T Cell Responses Correlate with Self-Reported Disease Severity and Neutralizing Antibody Responses Predict Protection against SARS-CoV-2 Breakthrough Infection
Abstract
:1. Introduction
2. Methods
2.1. Study Design
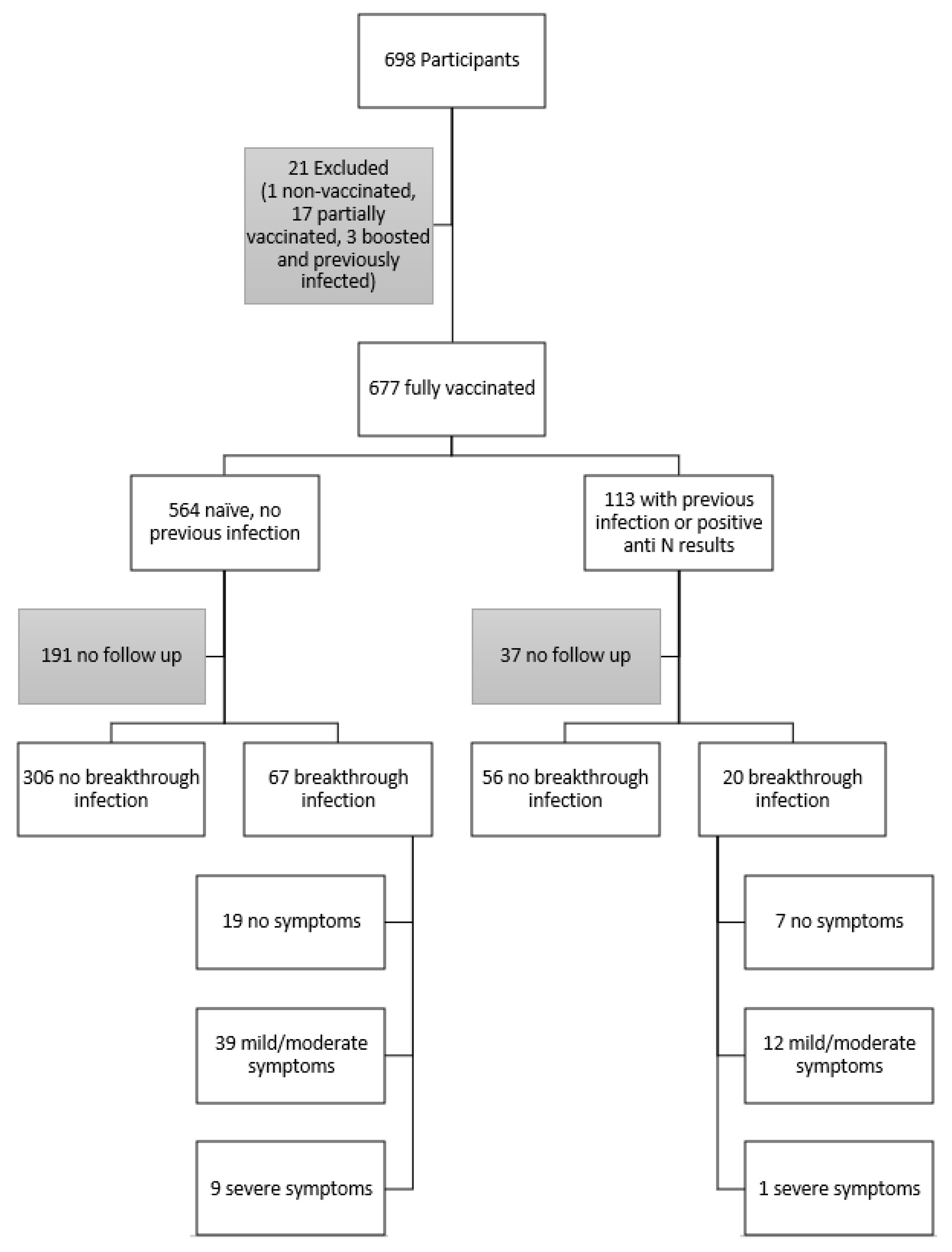
2.2. Inclusion and Exclusion Criteria
2.3. Specimen Collection and Storage
2.4. Laboratory Analysis
2.4.1. Anti-SARS-CoV-2 Nucleocapsid (Anti-N) Antibody Assay
2.4.2. SARS-CoV-2 NAb Assay
2.4.3. QuantiFERON SARS-CoV-2 Assay
2.5. Data Analysis
3. Results
3.1. Participant Demographics and Characterization
3.2. Follow-Up Survey
3.3. Post-Vaccination NAb and T Cell Responses at the Time of Sample Collection
3.4. NAb and T Cell Responses and Breakthrough Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Keeton, R.; Tincho, M.B.; Ngomti, A.; Baguma, R.; Benede, N.; Suzuki, A.; Khan, K.; Cele, S.; Bernstein, M.; Karim, F.; et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022, 603, 488–492. [Google Scholar] [CrossRef]
- Gao, Y.; Cai, C.; Grifoni, A.; Müller, T.R.; Niessl, J.; Olofsson, A.; Humbert, M.; Hansson, L.; Österborg, A.; Bergman, P.; et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022, 28, 472–476. [Google Scholar] [CrossRef]
- Vardhana, S.; Baldo, L.; Morice, W.G.; Wherry, E.J. Understanding T cell responses to COVID-19 is essential for informing public health strategies. Sci. Immunol. 2022, 7, eabo1303. [Google Scholar] [CrossRef]
- Feng, C.; Shi, J.; Fan, Q.; Wang, Y.; Huang, H.; Chen, F.; Tang, G.; Li, Y.; Li, P.; Li, J.; et al. Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat. Commun. 2021, 12, 4984. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Zhang, Y.; Tang, G.; Luo, Y.; Liu, W.; Cheng, C.; Jiang, Y.; Xiong, Z.; Wu, S.; Sun, Z.; et al. Immunologic memory to SARS-CoV-2 in convalescent COVID-19 patients at 1 year postinfection. J. Allergy Clin. Immunol. 2021, 148, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef]
