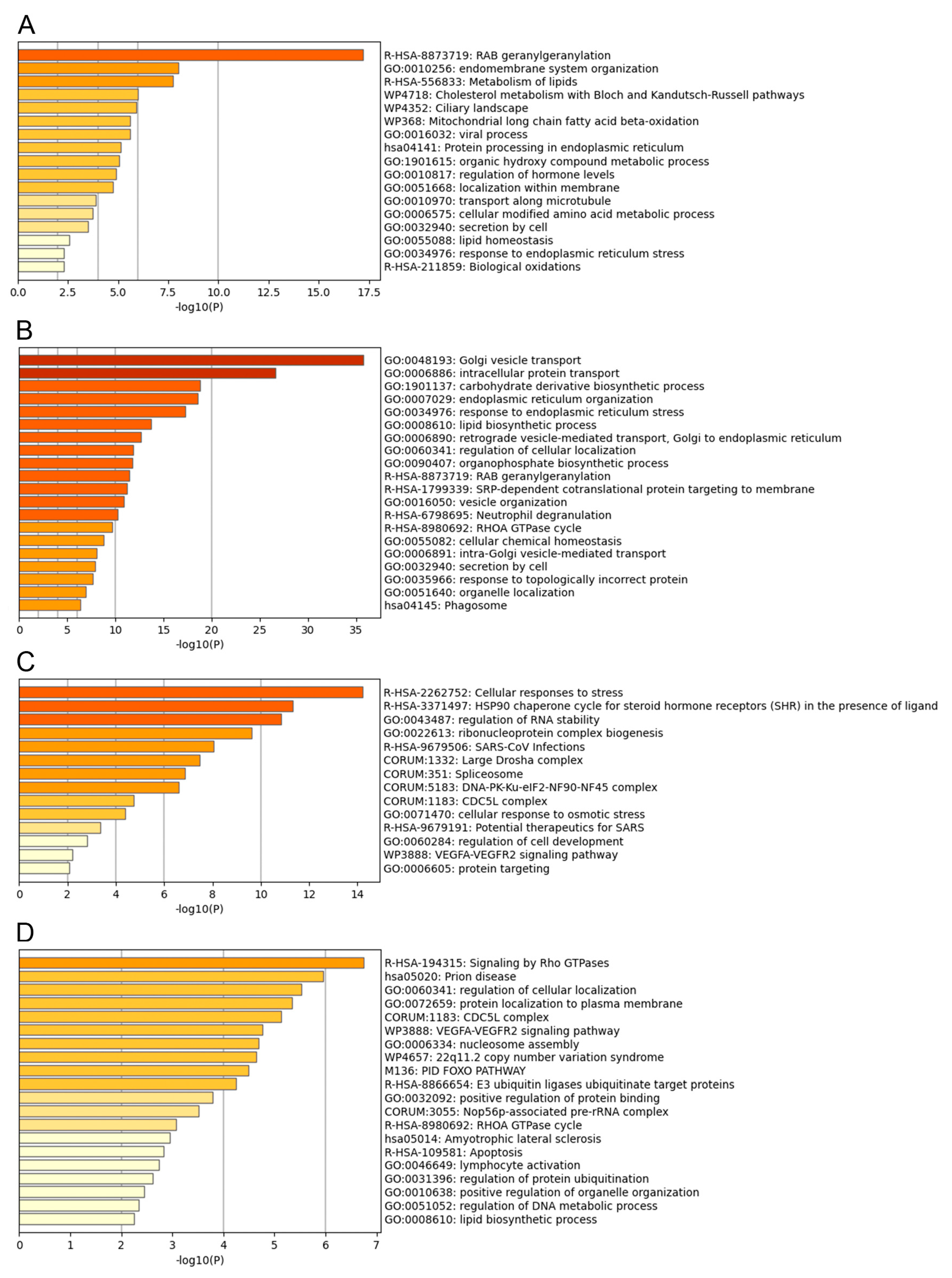
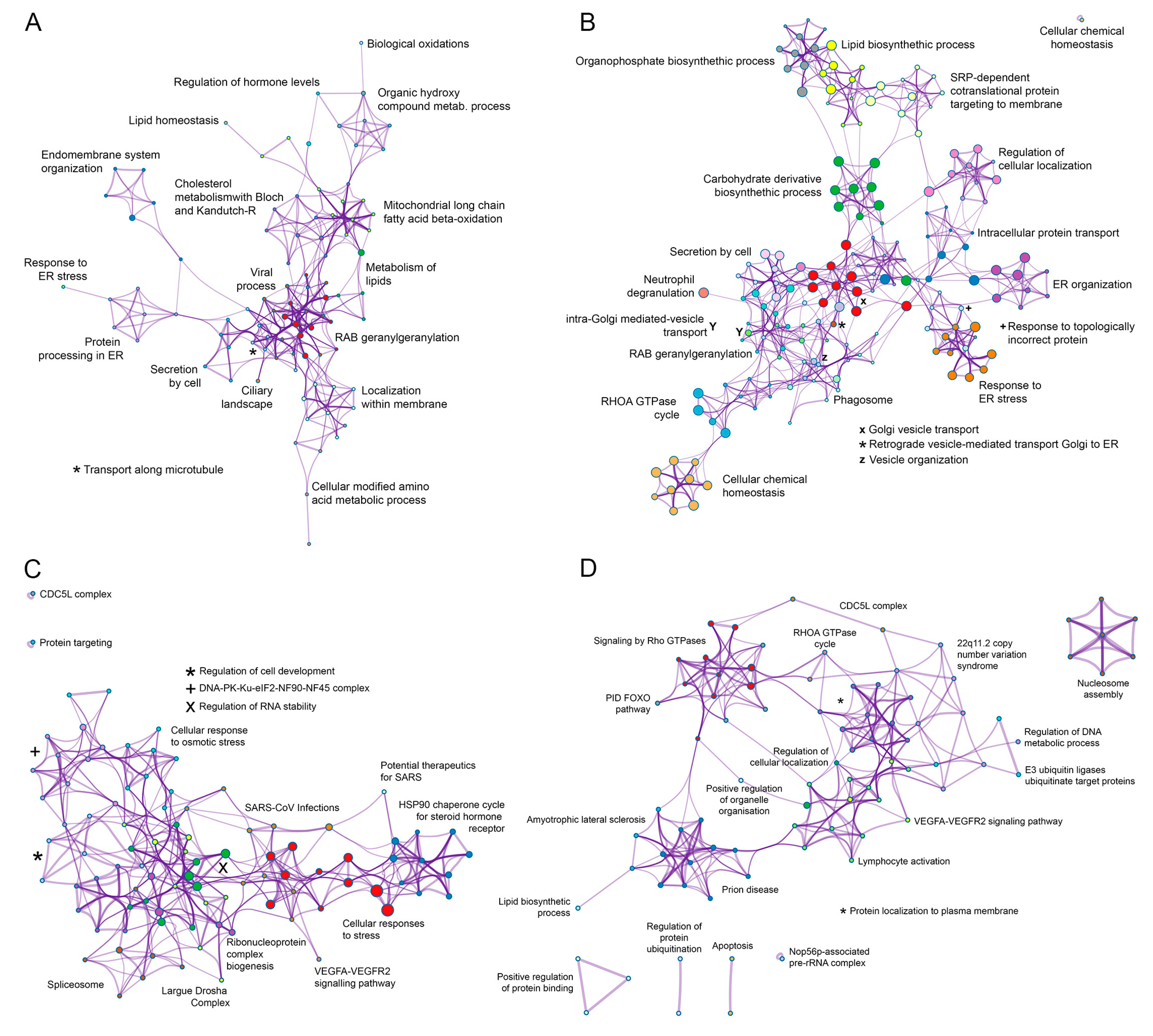
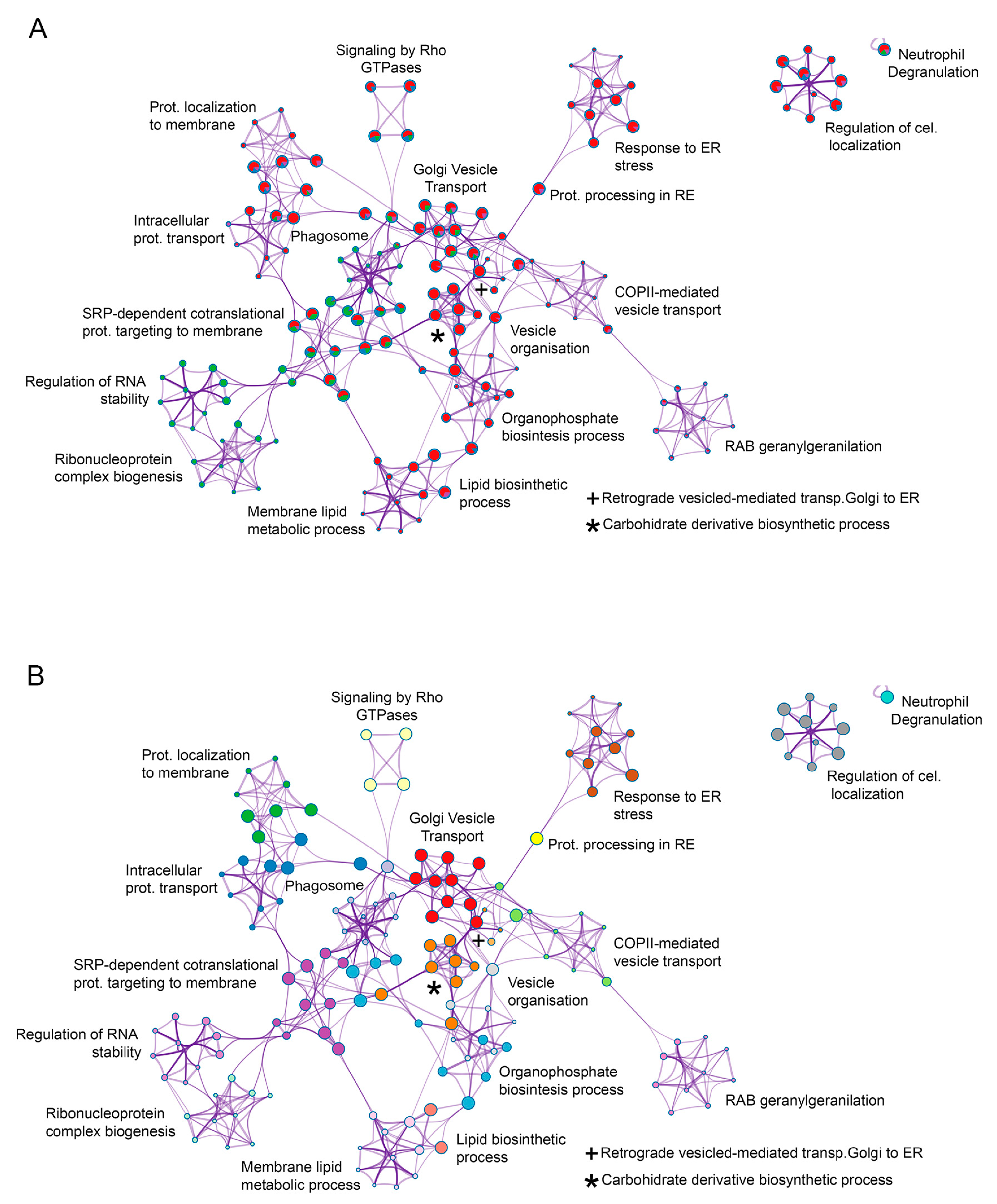
3.3. Functional Analysis of the ASFV Interactome Proteins
3.4. Common Interactomes and Pathways among ASFV Viral Proteins
3.5. Validation of MGF360-15R, P34, E248R and E199L Interaction Partners through Immunoprecipitation Analysis and Western Blot
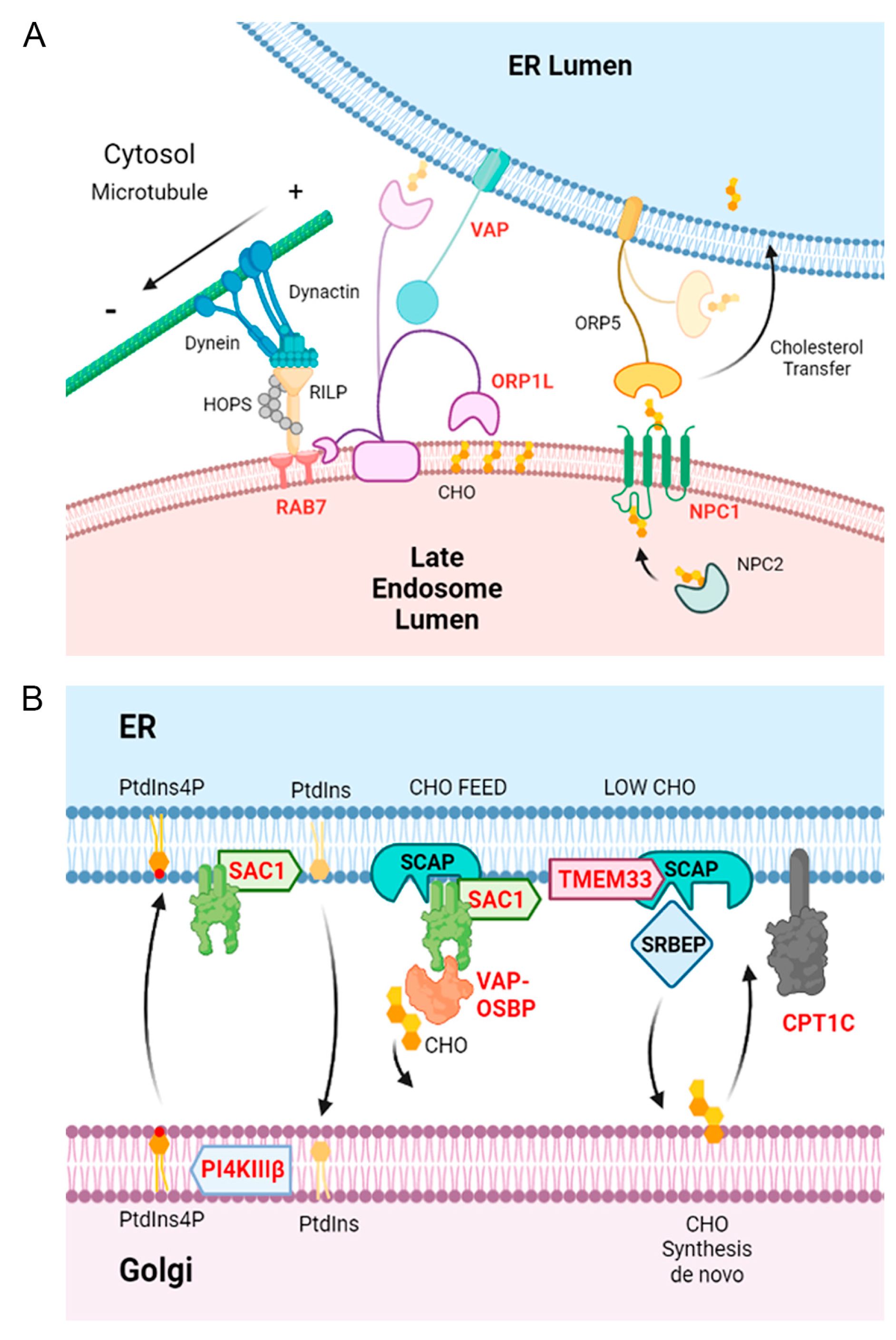
3.6. Inhibition of Lipid Metabolism and Other Significant Pathways in Vero Cells and Macrophages

4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gonzales, W.; Moreno, C.; Duran, U.; Henao, N.; Bencosme, M.; Lora, P.; Reyes, R.; Nunez, R.; De Gracia, A.; Perez, A.M. African swine fever in the Dominican Republic. Transbound. Emerg. Dis. 2021, 68, 3018–3019. [Google Scholar] [CrossRef]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Nguyen, C.T.; Chu, N.T.; Nguyen, H.T.; Dang, H.V. Genetic characterization of African swine fever viruses circulating in North Central region of Vietnam. Transbound. Emerg. Dis. 2021, 68, 1697–1699. [Google Scholar] [CrossRef]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Alonso, C.; Miskin, J.; Hernaez, B.; Fernandez-Zapatero, P.; Soto, L.; Canto, C.; Rodriguez-Crespo, I.; Dixon, L.; Escribano, J.M. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001, 75, 9819–9827. [Google Scholar] [CrossRef]
- Cuesta-Geijo, M.A.; Garcia-Dorival, I.; Del Puerto, A.; Urquiza, J.; Galindo, I.; Barrado-Gil, L.; Lasala, F.; Cayuela, A.; Sorzano, C.O.S.; Gil, C.; et al. New insights into the role of endosomal proteins for African swine fever virus infection. PLoS Pathog. 2022, 18, e1009784. [Google Scholar] [CrossRef]
- Hernaez, B.; Escribano, J.M.; Alonso, C. African swine fever virus protein p30 interaction with heterogeneous nuclear ribonucleoprotein K (hnRNP-K) during infection. FEBS Lett. 2008, 582, 3275–3280. [Google Scholar] [CrossRef]
- Munoz-Moreno, R.; Cuesta-Geijo, M.A.; Martinez-Romero, C.; Barrado-Gil, L.; Galindo, I.; Garcia-Sastre, A.; Alonso, C. Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PLoS ONE 2016, 11, e0154366. [Google Scholar] [CrossRef]
- Chen, X.; Chen, X.; Liang, Y.; Xu, S.; Weng, Z.; Gao, Q.; Huang, Z.; Zhang, G.; Gong, L. Interaction network of African swine fever virus structural protein p30 with host proteins. Front. Microbiol. 2022, 13, 971888. [Google Scholar] [CrossRef]
