Host-Specific Interplay between Foot-and-Mouth Disease Virus 3D Polymerase and the Type-I Interferon Pathway
Abstract
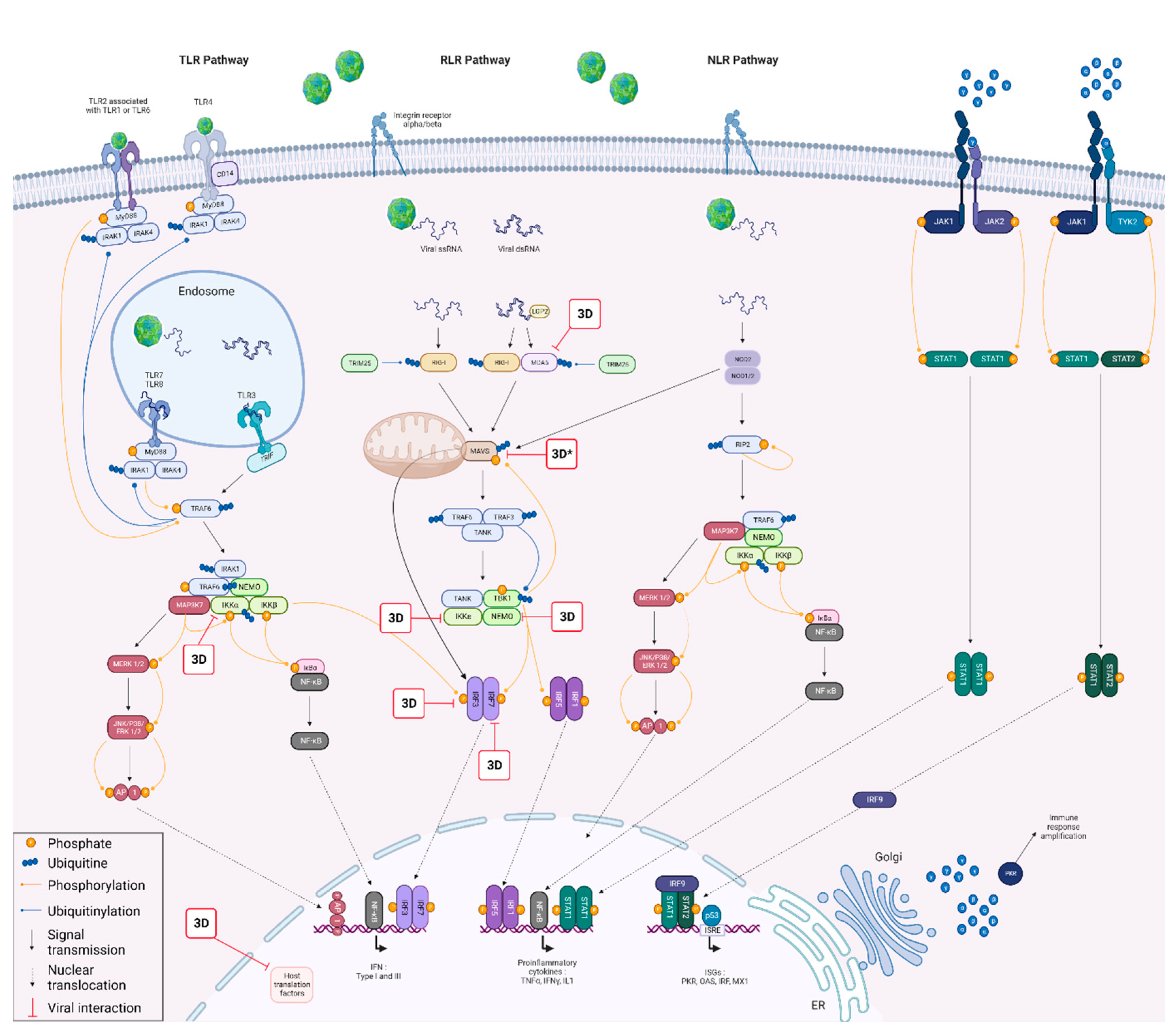
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Plasmid Libraries
2.2.1. Viral Plasmids Library
2.2.2. Cattle, Sheep, Goat and Swine Plasmid Libraries
2.3. Protein–Protein Interaction (PPI) Assays
2.3.1. Nanoluciferase-2-Hybrid Complementation Assay
2.3.2. GST Pull-Down Assays
2.4. Luciferase Reporter Assays
3. Results
3.1. NanoLuc-2-Hybrid Assays for Binary Protein–Protein Interactions FMDV/Host Screening
3.1.1. Evidence of PPI between FMDV Viral Proteins and Cattle, Sheep, Goat and Swine IFN Proteins
3.1.2. Screening Comparison between the Different Protein Libraries Derived from Four FMD-Susceptible Species
3.2. FMDV 3Dpol Inhibits the IFN Pathway Induction Phase
3.2.1. 3Dpol Effects on the IFN Pathway Induction Phase
3.2.2. 3Dpol Effects on the IFN Pathway Amplification Phase
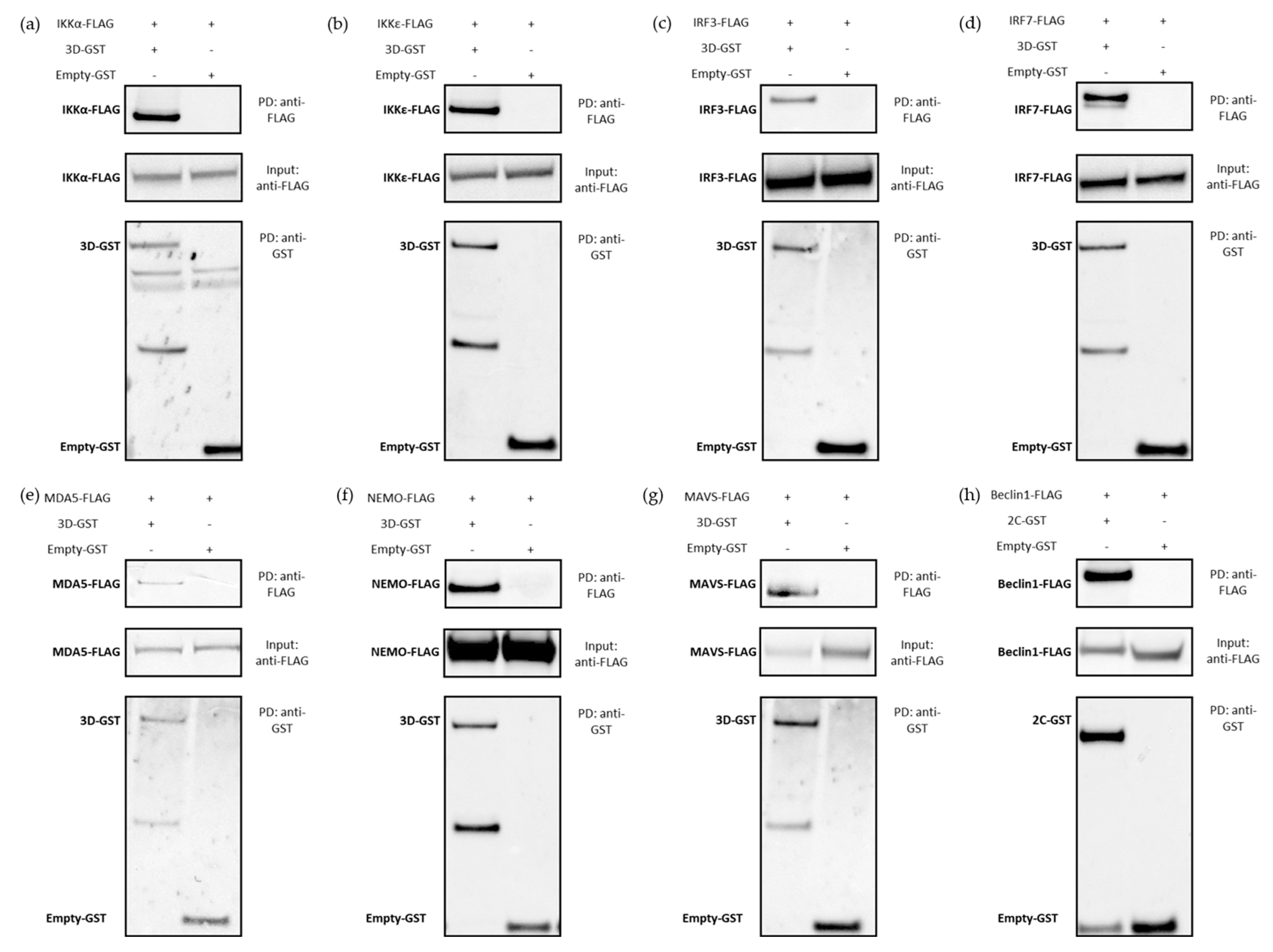
3.3. Biochemical Assessment of Candidate Protein–Protein Interaction by GST-Pull Down
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamal, S.M.; Belsham, G.J. Foot-and-mouth disease: Past, present and future. Vet. Res. 2013, 44, 116. [Google Scholar] [CrossRef] [PubMed]
- Knowles, N.; Samuel, A. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003, 91, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Belsham, G.J.; Kristensen, T.; Jackson, T. Foot-and-mouth disease virus: Prospects for using knowledge of virus biology to improve control of this continuing global threat. Virus Res. 2020, 281, 197909. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, J.M.; Henry, T.M.; O’Donnell, V.K.; Gregory, J.B.; Mason, P.W. Role of Nonstructural Proteins 3A and 3B in Host Rangeand Pathogenicity of Foot-and-Mouth DiseaseVirus. J. Virol. 2003, 77, 13017–13027. [Google Scholar] [CrossRef] [PubMed]
- Arzt, J.; Juleff, N.; Zhang, Z.; Rodriguez, L.L. The Pathogenesis of Foot-and-Mouth Disease I: Viral Pathways in Cattle. Transbound. Emerg. Dis. 2011, 58, 291–304. [Google Scholar] [CrossRef]
- Arzt, J.; Baxt, B.; Grubman, M.J.; Jackson, T.; Juleff, N.; Rhyan, J.; Rieder, E.; Waters, R.; Rodriguez, L.L. The Pathogenesis of Foot-and-Mouth Disease II: Viral Pathways in Swine, Small Ruminants, and Wildlife; Myotropism, Chronic Syndromes, and Molecular Virus-Host Interactions. Transbound. Emerg. Dis. 2011, 58, 305–326. [Google Scholar] [CrossRef]
- World Organization for Animal Health (OIE). Chapter 3.1.8: Foot and Mouth Disease (Infection with Foot and Mouth Disease Virus). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organization for Animal Health (OIE): Paris, France, 2021; Available online: https://www.oie.int/fileadmin/home/eng/health_standards/tahm/3.01.08_fmd.pdf (accessed on 11 April 2022).
- Stenfeldt, C.; Arzt, J. The Carrier Conundrum; A Review of Recent Advances and Persistent Gaps Regarding the Carrier State of Foot-and-Mouth Disease Virus. Pathogens 2020, 9, 167. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Belsham, G.J. Detection of foot-and-mouth disease virus RNA in pharyngeal epithelium biopsy samples obtained from infected cattle: Investigation of possible sites of virus replication and persistence. Vet. Microbiol. 2012, 154, 230–239. [Google Scholar] [CrossRef]