- Kent, S.J.; Khoury, D.S.; Reynaldi, A.; Juno, J.A.; Wheatley, A.K.; Stadler, E.; Wherry, E.J.; Triccas, J.; Sasson, S.C.; Cromer, D.; et al. Disentangling the relative importance of T cell responses in COVID-19: Leading actors or supporting cast? Nat. Rev. Immunol. 2022, 22, 387–397. [Google Scholar] [CrossRef]
- Tartof, S.Y.; Slezak, J.M.; Puzniak, L.; Hong, V.; Frankland, T.B.; Ackerson, B.K.; Takhar, H.S.; Ogun, O.A.; Simmons, S.R.; Zamparo, J.M.; et al. Effectiveness of a third dose of BNT162b2 mRNA COVID-19 vaccine in a large US health system: A retrospective cohort study. Lancet Reg. Health-Am. 2022, 9, 100198. [Google Scholar] [CrossRef]
- Pfizer and BioNTech Provide Update on Omicron Variant|Pfizer. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-provide-update-omicron-variant (accessed on 18 October 2022).
- Danza, P.; Koo, T.H.; Haddix, M.; Fisher, R.; Traub, E.; Oyong, K.; Balter, S. SARS-CoV-2 Infection and Hospitalization Among Adults Aged ≥18 Years, by Vaccination Status, Before and During SARS-CoV-2 B.1.1.529 (Omicron) Variant Predominance—Los Angeles County, California, 7 November 2021–8 January 2022. MMWR. Morb. Mortal. Wkly. Rep. 2022, 71, 177–181. [Google Scholar] [CrossRef]
- Melanson, S.E.F.; Zhao, Z.; Kumanovics, A.; Love, T.; Meng, Q.H.; Wu, A.H.B.; Apple, F.; Ondracek, C.R.; Schulz, K.M.; Wiencek, J.R.; et al. Tolerance for three commonly administered COVID-19 vaccines by healthcare professionals. Front. Public Health 2022, 10, 975781. [Google Scholar] [CrossRef]
- Vaccine Adverse Event Reporting System (VAERS). Available online: https://vaers.hhs.gov/index.html (accessed on 18 October 2022).