- Hao, Y.; Yang, J.; Yang, B.; Zhang, T.; Shi, X.; Yang, X.; Zhang, D.; Zhao, D.; Yan, W.; Chen, L.; et al. Identification and analysis of the interaction network of African swine fever virus D1133L with host proteins. Front. Microbiol. 2022, 13, 1037346. [Google Scholar] [CrossRef]
- Matamoros, T.; Alejo, A.; Rodriguez, J.M.; Hernaez, B.; Guerra, M.; Fraile-Ramos, A.; Andres, G. African Swine Fever Virus Protein pE199L Mediates Virus Entry by Enabling Membrane Fusion and Core Penetration. mBio 2020, 11, e03340-21. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Eisenhaber, F. Myristoylation of viral and bacterial proteins. Trends Microbiol. 2004, 12, 178–185. [Google Scholar] [CrossRef]
- Cuesta-Geijo, M.A.; Galindo, I.; Hernaez, B.; Quetglas, J.I.; Dalmau-Mena, I.; Alonso, C. Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry. PLoS ONE 2012, 7, e48853. [Google Scholar] [CrossRef]
- Carrascosa, A.L.; Santarén, J.F.; Viñuela, E. Production and titration of African swine fever virus in porcine alveolar macrophages. J. Virol. Methods 1982, 3, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dorival, I.; Wu, W.; Armstrong, S.D.; Barr, J.N.; Carroll, M.W.; Hewson, R.; Hiscox, J.A. Elucidation of the Cellular Interactome of Ebola Virus Nucleoprotein and Identification of Therapeutic Targets. J. Proteome Res. 2016, 15, 4290–4303. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dorival, I.; Wu, W.; Dowall, S.; Armstrong, S.; Touzelet, O.; Wastling, J.; Barr, J.N.; Matthews, D.; Carroll, M.; Hewson, R.; et al. Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J. Proteome Res. 2014, 13, 5120–5135. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Aljabr, W.; Armstrong, S.; Rickett, N.Y.; Pollakis, G.; Touzelet, O.; Cloutman-Green, E.; Matthews, D.A.; Hiscox, J.A. High Resolution Analysis of Respiratory Syncytial Virus Infection In Vivo. Viruses 2019, 11, 926. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Zar, J.H. Biostatistical Analysis, 4th ed.; Prentice Hall International Prentice-Hall International (UK): Upper Saddle River, NJ, USA; London, UK, 1999. [Google Scholar]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Enjuanes, L.; Carrascosa, A.L.; Moreno, M.A.; Vinuela, E. Titration of African swine fever (ASF) virus. J. Gen. Virol. 