- Stenfeldt, C.; Eschbaumer, M.; Rekant, S.I.; Pacheco, J.M.; Smoliga, G.R.; Hartwig, E.J.; Rodriguez, L.L.; Arzt, J. The Foot-and-Mouth Disease Carrier State Divergence in Cattle. J. Virol. 2016, 90, 6344–6364. [Google Scholar] [CrossRef]
- Arzt, J.; Belsham, G.J.; Lohse, L.; Bøtner, A.; Stenfeldt, C. Transmission of Foot-and-Mouth Disease from Persistently Infected Carrier Cattle to Naive Cattle via Transfer of Oropharyngeal Fluid. Msphere 2018, 3, e00365-18. [Google Scholar] [CrossRef]
- Fish, I.; Stenfeldt, C.; Spinard, E.; Medina, G.N.; Azzinaro, P.A.; Bertram, M.R.; Holinka, L.; Smoliga, G.R.; Hartwig, E.J.; Santos, T.D.L.; et al. Foot-and-Mouth Disease Virus Interserotypic Recombination in Superinfected Carrier Cattle. Pathogens 2022, 11, 644. [Google Scholar] [CrossRef]
- Childs, K.; Jackson, B.; Harvey, Y.; Seago, J. Trans-Encapsidation of Foot-and-Mouth Disease Virus Genomes Facilitates Escape from Neutralizing Antibodies. Viruses 2022, 14, 1161. [Google Scholar] [CrossRef] [PubMed]
- Van Bekkum: Observations on the Carrier State of-Google Scholar. Available online: https://scholar.google.com/scholar_lookup?title=Observations+on+the+carrier+state+of+cattle+exposed+to+foot-and-mouth+disease+virus&author=van+Bekkum+J.+G.&author=Frenkel+H.+S.&author=Frederiks+H.+H.+J.&author=Frenkel+S.&publication+year=1959&journal=Tijdschr.+Diergeneesk.&volume=84 (accessed on 19 January 2022).