- QuantiFERON ELISA Instructions for Use (Handbook)—QIAGEN. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=0eb3ad62-171e-4106-af9a-fd28d763ff66&lang=en (accessed on 18 October 2022).
- QuantiFERON SARS-CoV-2 Extended Set Blood Collection Tubes Instructions for Use (Handbook)—QIAGEN. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=60729925-a7b2-4cf3-8d28-52e4106af16b&lang=en (accessed on 18 October 2022).
- Platelia SARS-CoV-2 Total Ab-Instructions for Use. Available online: https://commerce.bio-rad.com/webroot/web/pdf/inserts/CDG/en/16008267_2020_04_EN.pdf (accessed on 18 October 2022).
- COVID-19 Detection|cPassTM Kit Technology. Available online: https://www.genscript.com/covid-19-detection-cpass.html (accessed on 18 October 2022).
- Jaganathan, S.; Stieber, F.; Rao, S.N.; Nikolayevskyy, V.; Manissero, D.; Allen, N.; Boyle, J.; Howard, J. Preliminary Evaluation of QuantiFERON SARS-CoV-2 and QIAreach Anti-SARS-CoV-2 Total Test in Recently Vaccinated Individuals. Infect. Dis. Ther. 2021, 10, 2765–2776. [Google Scholar] [CrossRef]
- VAERS|Vaccine Safety|CDC. Available online: https://www.cdc.gov/vaccinesafety/ensuringsafety/monitoring/vaers/index.html (accessed on 18 October 2022).
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2020, 590, 630–634. [Google Scholar] [CrossRef]
- Pantaleo, G.; Correia, B.; Fenwick, C.; Joo, V.S.; Perez, L. Antibodies to combat viral infections: Development strategies and progress. Nat. Rev. Drug Discov. 2022, 21, 676–696. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Zeng, C.; Carlin, C.; Lozanski, G.; Saif, L.J.; Oltz, E.M.; Gumina, R.J.; Liu, S.-L. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022, 14, eabn8057. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef]
- Apostolidis, S.A.; Kakara, M.; Painter, M.M.; Goel, R.R.; Mathew, D.; Lenzi, K.; Rezk, A.; Patterson, K.R.; Espinoza, D.A.; Kadri, J.C.; et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 2021, 27, 1990–2001. [Google Scholar] [CrossRef]
- Bange, E.M.; Han, N.A.; Wileyto, P.; Kim, J.Y.; Gouma, S.; Robinson, J.; Greenplate, A.R.; Hwee, M.A.; Porterfield, F.; Owoyemi, O.; et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021, 27, 1280–1289. [Google Scholar] [CrossRef]
- Baum, A.; Ajithdoss, D.; Copin, R.; Zhou, A.; Lanza, K.; Negron, N.; Ni, M.; Wei, Y.; Mohammadi, K.; Musser, B.; et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 2020, 370, 1110–1115. [Google Scholar] [CrossRef]
- Sormani, M.P.; Inglese, M.; Schiavetti, I.; Carmisciano, L.; Laroni, A.; Lapucci, C.; Da Rin, G.; Serrati, C.; Gandoglia, I.; Tassinari, T.; et al. Effect of SARS-CoV-2 mRNA vaccination in MS patients treated with disease modifying therapies. Ebiomedicine 2021, 72, 103581. [Google Scholar] [CrossRef]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, O.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: A retrospective cohort study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, J.; McMahan, K.; Jacob-Dolan, C.; He, X.; Giffin, V.; Wu, C.; Sciacca, M.; Powers, O.; Nampanya, F.; et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 2022, 7, eabq7647. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Variant Data|Department of Health. Available online: https://coronavirus.health.ny.gov/covid-19-variant-data (accessed on 18 October 2022).