1976, 32, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Hernaez, B.; Escribano, J.M.; Alonso, C. Visualization of the African swine fever virus infection in living cells by incorporation into the virus particle of green fluorescent protein-p54 membrane protein chimera. Virology 2006, 350, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava-Ranjan, P.; Flint, M.; Bergeron, E.; McElroy, A.K.; Chatterjee, P.; Albarino, C.G.; Nichol, S.T.; Spiropoulou, C.F. Statins Suppress Ebola Virus Infectivity by Interfering with Glycoprotein Processing. mBio 2018, 9, e00660-18. [Google Scholar] [CrossRef]
- Osuna-Ramos, J.F.; Reyes-Ruiz, J.M.; Del Ángel, R.M. The Role of Host Cholesterol During Flavivirus Infection. Front. Cell. Infect. Microbiol. 2018, 8, 388. [Google Scholar] [CrossRef] [PubMed]
- Abosheasha, M.A.; El-Gowily, A.H. Superiority of cilostazol among antiplatelet FDA-approved drugs against COVID 19 M(pro) and spike protein: Drug repurposing approach. Drug Dev. Res. 2021, 82, 217–229. [Google Scholar] [CrossRef]
- Schror, K. The pharmacology of cilostazol. Diabetes Obes. Metab. 2002, 4 (Suppl. S2), S14–S19. [Google Scholar] [CrossRef]
- Favari, E.; Zanotti, I.; Zimetti, F.; Ronda, N.; Bernini, F.; Rothblat, G.H. Probucol inhibits ABCA1-mediated cellular lipid efflux. Arter. Thromb. Vasc. Biol. 2004, 24, 2345–2350. [Google Scholar] [CrossRef]
- Morkholt, A.S.; Oklinski, M.K.; Larsen, A.; Bockermann, R.; Issazadeh-Navikas, S.; Nieland, J.G.K.; Kwon, T.H.; Corthals, A.; Nielsen, S.; Nieland, J.D.V. Pharmacological inhibition of carnitine palmitoyl transferase 1 inhibits and reverses experimental autoimmune encephalitis in rodents. PLoS ONE 2020, 15, e0234493. [Google Scholar] [CrossRef]
- Matsuda, D.; Namatame, I.; Ohshiro, T.; Ishibashi, S.; Omura, S.; Tomoda, H. Anti-atherosclerotic activity of triacsin C, an acyl-CoA synthetase inhibitor. J. Antibiot. 2008, 61, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Trinkle-Mulcahy, L. Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics 2012, 12, 1623–1638. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008, 183, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Munday, D.; Bickerton, E.; Britton, P.; Rodgers, M.A.; Whitehouse, A.; Zhou, E.M.; Hiscox, J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013, 87, 9486–9500. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Tran, K.C.; Teng, M.N.; Heesom, K.J.; Matthews, D.A.; Barr, J.N.; Hiscox, J.A. The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 2012, 86, 7777–7789. [Google Scholar] [CrossRef]
- Altan-Bonnet, N.; Balla, T. Phosphatidylinositol 4-kinases: Hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 2012, 37, 293–302. [Google Scholar] [CrossRef]
- Dorobantu, C.M.; Albulescu, L.; Harak, C.; Feng, Q.; van Kampen, M.; Strating, J.R.; Gorbalenya, A.E.; Lohmann, V.; van der Schaar, H.M.; van Kuppeveld, F.J. Modulation of the Host Lipid Landscape to Promote RNA Virus Replication: The Picornavirus Encephalomyocarditis Virus Converges on the Pathway Used by Hepatitis C Virus. PLoS Pathog. 2015, 11, e1005185. [Google Scholar] [CrossRef]
- Wong, M.T.; Chen, S.S. Hepatitis C Virus Subverts Human Choline Kinase-alpha To Bridge Phosphatidylinositol-4-Kinase IIIalpha (PI4KIIIalpha) and NS5A and Upregulates PI4KIIIalpha Activation, Thereby Promoting the Translocation of the Ternary Complex to the Endoplasmic Reticulum for Viral Replication. J. Virol. 2017, 91, e00355-17. [Google Scholar] [CrossRef]
- Cabukusta, B.; Berlin, I.; van Elsland, D.M.; Forkink, I.; Spits, M.; de Jong, A.W.M.; Akkermans, J.; Wijdeven, R.H.M.; Janssen, G.M.C.; van Veelen, P.A.; et al. Human VAPome Analysis Reveals MOSPD1 and MOSPD3 as Membrane Contact Site Proteins Interacting with FFAT-Related FFNT Motifs. Cell Rep. 2020, 33, 108475. [Google Scholar] [CrossRef]
- Cuesta-Geijo, M.A.; Barrado-Gil, L.; Galindo, I.; Munoz-Moreno, R.; Alonso, C. Redistribution of Endosomal Membranes to the African Swine Fever Virus Replication Site. Viruses 2017, 9, 133. [Google Scholar] [CrossRef]
- Laliberte, J.P.; Weisberg, A.S.; Moss, B. The membrane fusion step of vaccinia virus entry is cooperatively mediated by multiple viral proteins and host cell components. PLoS Pathog. 2011, 7, e1002446. [Google Scholar] [CrossRef] [PubMed]
- Schin, A.M.; Diesterbeck, U.S.; Moss, B. Insights into the Organization of the Poxvirus Multicomponent Entry-Fusion Complex from Proximity Analyses in Living Infected Cells. J. Virol. 2021, 95, e0085221. [Google Scholar] [CrossRef]