- Burrows, R. The persistence of foot-and-mouth disease virus in sheep. Epidemiol. Infect. 1968, 66, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Pacheco, J.M.; Smoliga, G.R.; Bishop, E.; Pauszek, S.J.; Hartwig, E.J.; Rodriguez, L.L.; Arzt, J. Detection of Foot-and-mouth Disease Virus RNA and Capsid Protein in Lymphoid Tissues of Convalescent Pigs Does Not Indicate Existence of a Carrier State. Transbound. Emerg. Dis. 2014, 63, 152–164. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, J.C.; Martínez-Salas, E.; Diez, J.; Villaverde, A.; Gebauer, F.; Rocha, E.; Dávila, M.; Domingo, E. Coevolution of cells and viruses in a persistent infection of foot-and-mouth disease virus in cell culture. J. Virol. 1988, 62, 2050–2058. [Google Scholar] [CrossRef]
- O’Donnell, V.; Pacheco, J.; Larocco, M.; Gladue, D.; Pauszek, S.; Smoliga, G.; Krug, P.; Baxt, B.; Borca, M.; Rodriguez, L. Virus–host interactions in persistently FMDV-infected cells derived from bovine pharynx. Virology 2014, 468, 185–196. [Google Scholar] [CrossRef]
- Kopliku, L.; Relmy, A.; Romey, A.; Gorna, K.; Zientara, S.; Bakkali-Kassimi, L.; Blaise-Boisseau, S. Establishment of persistent foot-and-mouth disease virus (FMDV) infection in MDBK cells. Arch. Virol. 2015, 160, 2503–2516. [Google Scholar] [CrossRef]
- Eschbaumer, M.; Stenfeldt, C.; Smoliga, G.R.; Pacheco, J.M.; Rodriguez, L.L.; Li, R.W.; Zhu, J.; Arzt, J. Transcriptomic Analysis of Persistent Infection with Foot-and-Mouth Disease Virus in Cattle Suggests Impairment of Apoptosis and Cell-Mediated Immunity in the Nasopharynx. PLoS ONE 2016, 11, e0162750. [Google Scholar] [CrossRef]
- Medina, G.N.; Segundo, F.D.-S.; Stenfeldt, C.; Arzt, J.; Santos, T.D.L. The Different Tactics of Foot-and-Mouth Disease Virus to Evade Innate Immunity. Front. Microbiol. 2018, 9, 2644. [Google Scholar] [CrossRef]
- Hägglund, S.; Laloy, E.; Näslund, K.; Pfaff, F.; Eschbaumer, M.; Romey, A.; Relmy, A.; Rikberg, A.; Svensson, A.; Huet, H.; et al. Model of persistent foot-and-mouth disease virus infection in multilayered cells derived from bovine dorsal soft palate. Transbound. Emerg. Dis. 2019, 67, 133–148. [Google Scholar] [CrossRef]
- Pfaff, F.; Hägglund, S.; Zoli, M.; Blaise-Boisseau, S.; Laloy, E.; Koethe, S.; Zühlke, D.; Riedel, K.; Zientara, S.; Bakkali-Kassimi, L.; et al. Proteogenomics Uncovers Critical Elements of Host Response in Bovine Soft Palate Epithelial Cells Following In Vitro Infection with Foot-And-Mouth Disease Virus. Viruses 2019, 11, 53. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, S.-Q.; Guo, H.-C. Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol. J. 2016, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, C.; Yang, F.; Cao, W.; Zhu, Z.; Zheng, H. Virus–Host Interactions in Foot-and-Mouth Disease Virus Infection. Front. Immunol. 2021, 12, 571509. [Google Scholar] [CrossRef] [PubMed]
- Sarry, M.; Vitour, D.; Zientara, S.; Kassimi, L.B.; Blaise-Boisseau, S. Foot-and-Mouth Disease Virus: Molecular Interplays with IFN Response and the Importance of the Model. Viruses 2022, 14, 2129. [Google Scholar] [CrossRef]
- Rai, D.K.; Lawrence, P.; Kloc, A.; Schafer, E.; Rieder, E. Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections. Virol. J. 2015, 12, 1–17. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zeng, Q.; Zheng, H.; Xue, Q.; Cai, X. The DEAD-Box RNA Helicase DDX1 Interacts with the Viral Protein 3D and Inhibits Foot-and-Mouth Disease Virus Replication. Virol. Sin. 2019, 34, 610–617. [Google Scholar] [CrossRef]
- Robertson, B.; Morgan, D.; Moore, D.; Grubman, M.; Card, J.; Fischer, T.; Weddell, G.; Dowbenko, D.; Yansura, D. Identification of amino acid and nucleotide sequence of thefoot-and-mouth disease virus RNA polymerase. Virology 1983, 126, 614–623. [Google Scholar] [CrossRef]