| Demographic # | Blood Draw (n = 698) | No Known Previous Infection and Negative Anti-N | Self-Reported Previous Infection or Positive Anti-N (n = 113) | p Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Pfizer (n = 314) | Moderna (n = 181) | Johnson & Johnson (n = 38) | Booster (n = 31) | Positive Anti-N * (n = 46) | |||||
| Sex | Male, N (%) | 328 (46.9) | 141 (44.9) | 89 (49.2) | 17 (44.7) | 12 (38.7) | 60 (53.1) | 28 (60.9) | 0.48 |
| Age | Median (IQR) | 50 (40–59) | 50 (39–58) | 50 (40–60) | 49 (38–58) | 50 (40–66) | 49 (40–58) | 49 (40–58) | 0.66 |
| Race | Caucasian, N (%) | 528 (75.6) | 239 (76.1) | 137 (75.7) | 30 (78.9) | 25 (80.6) | 89 (78.8) | 37 (80.4) | 0.86 |
| African American /Black, N (%) | 41 (5.8) | 20 (6.4) | 10 (5.5) | 3 (7.9) | 0 (0.0) | 6 (5.3) | 4 (8.7) | ||
| Asian, N (%) | 67 (9.5) | 30 (9.6) | 18 (9.9) | 3 (7.9) | 2 (6.5) | 9 (8.0) | 4 (8.7) | ||
| Native Hawaiian /Pacific Islander, N (%) | 6 (0.8) | 3 (1.0) | 3 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| Unknown/Other /Prefer Not to Say, N (%) | 56 (8.0) | 22 (7.0) | 13 (7.2) | 2 (5.2) | 4 (12.9) | 9 (8.0) | 1 (2.2) | ||
| Ethnicity | Hispanic and/or Latino, N (%) | 92 (13.1) | 33 (10.5) | 22 (12.2) | 6 (15.8) | 5 (16.1) | 22 (19.5) | 5 (10.9) | 0.56 |
| Non-Hispanic /Non-Latino, N (%) | 575 (82.3) | 268 (85.4) | 151 (83.4) | 30 (78.9) | 25 (80.6) | 87 (77.0) | 38 (82.6) | ||
| Prefer not to reply, N (%) | 31 (4.4) | 13 (4.1) | 8 (4.4) | 2 (5.3) | 1 (3.2) | 4 (3.5) | 3 (6.5) | ||
| 449 of the 677 Completed the Follow-Up Questionnaire | No Breakthrough Infection (N = 362) | Breakthrough Infection (N = 87) | Among Those with Breakthrough | Uni * p Value | Multi p Value | ||
|---|---|---|---|---|---|---|---|
| No Limitation (n = 26) | Mild-Moderate Limitations (n = 51) | Severe Limitations (n = 10) | |||||
| NAb (%) | 88 (69–95) | 83 (57–96) | 87.5 (70.5–97) | 82 (55.5–93.5) | 72 (53–85.8) | 0.475 | 0.405 |
| NAb (UmL) | 194.9 (86.3–300) | 151.4 (58.7–300) | 185.7 (74.8–300) | 148.4 (50.4–285.8) | 91.6 (46.9–198.9) | 0.285 | 0.197 |
| Ag1, median (IQR) | 0.11 (0.04–0.30) | 0.11 (0.03–0.30) | 0.09 (0.02–0.28) | 0.14 (0.04–0.30) | 0.11 (0.01–0.41) | 0.547 | 0.572 |
| Ag2, median (IQR) | 0.16 (0.06–0.40) | 0.16 (0.05–0.45) | 0.15 (0.05–0.36) | 0.20 (0.05–0.47) | 0.08 (0.01–0.19) | 0.629 | 0.621 |
| Ag3, median (IQR) | 0.17 (0.06–0.50) | 0.18 (0.06–0.48) | 0.17 (0.05–0.51) | 0.19 (0.07–0.48) | 0.17 (0.01–0.29) | 0.873 | 0.874 |