- Jongsma, M.L.M.; Bakker, N.; Neefjes, J. Choreographing the motor-driven endosomal dance. J. Cell Sci. 2023, 136, jcs259689. [Google Scholar] [CrossRef] [PubMed]
- Jordens, I.; Marsman, M.; Kuijl, C.; Neefjes, J. Rab proteins, connecting transport and vesicle fusion. Traffic 2005, 6, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Hoffenberg, S.; Liu, X.; Nikolova, L.; Hall, H.S.; Dai, W.; Baughn, R.E.; Dickey, B.F.; Barbieri, M.A.; Aballay, A.; Stahl, P.D.; et al. A novel membrane-anchored Rab5 interacting protein required for homotypic endosome fusion. J. Biol. Chem. 2000, 275, 24661–24669. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Cantalupo, G.; Alifano, P.; Roberti, V.; Bruni, C.B.; Bucci, C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001, 20, 683–693. [Google Scholar] [CrossRef]
- Johansson, M.; Lehto, M.; Tanhuanpaa, K.; Cover, T.L.; Olkkonen, V.M. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol. Biol. Cell 2005, 16, 5480–5492. [Google Scholar] [CrossRef]
- Strunnikova, N.V.; Barb, J.; Sergeev, Y.V.; Thiagarajasubramanian, A.; Silvin, C.; Munson, P.J.; Macdonald, I.M. Loss-of-function mutations in Rab escort protein 1 (REP-1) affect intracellular transport in fibroblasts and monocytes of choroideremia patients. PLoS ONE 2009, 4, e8402. [Google Scholar] [CrossRef]
- Esposito, G.; De Falco, F.; Tinto, N.; Testa, F.; Vitagliano, L.; Tandurella, I.C.; Iannone, L.; Rossi, S.; Rinaldi, E.; Simonelli, F.; et al. Comprehensive mutation analysis (20 families) of the choroideremia gene reveals a missense variant that prevents the binding of REP1 with Rab geranylgeranyl transferase. Hum. Mutat. 2011, 32, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Oehlke, O.; Martin, H.W.; Osterberg, N.; Roussa, E. Rab11b and its effector Rip11 regulate the acidosis-induced traffic of V-ATPase in salivary ducts. J. Cell Physiol. 2011, 226, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Knudsen, G.M.; Betegon, M.; Burlingame, A.L.; DeRisi, J.L. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. mBio 2013, 4, e00098-00013. [Google Scholar] [CrossRef]