- Domingo, E.; Escarmís, C.; Sevilla, N.; Moya, A.; Elena, S.F.; Quer, J.; Novella, I.S.; Holland, J.J.; Shi, J.; Sun, J.; et al. Basic concepts in RNA virus evolution. FASEB J. 1996, 10, 859–864. [Google Scholar] [CrossRef]
- Ferrer-Orta, C.; Ferrero, D.; Verdaguer, N. RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms. Viruses 2015, 7, 4438–4460. [Google Scholar] [CrossRef]
- Nayak, A.; Goodfellow, I.G.; Belsham, G.J. Factors Required for the Uridylylation of the Foot-and-Mouth Disease Virus 3B1, 3B2, and 3B3 Peptides by the RNA-Dependent RNA Polymerase (3D pol ) In Vitro. J. Virol. 2005, 79, 7698–7706. [Google Scholar] [CrossRef]
- Loundras, E.-A.; Streetley, J.; Herod, M.R.; Thompson, R.; Harris, M.; Bhella, D.; Stonehouse, N.J. Higher-order structures of the foot-and-mouth disease virus RNA-dependent RNA polymerase required for genome replication. Commun. Biol. 2022, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lagrée, A.-C.; Fasani, F.; Rouxel, C.; Pivet, M.; Pourcelot, M.; Fablet, A.; Romey, A.; Caignard, G.; Vitour, D.; Blaise-Boisseau, S.; et al. Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections. Int. J. Mol. Sci. 2020, 21, 5249. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.G.; Olivet, J.; Cassonnet, P.; Vidalain, P.-O.; Luck, K.; Lambourne, L.; Spirohn, K.; Lemmens, I.; Dos Santos, M.; Demeret, C.; et al. Maximizing binary interactome mapping with a minimal number of assays. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Coutant, E.P.; Goyard, S.; Hervin, V.; Gagnot, G.; Baatallah, R.; Jacob, Y.; Rose, T.; Janin, Y.L. Gram-scale synthesis of luciferins derived from coelenterazine and original insights into their bioluminescence properties. Org. Biomol. Chem. 2019, 17, 3709–3713. [Google Scholar] [CrossRef] [PubMed]
- Coutant, E.P.; Gagnot, G.; Hervin, V.; Baatallah, R.; Goyard, S.; Jacob, Y.; Rose, T.; Janin, Y.L. Bioluminescence Profiling of NanoKAZ/NanoLuc Luciferase Using a Chemical Library of Coelenterazine Analogues. Chem. Eur. J. 2019, 26, 948–958. [Google Scholar] [CrossRef]
- Li, X.; Leung, S.; Qureshi, S.; Darnell, J.E.; Stark, G.R. Formation of STAT1-STAT2 Heterodimers and Their Role in the Activation of IRF-1 Gene Transcription by Interferon-α. J. Biol. Chem. 1996, 271, 5790–5794. [Google Scholar] [CrossRef]
- Cassonnet, P.; Rolloy, C.; Neveu, G.; Vidalain, P.-O.; Chantier, T.; Pellet, J.; Jones, L.; Muller, M.; Demeret, C.; Gaud, G.; et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods 2011, 8, 990–992. [Google Scholar] [CrossRef]
- Morel, J.; Sedano, L.; Lejal, N.; Da Costa, B.; Batsché, E.; Muchardt, C.; Delmas, B. The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription. Viruses 2022, 14, 518. [Google Scholar] [CrossRef]
- Mendoza, J.-A.; Jacob, Y.; Cassonnet, P.; Favre, M. Human Papillomavirus Type 5 E6 Oncoprotein Represses the Transforming Growth Factor β Signaling Pathway by Binding to SMAD3. J. Virol. 2006, 80, 12420–12424. [Google Scholar] [CrossRef]
- Gladue, D.P.; O’Donnell, V.; Baker-Branstetter, R.; Holinka, L.G.; Pacheco, J.M.; Fernandez-Sainz, I.; Lu, Z.; Brocchi, E.; Baxt, B.; Piccone, M.E.; et al. Foot-and-Mouth Disease Virus Nonstructural Protein 2C Interacts with Beclin1, Modulating Virus Replication. J. Virol. 2012, 86, 12080–12090. [Google Scholar] [CrossRef]