| Sum, median (IQR) | 0.46 (0.16–1.24) | 0.51 (0.12–1.10) | 0.47 (0.11–1.05) | 0.54 (0.15–1.11) | 0.37 (0.04–0.99) | 0.766 | 0.762 |
| Univariate | ||||||
| COVID-19 | n | NAb (>95 U/mL) | sum Ag(>0.385 IU/mL) | Ag1 (>0.435 IU/mL) | Ag2 (>0.120 IU/mL) | Ag3 (>0.055 IU/mL) |
| No breakthrough | 362 | 1.78 | 0.88 | 1.15 | 0.89 | 1.22 |
| No limitations | 26 | 1.07 | 0.95 | 0.84 | 0.93 | 0.90 |
| Mild-moderate limitations | 51 | 1.03 | 1.15 | 1.04 | 1.23 | 1.22 |
| Severe limitations | 10 | 0.91 | 0.92 | 1.14 | 0.87 | 0.91 |
| Hospitalizations | 0 | - | - | - | - | - |
| Multivariate | ||||||
| COVID-19 | n | NAb (>95 U/mL) | sum Ag(>0.385 IU/mL) | Ag1 (>0.435 IU/mL) | Ag2 (>0.120 IU/mL) | Ag3 (>0.055 IU/mL) |
| No breakthrough | 362 | 1.95 | 0.91 | 1.21 | 0.92 | 1.27 |
| No limitations | 26 | 1.04 | 0.91 | 0.85 | 0.90 | 0.88 |
| Mild-moderate limitations | 51 | 1.08 | 1.23 | 1.03 | 1.31 | 1.28 |
| Severe limitations | 10 | 0.90 | 0.89 | 1.14 | 0.85 | 0.89 |
| Hospitalizations | 0 | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Kumanovics, A.; Love, T.; Melanson, S.E.F.; Meng, Q.H.; Wu, A.H.B.; Wiencek, J.; Apple, F.S.; Ondracek, C.R.; Koch, D.D.; et al. T Cell Responses Correlate with Self-Reported Disease Severity and Neutralizing Antibody Responses Predict Protection against SARS-CoV-2 Breakthrough Infection. Viruses 2023, 15, 709. https://doi.org/10.3390/v15030709
Zhao Z, Kumanovics A, Love T, Melanson SEF, Meng QH, Wu AHB, Wiencek J, Apple FS, Ondracek CR, Koch DD, et al. T Cell Responses Correlate with Self-Reported Disease Severity and Neutralizing Antibody Responses Predict Protection against SARS-CoV-2 Breakthrough Infection. Viruses. 2023; 15(3):709. https://doi.org/10.3390/v15030709
Chicago/Turabian StyleZhao, Zhen, Attila Kumanovics, Tanzy Love, Stacy E. F. Melanson, Qing H. Meng, Alan H. B. Wu, Joesph Wiencek, Fred S. Apple, Caitlin R. Ondracek, David D. Koch, and et al. 2023. "T Cell Responses Correlate with Self-Reported Disease Severity and Neutralizing Antibody Responses Predict Protection against SARS-CoV-2 Breakthrough Infection" Viruses 15, no. 3: 709. https://doi.org/10.3390/v15030709
APA StyleZhao, Z., Kumanovics, A., Love, T., Melanson, S. E. F., Meng, Q. H., Wu, A. H. B., Wiencek, J., Apple, F. S., Ondracek, C. R., Koch, D. D., Christenson, R. H., & Zhang, Y. V. (2023). T Cell Responses Correlate with Self-Reported Disease Severity and Neutralizing Antibody Responses Predict Protection against SARS-CoV-2 Breakthrough Infection. Viruses, 15(3), 709. https://doi.org/10.3390/v15030709