- de la Mora, E.; Dezi, M.; Di Cicco, A.; Bigay, J.; Gautier, R.; Manzi, J.; Polidori, J.; Castano-Diez, D.; Mesmin, B.; Antonny, B.; et al. Nanoscale architecture of a VAP-A-OSBP tethering complex at membrane contact sites. Nat. Commun. 2021, 12, 3459. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Neefjes, J. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 2017, 47, 1–8. [Google Scholar] [CrossRef]
- Gennerich, A.; Vale, R.D. Walking the walk: How kinesin and dynein coordinate their steps. Curr. Opin. Cell Biol. 2009, 21, 59–67. [Google Scholar] [CrossRef]
- van der Kant, R.; Fish, A.; Janssen, L.; Janssen, H.; Krom, S.; Ho, N.; Brummelkamp, T.; Carette, J.; Rocha, N.; Neefjes, J. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J. Cell Sci. 2013, 126, 3462–3474. [Google Scholar] [CrossRef]
- Rocha, N.; Kuijl, C.; van der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 2009, 185, 1209–1225. [Google Scholar] [CrossRef]
- Venditti, R.; Masone, M.C.; Rega, L.R.; Di Tullio, G.; Santoro, M.; Polishchuk, E.; Serrano, I.C.; Olkkonen, V.M.; Harada, A.; Medina, D.L.; et al. The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1. J. Cell Biol. 2019, 218, 783–797. [Google Scholar] [CrossRef]
- Hamamoto, I.; Nishimura, Y.; Okamoto, T.; Aizaki, H.; Liu, M.; Mori, Y.; Abe, T.; Suzuki, T.; Lai, M.M.; Miyamura, T.; et al. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 2005, 79, 13473–13482. [Google Scholar] [CrossRef]
- Wakana, Y.; Hayashi, K.; Nemoto, T.; Watanabe, C.; Taoka, M.; Angulo-Capel, J.; Garcia-Parajo, M.F.; Kumata, H.; Umemura, T.; Inoue, H.; et al. The ER cholesterol sensor SCAP promotes CARTS biogenesis at ER-Golgi membrane contact sites. J. Cell Biol. 2021, 220, e202002150. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Song, J.; He, Y.; Liu, Y.; Liu, Z.; Sun, W.; Hu, W.; Lei, Q.Y.; Hu, X.; Chen, Z.; et al. CRISPR/Cas9 Screens Reveal that Hexokinase 2 Enhances Cancer Stemness and Tumorigenicity by Activating the ACSL4-Fatty Acid beta-Oxidation Pathway. Adv. Sci. 2022, 9, e2105126. [Google Scholar] [CrossRef]
- Reeves, A.R.; Sansbury, B.E.; Pan, M.; Han, X.; Spite, M.; Greenberg, A.S. Myeloid-Specific Deficiency of Long-Chain Acyl CoA Synthetase 4 Reduces Inflammation by Remodeling Phospholipids and Reducing Production of Arachidonic Acid-Derived Proinflammatory Lipid Mediators. J. Immunol. 2021, 207, 2744–2753. [Google Scholar] [CrossRef] [PubMed]