- Chauveau, E.; Doceul, V.; Lara, E.; Breard, E.; Sailleau, C.; Vidalain, P.-O.; Meurs, E.F.; Dabo, S.; Schwartz-Cornil, I.; Zientara, S.; et al. NS3 of Bluetongue Virus Interferes with the Induction of Type I Interferon. J. Virol. 2013, 87, 8241–8246. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lei, C.; Xu, Z.; Yang, F.; Liu, H.; Zhu, Z.; Li, S.; Liu, X.; Shu, H.; Zheng, H. Foot-and-mouth disease virus non-structural protein 3A inhibits the interferon-β signaling pathway. Sci. Rep. 2016, 6, 21888. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, L.; Liu, L.; Zhong, H.; Chen, Q.; Luo, R.; Liu, X.; Zhang, Z.; Chen, H.; Xiao, S. Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-λ1 pathway. Mol. Immunol. 2011, 49, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, L.; Li, P.; Sun, L.; Fan, J.; Zhang, Q.; Luo, R.; Liu, X.; Li, K.; Chen, H.; et al. The Leader Proteinase of Foot-and-Mouth Disease Virus Negatively Regulates the Type I Interferon Pathway by Acting as a Viral Deubiquitinase. J. Virol. 2011, 85, 3758–3766. [Google Scholar] [CrossRef]
- Pulido, M.R.; Martínez-Salas, E.; Sobrino, F.; Sáiz, M. MDA5 cleavage by the Leader protease of foot-and-mouth disease virus reveals its pleiotropic effect against the host antiviral response. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Kim, H.; Kim, A.-Y.; Choi, J.; Park, S.; Park, S.; Kim, J.-S.; Lee, S.-I.; Park, J.-H.; Park, C.-K.; Ko, Y.-J. Foot-and-Mouth Disease Virus Evades Innate Immune Response by 3C-Targeting of MDA5. Cells 2021, 10, 271. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.; Li, K.; Zhong, H.; Fan, J.; Ouyang, C.; Zhang, H.; Duan, E.; Luo, R.; Zhang, Z.; et al. Foot-and-Mouth Disease Virus 3C Protease Cleaves NEMO To Impair Innate Immune Signaling. J. Virol. 2012, 86, 9311–9322. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.; Luo, R.; Ye, R.; Fang, Y.; Xie, L.; Chen, H.; Xiao, S. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 2010, 399, 72–78. [Google Scholar] [CrossRef]
- Ma, X.; Luo, Z.; Song, R.; Nian, X.; Choudhury, S.M.; Ru, Y.; Yang, F.; Zhang, Y.; Zeng, Z.; Cao, W.; et al. The Foot-and-Mouth Disease Virus Lb Protease Cleaves Intracellular Transcription Factors STAT1 and STAT2 to Antagonize IFN-β–Induced Signaling. J. Immunol. 2023, 210, 283–296. [Google Scholar] [CrossRef]
- Li, M.; Xin, T.; Gao, X.; Wu, J.; Wang, X.; Fang, L.; Sui, X.; Zhu, H.; Cui, S.; Guo, X. Foot-and-mouth disease virus non-structural protein 2B negatively regulates the RLR-mediated IFN-β induction. Biochem. Biophys. Res. Commun. 2018, 504, 238–244. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, G.; Yang, F.; Cao, W.; Mao, R.; Du, X.; Zhang, X.; Li, C.; Li, D.; Zhang, K.; et al. Foot-and-Mouth Disease Virus Viroporin 2B Antagonizes RIG-I-Mediated Antiviral Effects by Inhibition of Its Protein Expression. J. Virol. 2016, 90, 11106–11121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, Z.; Wang, C.; Yang, F.; Cao, W.; Li, P.; Du, X.; Zhao, F.; Liu, X.; Zheng, H. Foot-and-Mouth Disease Virus 3B Protein Interacts with Pattern Recognition Receptor RIG-I to Block RIG-I–Mediated Immune Signaling and Inhibit Host Antiviral Response. J. Immunol. 2020, 205, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Bi, J.; Liu, J.; Liu, X.; Wu, X.; Jiang, P.; Yoo, D.; Zhang, Y.; Wu, J.; Wan, R.; et al. 3C pro of Foot-and-Mouth Disease Virus Antagonizes the Interferon Signaling Pathway by Blocking STAT1/STAT2 Nuclear Translocation. J. Virol. 2014, 88, 4908–4920. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, Z.; Du, X.; Cao, W.; Yang, F.; Zhang, X.; Feng, H.; Li, D.; Zhang, K.; Liu, X.; et al. Foot-and-mouth disease virus induces lysosomal degradation of host protein kinase PKR by 3C proteinase to facilitate virus replication. Virology 2017, 509, 222–231. [Google Scholar] [CrossRef]
- Visser, L.J.; Aloise, C.; Swatek, K.N.; Medina, G.N.; Olek, K.M.; Rabouw, H.H.; de Groot, R.J.; Langereis, M.A.; Santos, T.D.L.; Komander, D.; et al. Dissecting distinct proteolytic activities of FMDV Lpro implicates cleavage and degradation of RLR signaling proteins, not its deISGylase/DUB activity, in type I interferon suppression. PLoS Pathog. 2020, 16, e1008702. [Google Scholar] [CrossRef]
- Ekanayaka, P.; Lee, S.-Y.; Herath, T.U.B.; Kim, J.-H.; Kim, T.-H.; Lee, H.; Chathuranga, K.; Chathuranga, W.A.G.; Park, J.-H.; Lee, J.-S. Foot-and-mouth disease virus VP1 target the MAVS to inhibit type-I interferon signaling and VP1 E83K mutation results in virus attenuation. PLoS Pathog. 2020, 16, e1009057. [Google Scholar] [CrossRef]
- Li, D.; Yang, W.; Yang, F.; Liu, H.; Zhu, Z.; Lian, K.; Lei, C.; Li, S.; Liu, X.; Zheng, H.; et al. The VP3 structural protein of foot-and-mouth disease virus inhibits the IFN-β signaling pathway. FASEB J. 2016, 30, 1757–1766. [Google Scholar] [CrossRef]
- Park, J.-H.; Tark, D.; Lee, K.-N.; Lee, S.-Y.; Ko, M.-K.; Lee, H.-S.; Kim, S.-M.; Ko, Y.-J.; Seo, M.-G.; Chun, J.-E.; et al. Novel foot-and-mouth disease virus in Korea, July-August 2014. Clin. Exp. Vaccine Res. 2016, 5, 83–87. [Google Scholar] [CrossRef]
- Núñez, J.I.; Baranowski, E.; Molina, N.; Ruiz-Jarabo, C.M.; Sánchez, C.; Domingo, E.; Sobrino, F. A Single Amino Acid Substitution in Nonstructural Protein 3A Can Mediate Adaptation of Foot-and-Mouth Disease Virus to the Guinea Pig. J. Virol. 2001, 75, 3977–3983. [Google Scholar] [CrossRef]
- Banninger, G.; Reich, N.C. STAT2 Nuclear Trafficking. J. Biol. Chem. 2004, 279, 39199–39206. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, C.; Du, X.; Wang, G.; Cao, W.; Yang, F.; Feng, H.; Zhang, X.; Shi, Z.; Liu, H.; et al. Foot-and-mouth disease virus infection inhibits LGP2 protein expression to exaggerate inflammatory response and promote viral replication. Cell Death Dis. 2017, 8, e2747. [Google Scholar] [CrossRef] [PubMed]
- Maree, F.; de Klerk-Lorist, L.-M.; Gubbins, S.; Zhang, F.; Seago, J.; Pérez-Martín, E.; Reid, L.; Scott, K.; van Schalkwyk, L.; Bengis, R.; et al. Differential Persistence of Foot-and-Mouth Disease Virus in African Buffalo Is Related to Virus Virulence. J. Virol. 2016, 90, 5132–5140. [Google Scholar] [CrossRef] [PubMed]
- Kloc, A.; Rai, D.K.; Rieder, E. The Roles of Picornavirus Untranslated Regions in Infection and Innate Immunity. Front. Microbiol. 2018, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Drahos, J.; Racaniello, V.R. Cleavage of IPS-1 in Cells Infected with Human Rhinovirus. J. Virol. 2009, 83, 11581–11587. [Google Scholar] [CrossRef]
- Cao, X.; Xue, Y.-J.; Du, J.-L.; Xu, Q.; Yang, X.-C.; Zeng, Y.; Wang, B.-B.; Wang, H.-Z.; Liu, J.; Cai, K.-Z.; et al. Induction and Suppression of Innate Antiviral Responses by Hepatitis A Virus. Front. Microbiol. 2018, 9, 1865. [Google Scholar] [CrossRef]
- Qian, S.; Fan, W.; Liu, T.; Wu, M.; Zhang, H.; Cui, X.; Zhou, Y.; Hu, J.; Wei, S.; Chen, H.; et al. Seneca Valley Virus Suppresses Host Type I Interferon Production by Targeting Adaptor Proteins MAVS, TRIF, and TANK for Cleavage. J. Virol. 2017, 91, e00823-17. [Google Scholar] [CrossRef]
- Yang, F.; Yamashita, J.; Tang, E.; Wang, H.-L.; Guan, K.; Wang, C.-Y. The Zinc Finger Mutation C417R of I-κB Kinase γ Impairs Lipopolysaccharide- and TNF-Mediated NF-κB Activation through Inhibiting Phosphorylation of the I-κB Kinase β Activation Loop. J. Immunol. 2004, 172, 2446–2452. [Google Scholar] [CrossRef]
- Ji, L.; Yang, E.; He, S.; Jin, Y.; Chen, D. Enterovirus 2C Protein Suppresses IKKα Phosphorylation by Recruiting IKKβ and IKKα into Viral Inclusion Bodies. Viral Immunol. 2021, 34, 218–226. [Google Scholar] [CrossRef]
- Huang, L.; Xiong, T.; Yu, H.; Zhang, Q.; Zhang, K.; Li, C.; Hu, L.; Zhang, Y.; Zhang, L.; Liu, Q.; et al. Encephalomyocarditis virus 3C protease attenuates type I interferon production through disrupting the TANK–TBK1–IKKε–IRF3 complex. Biochem. J. 2017, 474, 2051–2065. [Google Scholar] [CrossRef]