- Quetglas, J.I.; Hernaez, B.; Galindo, I.; Munoz-Moreno, R.; Cuesta-Geijo, M.A.; Alonso, C. Small rho GTPases and cholesterol biosynthetic pathway intermediates in African swine fever virus infection. J. Virol. 2012, 86, 1758–1767. [Google Scholar] [CrossRef]
- Sierra, A.Y.; Gratacos, E.; Carrasco, P.; Clotet, J.; Urena, J.; Serra, D.; Asins, G.; Hegardt, F.G.; Casals, N. CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. J. Biol. Chem. 2008, 283, 6878–6885. [Google Scholar] [CrossRef] [PubMed]
- Palomo-Guerrero, M.; Fado, R.; Casas, M.; Perez-Montero, M.; Baena, M.; Helmer, P.O.; Dominguez, J.L.; Roig, A.; Serra, D.; Hayen, H.; et al. Sensing of nutrients by CPT1C regulates late endosome/lysosome anterograde transport and axon growth. Elife 2019, 8, e51063. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, G.; Wang, Y.; Cai, L.; Qian, K.; Ju, L.; Liu, X.; Xiao, Y.; Wang, X. Fatty acid oxidation inhibitor etomoxir suppresses tumor progression and induces cell cycle arrest via PPARgamma-mediated pathway in bladder cancer. Clin. Sci. 2019, 133, 1745–1758. [Google Scholar] [CrossRef]
- Reinisch, K.M.; Prinz, W.A. Mechanisms of nonvesicular lipid transport. J. Cell Biol. 2021, 220, e202012058. [Google Scholar] [CrossRef]
- Casas, M.; Fado, R.; Dominguez, J.L.; Roig, A.; Kaku, M.; Chohnan, S.; Sole, M.; Unzeta, M.; Minano-Molina, A.J.; Rodriguez-Alvarez, J.; et al. Sensing of nutrients by CPT1C controls SAC1 activity to regulate AMPA receptor trafficking. J. Cell Biol. 2020, 219, e201912045. [Google Scholar] [CrossRef]
- Urade, T.; Yamamoto, Y.; Zhang, X.; Ku, Y.; Sakisaka, T. Identification and characterization of TMEM33 as a reticulon-binding protein. Kobe J. Med. Sci. 2014, 60, E57–E65. [Google Scholar]
- Yamamoto, Y.; Yoshida, A.; Miyazaki, N.; Iwasaki, K.; Sakisaka, T. Arl6IP1 has the ability to shape the mammalian ER membrane in a reticulon-like fashion. Biochem. J. 2014, 458, 69–79. [Google Scholar] [CrossRef]
- Ikonen, E. Mechanisms of cellular cholesterol compartmentalization: Recent insights. Curr. Opin. Cell Biol. 2018, 53, 77–83. [Google Scholar] [CrossRef]
- Galindo, I.; Garaigorta, U.; Lasala, F.; Cuesta-Geijo, M.A.; Bueno, P.; Gil, C.; Delgado, R.; Gastaminza, P.; Alonso, C. Antiviral drugs targeting endosomal membrane proteins inhibit distant animal and human pathogenic viruses. Antivir. Res. 2021, 186, 104990. [Google Scholar] [CrossRef]