- Kuo, R.-L.; Chen, C.-J.; Wang, R.Y.L.; Huang, H.-I.; Lin, Y.-H.; Tam, E.-H.; Tu, W.-J.; Wu, S.-E.; Shih, S.-R. Role of Enteroviral RNA-Dependent RNA Polymerase in Regulation of MDA5-Mediated Beta Interferon Activation. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Wang, L.-C.; Chen, S.-O.; Chang, S.-P.; Lee, Y.-P.; Yu, C.-K.; Chen, C.-L.; Tseng, P.-C.; Hsieh, C.-Y.; Chen, S.-H.; Lin, C.-F. Enterovirus 71 Proteins 2A and 3D Antagonize the Antiviral Activity of Gamma Interferon via Signaling Attenuation. J. Virol. 2015, 89, 7028–7037. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.E.; Schlegel, A.; Kirkegaard, K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2296–2301. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Pacheco, J.M.; LaRocco, M.; Burrage, T.; Jackson, W.; Rodriguez, L.L.; Borca, M.V.; Baxt, B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 2011, 410, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, J.; Yang, W.; He, Y.; Ru, Y.; Fu, S.; Li, L.; Liu, X.; Zheng, H. Poly (rC) binding protein 2 interacts with VP0 and increases the replication of the foot-and-mouth disease virus. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Stenfeldt, C.; Diaz-San Segundo, F.; de Los Santos, T.; Rodriguez, L.L.; Earzt, J. The Pathogenesis of Foot-and-Mouth Disease in Pigs. Front. Vet. Sci. 2016, 3, 41. [Google Scholar] [CrossRef]


| Viral Protein | Cellular Target | Direct or Indirect InterAction? | Study Model | Reference | Found by NanoLuc Approach (Cattle) |
|---|---|---|---|---|---|
| 2B | MDA5 | direct | Human | [52] | No |
| RIG-I | direct | Swine | [53] | No | |
| 3A | MAVS | direct | Human | [44] | No |
| MDA5 | direct | Human | [44] | Yes | |
| RIG-I | direct | Human | [44] | Yes | |
| 3B | RIG-I | direct | Swine | [54] | No |
| 3C | MDA5 | direct | Swine | [48] | Yes |
| STAT1/2 | co-factor(s) | Human | [55] | No | |
| NEMO | direct | Swine | [49] | Yes | |
| PKR | indirect | Swine | [56] | No | |
| Lbpro | MDA5 | direct | Human | [47] | Yes |
| STAT1/2 | direct | Swine | [51] | Yes | |
| TRAF3 | direct | Human | [46] | No | |
| TBK1 | direct | Human | [46] | No | |
| RIG-I | direct | Human | [46] | No | |
| Labpro | IRF3 | direct | Swine | [50] | No* |
| IRF7 | direct | Swine | [50] | No* | |
| TBK1 | direct | Swine | [57] | No | |
| MAVS | N.A. | Swine | [57] | No | |
| VP1 | MAVS | direct | N.A. | [58] | No |
| VP3 | MAVS | direct | Human | [59] | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarry, M.; Caignard, G.; Dupré, J.; Zientara, S.; Vitour, D.; Bakkali Kassimi, L.; Blaise-Boisseau, S. Host-Specific Interplay between Foot-and-Mouth Disease Virus 3D Polymerase and the Type-I Interferon Pathway. Viruses 2023, 15, 666. https://doi.org/10.3390/v15030666
Sarry M, Caignard G, Dupré J, Zientara S, Vitour D, Bakkali Kassimi L, Blaise-Boisseau S. Host-Specific Interplay between Foot-and-Mouth Disease Virus 3D Polymerase and the Type-I Interferon Pathway. Viruses. 2023; 15(3):666. https://doi.org/10.3390/v15030666
Chicago/Turabian StyleSarry, Morgan, Grégory Caignard, Juliette Dupré, Stephan Zientara, Damien Vitour, Labib Bakkali Kassimi, and Sandra Blaise-Boisseau. 2023. "Host-Specific Interplay between Foot-and-Mouth Disease Virus 3D Polymerase and the Type-I Interferon Pathway" Viruses 15, no. 3: 666. https://doi.org/10.3390/v15030666
APA StyleSarry, M., Caignard, G., Dupré, J., Zientara, S., Vitour, D., Bakkali Kassimi, L., & Blaise-Boisseau, S. (2023). Host-Specific Interplay between Foot-and-Mouth Disease Virus 3D Polymerase and the Type-I Interferon Pathway. Viruses, 15(3), 666. https://doi.org/10.3390/v15030666