- Carette, J.E.; Raaben, M.; Wong, A.C.; Herbert, A.S.; Obernosterer, G.; Mulherkar, N.; Kuehne, A.I.; Kranzusch, P.J.; Griffin, A.M.; Ruthel, G.; et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011, 477, 340–343. [Google Scholar] [CrossRef]
- Côté, M.; Misasi, J.; Ren, T.; Bruchez, A.; Lee, K.; Filone, C.M.; Hensley, L.; Li, Q.; Ory, D.; Chandran, K.; et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011, 477, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dorival, I.; Cuesta-Geijo, M.A.; Barrado-Gil, L.; Galindo, I.; Garaigorta, U.; Urquiza, J.; Puerto, A.D.; Campillo, N.E.; Martinez, A.; Gastaminza, P.; et al. Identification of Niemann-Pick C1 protein as a potential novel SARS-CoV-2 intracellular target. Antivir. Res. 2021, 194, 105167. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Leao, I.C.; Coleman, E.M.; Broughton, R.S.; Hildreth, J.E. Deficiency of niemann-pick type C-1 protein impairs release of human immunodeficiency virus type 1 and results in Gag accumulation in late endosomal/lysosomal compartments. J. Virol. 2009, 83, 7982–7995. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Dorival, I.; Cuesta-Geijo, M.Á.; Galindo, I.; del Puerto, A.; Barrado-Gil, L.; Urquiza, J.; Alonso, C. Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets. Viruses 2023, 15, 1098. https://doi.org/10.3390/v15051098
García-Dorival I, Cuesta-Geijo MÁ, Galindo I, del Puerto A, Barrado-Gil L, Urquiza J, Alonso C. Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets. Viruses. 2023; 15(5):1098. https://doi.org/10.3390/v15051098
Chicago/Turabian StyleGarcía-Dorival, Isabel, Miguel Ángel Cuesta-Geijo, Inmaculada Galindo, Ana del Puerto, Lucía Barrado-Gil, Jesús Urquiza, and Covadonga Alonso. 2023. "Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets" Viruses 15, no. 5: 1098. https://doi.org/10.3390/v15051098
APA StyleGarcía-Dorival, I., Cuesta-Geijo, M. Á., Galindo, I., del Puerto, A., Barrado-Gil, L., Urquiza, J., & Alonso, C. (2023). Elucidation of the Cellular Interactome of African Swine Fever Virus Fusion Proteins and Identification of Potential Therapeutic Targets. Viruses, 15(5), 1098. https://doi.org/10.3390/v15051098






